Abstract
The majority of normal cells in a human do not multiply continuously but are quiescent and devote most of their energy to gene transcription. When DNA damages in the transcribed strand of an active gene are bypassed by an RNA polymerase, they can miscode at the damaged site and produce mutant transcripts. This process known as transcriptional mutagenesis can lead to the production of mutant proteins that could be important in tumor development.
Nearly every aspect of cellular behavior and properties can be altered by the production of erroneous proteins. This situation holds true for cells of every living organism from the simplest prokaryote to the most complex metazoan species. Amino acid substitutions or deletions are at the origin of changes in protein structure and function that are responsible for a large variety of biological outcomes ranging from conferring advantageous growth ability for unicellular organisms to cell death and cancer in mammalian species1. DNA damage-induced mutation is an extensively documented route of mutagenesis for replicating cells and is due to an encounter between DNA polymerase and damaged bases which results in the insertion of a non-complementary nucleotide opposite to the lesion that gives rise to a permanent and heritable change in the DNA sequence2, 3. This replication-centric model for the initiation of mutagenesis has provided a plethora of information for understanding major routes of mutagenesis for organisms existing under conditions of cell growth and division and have contributed substantially to our understanding of a host of biological events including the origin of genetic variability, evolution, and the development of cancer4, 5.
However, the majority of cells living outside of the artificial, growth factor-rich environment of a laboratory do not undergo continuous cycles of replication and growth but are instead more likely to exist in a non-proliferative state6. For example, several organs of multicellular organisms, such as heart or brain, are comprised primarily of non-dividing cells in which lifespan is limited by functional degeneration of their normal physiology. Therefore, it follows that quiescent, non-replicating cells represent the largest proportion of an organism’s tissues that are exposed to exogenous and endogenous DNA damaging agents and it is these cells that are likely to be the origin of tumours. Therefore, the physiological maintenance of cells and organisms is likely to be largely dependent on the fidelity of both transcription and translation.
There are a number of possible pathways for generating erroneous proteins that do not involve DNA synthesis (Figure 1). At the level of translation, errors can occur through incorrect amino acids incorporation, slippage of the translational machinery or absence of tRNA modifications resulting in misreading of the mRNA7. Altogether, these errors occur once for every 1,000 to 10,000 codons translated which renders synthesis of a functional protein from an mRNA noticeably error prone. Nonetheless, lapses in translational or post-translational can functionally alter proteins and possibly change the physiology of the cell8, 9 and can be crucial for cancer development or progression10.
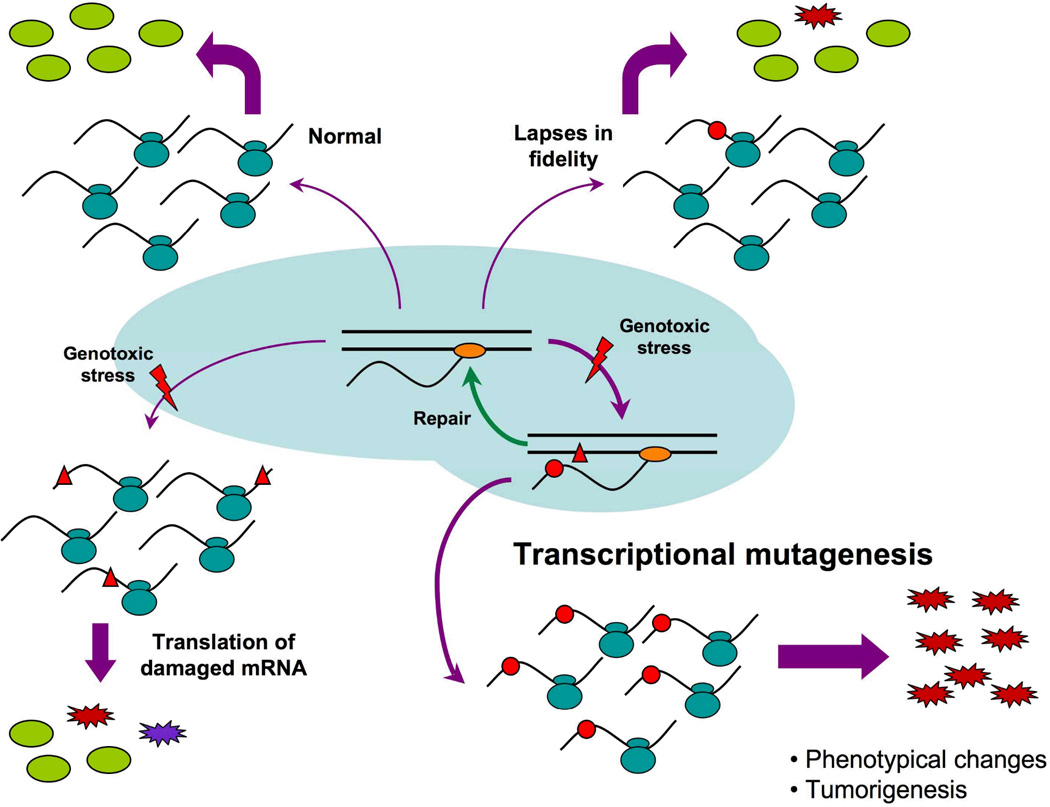
Figure 1. DNA replication-independent production of erroneous proteins.
Under normal conditions (top left), transcription in the nucleus (pale blue background) produces error-free mRNAs that are translated by ribosomes (double blue ovals) to normal proteins (green ovals) in the cytoplasm. In some cases (top right), lapses in RNA polymerase fidelity (orange ovals) can generate aberrant transcripts (red dot) that are translated into erroneous proteins (red jagged ovals). This random low frequency production of erroneous proteins can also be due to translational errors or to lapses in translation fidelity. When exposed to a genotoxic agent (red thunderbolt) (bottom left), RNA molecules in a cell may contain lesions (red triangles) that could induce the production of erroneous proteins during translation because of their potentially altered specificities of codon/anticodon pairing during tRNA selection. DNA is the other target for genotoxic stress (bottom right). RNA polymerase can bypass numerous unrepaired damaged nucleotides on the transcribed strand of a gene (red triangle) that can result in misincorporation events in the transcript sequence (red dots) as long as the DNA damage is not removed by one of the cellular DNA repair pathways (green arrow). Transcriptional mutagenesis results in the production of a primarily homogenous mutant transcript population that, in turn, lead to the production of high levels of erroneous proteins, all possessing the same mutant sequence, that could alter the phenotype of the cell.
At the level of transcription, some damaged ribonucleotides with altered pairing specificities can be incorporated into the nascent mRNA by the RNA polymerase (RNAP) thus leading to a mutant transcript that is translated into erroneous proteins11. Another possibility is direct damage to the transcript itself. In that case, damaged ribonucleotides can lead to altered specificities of codon-anticodon recognition such that incorrect amino acids can be incorporated. This latter event has been proposed to explain the etiology of some human diseases including neurodegenerative syndromes and development of several types of cancer12–14. In addition, lapses in RNA polymerase fidelity may also result in ribonucleotide misincorporations to produce mutant transcripts, although non-targeted misincorporation rates are reported to be to be quite low (10−4 to 10−5) in both prokaryotic and eukaryotic systems15–17.
The types of replication-independent pathways that are described above will produce erroneous proteins that have the potential to affect the normal physiology of a cell or to change the cellular phenotype7, 10. However, with these pathways, erroneous proteins are likely to have only a limited effect on the cell. Indeed, those errors are not targeted to a particular nucleotide of a specific mRNA or to a unique amino acid change in one protein and the amount of erroneous proteins that could be made by those randomly targeted errors is expected to be very low. Nonetheless, there is substantial evidence demonstrating that randomly produced, erroneous proteins could potentially have significant effects. A common example is the decrease of organism fitness when translational fidelity is disrupted with commonly used antibiotics that kill bacteria18, 19. Additionally, deficiencies in translational and transcriptional proofreading has the ability to alter cell morphology20 and lead to severe fitness defects21, 22.
Another route for producing erroneous proteins is transcriptional mutagenesis. This process involves the efficient bypass of a DNA damage by the transcriptional machinery accompanied by base misinsertion (Figure 1). Transcriptional mutagenesis has the potential for generating substantial levels of mutant transcripts and is likely to be the cause of the production of much greater levels of erroneous proteins compared to the other non-replication-dependent routes discussed above. This is due to the inherent targeting of the change in the transcript sequence for every RNA polymerase bypass event, eventually resulting in a pure population of identical, mutant transcripts (Figure 1). Each mutant transcript will, in turn, be translated multiple times and, if the bypass/miscoding event at the site of DNA damage changes a codon specificity, the entire translational output from that transcript population will contain the same amino acid change. Consequently, as long as the inducing nucleotide lesion is not repaired, a largely pure population of erroneous proteins, all with the same amino acid sequence, will be present in the cell with the potential to change its phenotype modify its physiology and may play an important role in the development of several types of cancer.
In this Perspective, we describe transcriptional mutagenesis in non-replicating, quiescent cells. We will then present evidence that transcription can be affected by nucleotide damage in transcribed DNA induced by a plethora of physical and chemical agents present in natural environments and are at the origin of transcriptional mutagenesis. We will also present evidence supporting the involvement of transcriptional mutagenesis in tumor development23.
The mechanism of transcriptional mutagenesis
In quiescent cells, an elongating RNAP is likely to encounter a DNA lesion on the template strand of actively transcribed genes frequently, which could result in either blockage of RNAP elongation or bypass of the lesion by the transcriptional machinery. Stalling or blockage of the RNAP at a lesion site activates the transcription-coupled repair (TCR) pathway (Box 1), which is a sub-pathway of the nucleotide excision repair (NER) pathway that is dedicated to the elimination of DNA damage from the transcribed strand of a gene (reviewed in24, 25). However, not all DNA lesions have the ability to stall or block an elongating transcription complex and such lesions, having modified pairing properties, are readily bypassed by RNAP with ribonucleotide misincorporation which can result in the generation of mutant transcripts with a specific codon-specificity change (reviewed in26, 27).
Box 1. Transcriptional mutagenesis vs. transcription-coupled repair.
It is well documented that RNAPII elongation complex arrest at sites of DNA damage represents a strong signal for apoptosis79–81. Therefore, cells have evolved a specialized mechanism called transcription-coupled repair (TCR), which involves the hierarchical recruitment of several proteins, whose role is to remove transcription-blocking DNA lesions from the genome24, 25. Transcriptional mutagenesis and TCR are therefore competing processes.
A major issue is to determine the proportion of specific DNA damage that can be bypassed by RNAPII causing transcriptional mutagenesis and the proportion that is repaired by TCR. This has been the subject of a number of studies addressing the fate of RNAPII encountering an oxidative DNA lesion such as 8-oxoguanine (8-oxoG). TCR was originally documented for DNA damage induced by UV light, but it has also been reported that it may act in a transcription-dependent manner upon oxidative DNA damage in E. coli33. Additionally, it was shown that cells from individuals with Cockayne syndrome (CS) were defective for both TCR and the repair of oxidative DNA lesions that could explain the non cancer-prone mutator phenotype of patients with CS82. However, a number of key papers addressing this question have been retracted83–85. Nonetheless, a link between repair of oxidative damage and transcription might exist as host cell reactivation of plasmids containing 8-oxoG is defective in CSA and CSB cells, two key proteins of TCR86. Additionally, after transfection into mouse embryonic fibroblasts, expression of the reporter gene from shuttle vectors containing a single 8-oxoG in the transcribed strand required CSB87. However, the same effect was not observed when 8-oxoG was located in a different sequence context and using a weaker promoter88. Moreover, measurements of transcriptional mutagenesis induced by 8-oxoG in different cell lines revealed that the magnitude of erroneous protein expression was the same in both TCR-proficient and –deficient cell lines but was dependent on the sequence context surrounding the lesion35, 37. These findings suggest that factors such as promoter strength, sequence context and position of the lesion with respect to the promoter may influence transcription past a single 8-oxoG by RNAPII.
How to study transcriptional mutagenesis?
A large number of studies investigating transcriptional mutagenesis have been carried out with defined, in vitro transcription systems containing purified components and/or various types of cell extracts necessary for initiation, elongation and termination of transcription on a DNA template. Central to the success of these studies were the availability of transcription templates containing single, well-characterized DNA lesions at defined locations. Although it is theoretically feasible to introduce many different types of DNA damage into transcription templates, such procedures are technically challenging and often result in poor yields of product. New methodologies which result in large quantities of damage-containing substrates were then developed28, 29. In vitro transcription studies have employed phage, bacterial, yeast and mammalian RNA polymerases to determine whether such enzymes can bypass DNA lesions or are stalled or blocked at such sites. Both multiple (template reutilization/recycling) and single round (one promoter-dependent elongation cycle per template-RNA polymerase elongation complex) transcription conditions have been explored. In several cases, sequencing analysis has been carried out on transcripts resulting from RNA polymerase bypass of lesions to determine the nature of the incorporation-misincorporation events (Table 1).
Table 1.
Insertion events at sites of DNA damage during replication and transcription
| DNA damage type | RNAP type | Nucleotide inserted | DNAP type | Nucleotide inserted |
|---|---|---|---|---|
| Uracil | Mammalian RNAPII Phage (SP6, T7) E. coli |
A,G38 A23, 41, 42 A33, 34, 40, 41, 43 |
Various | A4 |
| 5-Hydroxyuracil | Mammalian RNAPII | A48 | Human DNA polι E. coli PolI |
G96 A97 |
| Thymine glycol | Mammalian RNAPII | A48 | Mammalian DNA polλ and β E. coli PolI |
A98 Strong block99 |
| 5-Guanido-4-nitroimidazole | Mammalian RNAPII Phage(T7) |
C51 C>A>-1del>G>>U51 |
Human polα Human polβ E.coli PolI E. coli PolV |
A,G100 C100 C>A>G100 A101 |
| Dihydrouracil | Phage (SP6, T7) E. coli |
A,G23 A39 |
Human DNA polι E. coli PolI |
G96 A39 |
| 8-Oxoguanine |
Mammalian RNAPII Phage (T7) E. coli |
C,A, del35, 37, 38 A,C52 A,C,del33, 34, 40 |
Various mammalian E. coli PolI |
A,C102 A,C54, 55, 57–59 |
| Cyclobutane pyrimidine dimmer (TT) |
Mammalian RNAPII |
multiple nucleotide deletion36 |
Human polη | AA103 |
| 8,5'-Cyclo-2'-deoxyadenine | Mammalian RNAPII |
5'A, multiple nucleotide deletion36 |
Replicative (mammalian and bacterial) Polη |
Block44 T104 |
| 1,2-d(GpG) intrastrand cross-links | Mammalian RNAPII | AC, CC63 | Human Polη Human Polζ Human Polγ |
CC105 G?105 A?105 |
| O6-Methylguanine | Mammalian RNAPII E. coli Phage (T7) |
C>U64 U40 C,U64 |
Various | T57 |
| BPDE-Adenine(−) | Phage (T7) | A,G,del16 | Various | A106 |
| BPDE-Adenine(+) | Phage (T7) | A,G,del16 | Various | A106 |
| BPDE-Guanine adducts | Phage (T7) | C67 | E. coli PolI | A, del56 |
| AF-Guanine | Phage (T7) | C52 | Mammalian E. coli PolI |
A107 del108 |
| AAF-Guanine | Phage (T7) | C52 | Mammalian E. coli PolI |
A107 A108 |
| Abasic site | Mammalian RNAPII Phage (SP6, T7) E. coli |
C38 A41, 43 A34, 41, 43 |
Mammalian DNA polα E. coli PolI |
A109 A109 |
| Tetrahydrofuran | Phage (T7) | A,G52 | Mammalian DNA polα E. coli PolI |
A109 A109 |
| Single strand breaks/gaps | Phage (SP6, T7) E. coli |
del42, 69, 70 del, A34, 69 |
Various | Strong block4 |
In vivo studies of transcriptional mutagenesis are shown in bold type. Abbreviations: RNAP, RNA polymerase; RNAPII, RNA polymerase II; DNAP, DNA polymerase; AF-Guanine, N-(deoxyguanosin-8-yl)-2-aminofluorene; AAF-Guanine, N-(deoxyguanosin-8-yl)-2-acetylaminofluorene; BPDE-Adenine(−) and (+), (−) and (+)-anti-trans-benzo[a]pyrene-7,8-dihydro-9,10-diol epoxide N6-deoxyadenosine adducts, respectively; BPDE-Guanine adducts, (−)trans, (+)cis, and (−)cis-anti-N2-BPDE-deoxyguanosine adducts.
Although lesions that are bypassed by RNAP are likely to cause transcriptional mutagenesis, it was unknown whether such a situation would also occur in living cells due to the simultaneous presence of multiple DNA repair pathways that might occur instead of RNAP bypass (Box 1). Experimental systems devised to address this question in living cells had to meet two requirements28, 30–32. The first is utilization of a reporter plasmid expressing a protein whose activity can be biochemically determined. In most studies, a site-specific DNA lesion was positioned in the transcribed strand of the reporter gene such that if the damage is repaired, the resulting mRNA encodes a non-active protein. However, if the DNA lesion is misread during transcription, a fully active protein is produced and activity can be measured to ascertain the extent of the bypass. The second requirement for these in vivo systems is to ensure that the measured activity of the reporter protein is the result of transcription across the DNA lesion and not permanent fixation of potential base sequence changes via DNA replication into a heritable mutation. In prokaryotic systems, bacterial cells had to be maintained in a non-growth state, where transcription, but not DNA replication is occurring and was achieved by the use of growth static antibiotics such as novobiocin32–34. In eukaryotic systems this problem was eliminated by employing reporter vectors devoid of replication initiation sites35–37.
The case of uracil
RNAPs from different organisms, including mammals, are able to bypass many types of non-bulky DNA lesions that are typically substrates of the base excision repair (BER) pathway including uracil, the spontaneous deamination product of cytosine38–43. When a uracil is present on a transcribed strand, adenine is always inserted by prokaryotic RNAPs39–43, but mammalian RNAPII inserts either A or G into the transcripts44, partially reducing the ability of this lesion to cause transcriptional mutagenesis in mammalian cells as G should not be considered as mutagenic. The first published study of transcriptional mutagenesis in living cells confirmed that bacterial RNAP inserted an adenine residue opposite to a uracil present in the transcribed strand of a luciferase reporter gene32, an observation that was confirmed by other studies33, 34.
Oxidative base damages
Reactive oxygen species (ROS), including the O2•− radical, H2O2, and the OH• radical, may be the most important genotoxic agents because of their ubiquitous and continuous production as by-products of respiration45. In mammalian cells, they have been implicated in the etiology of a variety of pathophysiologies including aging and a large variety of cancers46. Genotoxicity of ROS results from their reaction with DNA to produce a plethora of damages that are repaired primarily via the BER pathway in both bacteria and eukaryotes47. Several oxidative base damages were tested for their ability to induce transcriptional mutagenesis including 5-hydroxyuracil48, thymine glycol48–50, 5-guanido-4-nitroimidazole51, dihydrouracil23, 39 and 8-oxoguanine (8-oxoG)38, 40, 52.
In some cases, certain types of oxidative damage do not cause transcriptional mutagenesis. For example, transcript sequencing has revealed human RNAPII incorporates the correct adenine is opposite 5-hydroxyuracil and thymine glycol48 and the correct cytosine opposite guanido-4-nitroimidazole51. However, other oxidative DNA lesions are prone to transcriptional mutagenesis as dihydrouracil, a cytosine damage, primarily directs the addition of A23, 39. Additionally, it was shown that in vitro bypass of 8-oxoG by prokaryotic RNAP incorrectly directs the incorporation of an adenine only about half the time, correctly coding for cytosine in other cases40, 52. However, when HeLa RNAPII is used, this A:C ratio seems to depend on the concentration of NTPs used in the reaction mixture38.
Transcriptional mutagenesis studies using a luciferase reporter gene in E. coli also confirmed that 8-oxoG was able to instruct the incorporation of the correct cytosine and the misincorporation of adenine during transcription, but a single nucleotide deletion corresponding to the lesion site was also identified in about one fourth of the mRNA population33. In mammalian cells, two studies were carried out with vectors containing an 8-oxoG at a defined position on the transcribed strand of the reporter gene. As with bacterial RNAP, mammalian RNAPII incorporates either cytosine, adenine or no nucleotide opposite to the lesion35, 37. The ratio of each incorporation event was also shown to vary depending on the sequence context flanking the 8-oxoG, the relative distance of the lesion to the promoter, the nature of the deoxyribonucleotide opposite to the lesion in the non-transcribed strand and, most importantly, on the availability of the DNA repair pathways capable of removing 8-oxoG from the DNA35, 37.
Structure-function analysis provides insight into the structural basis for how readily bypassed carcinogenic DNA lesions, such as 8-oxoG, can cause transcriptional mutagenesis. With yeast RNAPII, it was shown that misincorporation of adenine forms a Hoogsteen base pair at the active center and that, to achieve this pairing, 8-oxoG has to adopt the syn-conformation53. Similar to RNAPII, high fidelity DNA polymerases incorporate cytosine and adenine to various extents during opposite to 8-oxoG54–59 (Table 1) and cytosine incorporation occurs with the same type of nucleotide pairing and conformation60, 61.
DNA helix-distortive damages
The majority of transcriptional mutagenesis studies carried out to date involve the elucidation of misincorporation events occurring during transcription across lesions that do not cause significant distortion of the DNA backbone and are repaired primarily by the BER pathway. In contrast, almost all bulky, distortive lesions, primarily repaired by the NER pathway generally represent strong blocks to replication-associated DNA polymerases as well as elongating RNAPs in vitro and are likely to elicit TCR24, 25 (Box 1). However, in NER-deficient human cells, two helix-distorting lesions, UV light-induced cyclobutane pyrimidine dimers (CPDs) and the oxidative base damage 8,5'-cyclo-2'-deoxyadenosine (cyclo-dA), are strong but incomplete blocks to transcription62. Characterization of the transcripts produced from the in vivo RNAPII bypass of these lesions has led to the identification of new types of transcriptional mutagenesis events. When bypassed, cyclo-dA induces the incorporation of uridine opposite to the lesion followed by misincorporation of adenine opposite to the next nucleotide downstream to the lesion36 (5'A in Table 1). Additionally, rare transcripts containing multiple nucleotide deletions were also identified in transcripts resulting from the bypass of cyclo-dA and CPDs36. Although such mutant transcripts represent a small proportion of the total mRNA population, they were identified in cells from patients with cancer-prone or developmental disorder-associated DNA repair diseases (xeroderma pigmentosum and Cockayne Syndrome, respectively), thus raising the possibility of a link between transcriptional mutagenesis and the etiology of these diseases.
Another DNA helix-distortive lesion that was studied for its ability to induce transcriptional mutagenesis is 1,2-d(GpG) intrastrand crosslink formed by the anticancer drug cisplatin. When artificially placed on the template strand in proximity to the active site of RNAPII, this damage induces the misincorporation of adenine opposite the first guanine whereas the correct cytosine is incorporated opposite the second guanine63. However, transcriptional mutagenesis caused by cisplatin adducts might not be observed in living cells as the artificial conditions used for this study are not likely to occur in vivo.
DNA damages caused by chemical carcinogens
DNA lesions resulting from exposure to carcinogenic chemicals were also examined for their propensity to induce transcriptional mutagenesis. For example, the methylation product of guanine, O6-methylguanine (O6mG), directs the misincorporation of uracil by E. coli RNAP40. However with human RNAPII, cytosine and uracil are incorporated opposite to O6mG with a 3:1 ratio, respectively64, thus reducing the ability of the lesion to cause transcriptional mutagenesis in mammalian cells. Interestingly, computational modeling with yeast RNAPII showed that bypass of O6mG can only occur when the lesion adopts an anti-conformation with the methyl group in the proximal orientation and that accommodation of the misincorporated uracil involves specific hydrogen bonding64. As for 8-oxoG, this specific scheme of pairing during transcription is similar to the one used to explain thymine misincorporation during replication65.
Transcriptional bypass by prokaryotic RNAP has also been demonstrated, albeit with much lower efficiency, for several bulky adducts induced by carcinogenic chemicals such as benzo[a]pyrene or 2-acetylaminofluorene66. For example, N6-Benzo[a]pyrene diol epoxide (BPDE) adducts of both adenine and guanine are bypassed by T7 RNAP16, 67. While the adenine adducts direct the misincorporation of either A or G16, sequence analysis of bypassed G adducts indicates that nonmutagenic C is inserted67. However, this study also demonstrated that truncated transcripts resulting from arrest at this lesion contained mutagenic nucleotide insertions, indicating that in some cases RNAP arrest may result from the structural strain of incorrect base pairing opposite the lesion site. Guanine C-8 aminofluorene (AF-G) and acetylaminofluorene (AAF-G) adducts are also subject to some level of bypass, with the bulkier AAF-G moiety more effective at blocking the transcription machinery52. Both lesions were found to be nonmutagenic at the level of transcription, directing the correct incorporation of C52.
Abasic sites, strand breaks and other DNA repair intermediates
Deficiencies in one or several DNA repair pathways is a hallmark feature of cancerous or precancerous cells4. Consequently, presence of repair intermediates (such as abasic sites, single-strand breaks and gaps) in the transcribed strand of a gene might be more frequent in these type of cells. Abasic sites, as well as the abasic site analogue tetrahydrofuran, are efficiently bypassed by prokaryotic RNAPs34, 41, 43, 52. Adenine was most often incorporated opposite a template abasic site, although a small fraction of G was incorporated opposite to tetrahydrofuran by T7 RNAP41, 43, 52. This pattern of insertion events could be highly mutagenic at the level of transcription, given that depurination at G residues is the most frequent event leading to spontaneous abasic site formation68. However, the potential to elicit transcriptional mutagenesis by these lesions might be reduced in mammalian cells as purified HeLa RNAPII prefers the addition of cytosine opposite to abasic sites38.
Remarkably, single-strand breaks and gaps have also been demonstrated to be bypassed by prokaryotic RNAPs42, 69, 70. This event occurs with varying levels of efficiency and depends on the size of the template gap, the flanking DNA termini, and the type of RNAP involved. Small gaps are bypassed with higher efficiency than larger gaps70, termini containing hydroxyl groups are negotiated better than those containing phosphates or modified sugars42, and bacteriophage polymerases are more efficient at bypass than RNAP from E. coli69. Analysis of the transcripts generated by transcription across single-strand gaps indicates that they contain correctly templated nucleotides on both sides of the gap, but the site of the gap shows up as a deletion in the transcript (a 1-nt deletion for a 1-nt gap, a 2-nt deletion for a 2-nt gap, and so on)42, 69, 70.
Although in vivo studies brought confirmation of some results obtained in vitro, they also revealed surprising features regarding the occurrence of transcriptional mutagenesis in living cells as in some cases newly, unexpected events were identified by sequencing of the reporter gene mRNA. Indeed, recent studies in E. coli indicate that there are factors that facilitate RNAP bypass of strand breaks and gaps in vivo34. Sequence analysis of the luciferase reporter mRNA produced from the bypass of RNAP across the strand break revealed that adenine is incorporated opposite the gap and no deletion in the transcript was detected34, thus leading to the production of mRNA with a change in codon specificity but avoiding the production of mRNA with frameshift mutations.
Expression of erroneous proteins via transcriptional mutagenesis
An important potential physiological consequence of transcriptional mutagenesis is a transient phenotypic change due to the expression of a large pool of mutant proteins within the cell. It is clear that many DNA lesions have the ability to induce transcriptional mutagenesis and thus initiate a change in biological activity. However, it is likely that in most cases, the population of erroneous protein has to be sufficiently large to be able to induce such a change. It follows that infrequently bypassed lesions (such as CPDs) should be less capable of eliciting a phenotypic change compared to frequently bypassed lesions (such as 8-oxoG). Thus all in vivo studies to date investigating phenotypic changes caused by transcriptional mutagenesis have been conducted with replication-defective DNA constructs containing a single non-distorting lesion in the transcribed (template) strand of a reporter gene.
Using a luciferase transcriptional mutagenesis reporter-based expression system, erroneous transcriptional bypass of uracil, abasic sites or 8-oxoG gave rise to a phenotypic change caused by production of the reporter protein and displayed by the transient expression of active luciferase enzyme in both bacterial and mammalian systems32–35. In all of these studies, the bypass of the DNA lesion was enhanced by disruption of the genes encoding the major BER proteins responsible for the removal of these lesions in the cells, thereby prolonging the half-life of the DNA damage harbored in the template DNA. Consequently, the phenotypic change was more robust and prolonged over time in the DNA repair-deficient cell lines. These observations are relevant to the potential role of transcriptional mutagenesis to the development of cancer as several reports demonstrate that individuals expressing hypomorphic alleles of BER genes have higher risks of developing different types of cancer71–73.
Interestingly, high levels of active luciferase, produced via transcriptional mutagenesis, was detected for up to seven days following transfection of an 8-oxoG containing vector in DNA repair-proficient cells35. Given the fact that luciferase is not a very stable protein, as its half-life was estimated to be of no more than four hours in mammalian cells, transcriptional mutagenesis must therefore continue for a prolonged period of time even in human cells with normal DNA repair capacity.
Involvement of TM in tumor development
Activation of ERK phosphorylation following transcriptional mutagenesis events
Although very informative, luciferase based studies described above are not adapted for studying the potential of DNA lesions to cause transcriptional mutagenesis with a biologically relevant outcome. In this regard, replacement of the luiferase gene by a cellular cDNA gene in which erroneous transcription events can be detected through the translation of the encoded protein that elicits a measurable biological endpoint represented a major step forward for investigations of transcriptional mutagenesis in living cells. Such a system was used for the expression after transfection in mouse cells of the H-Ras oncogene in which an 8-oxoG replaced guanine in codon 6137. Bypass and misinsertion opposite to this lesion during transcription would result in the production of the constituvely active (dominant) Q61K mutant H-Ras protein. The occurrence of such a transcriptional mutagenesis-mediated event was followed in different cell lines by the activation of MAP kinase signaling components, that should have resulted in increased ERK phosphorylation. Despite the detection of mutant Ras transcript in DNA repair-proficient cells, the extent of transcriptional mutagenesis was not sufficient to induce a detectable increase in ERK phosphorylation. However, in cells lacking 8-oxoguanine DNA glycosylase (OGG1), the major 8-oxoG glycosylase that initiates BER, the number of H-Ras mutant transcripts was elevated four- to fivefold and ERK phosphorylation was readily detected and found to be significantly increased. However, it is currently not known whether or not such cells are capable of forming tumors in xenografts. Knowing that the vast majority of cancer cells are deficient in one or more DNA repair pathway74, including BER75, these observations strongly suggest that transcriptional mutagenesis could lead to the activation of an oncogenic pathway.
Retromutagenesis: a transcriptional mutagenesis-driven switch from non-growth to growth state
All of the observations outlined above suggest that bypassed carcinogenic lesions could lead to activation of oncogenic pathways. One key point for this hypothesis is that, as already discussed for 8-oxoG and O6mG, the majority of these types of DNA lesions cause the same types of misincorporations during both transcription and replication. In some cases transcriptional mutagenesis-inducing DNA damage can reside in the DNA for a prolonged period of time leading to the production of erroneous proteins35. It is conceivable that certain mutant proteins (such as H-Ras) produced via transcriptional mutagenesis activate an oncogenic, growth positive pathway, promoting re-entry into the cell cycle and increase the chance that the DNA replication machinery will now encounter the same DNA lesion that originally caused transcriptional mutagenesis. The nucleotide inserted opposite to the lesion by DNA polymerases will, in many cases, be equivalent to the one inserted during transcription. Such an event would ensure permanent retention of the growth initiating mutation in one daughter cell, resulting in a heritable change. This process has been termed retromutagenesis26, 76 (Figure 2) and can thus initiate a transition from non-growth state to a growth state that could account for the development of mammalian tumors at the stage where expansion of an abnormal cell population occurs. For example, as seen with the activation of H-Ras, transcriptional mutagenesis could lead to the acquisition of limitless replicative potential, a hallmark of cancer. Furthermore, production of certain mutant proteins could also perturb signaling pathways that are normally dedicated to responding to antigrowth signals or the induction of apoptosis.
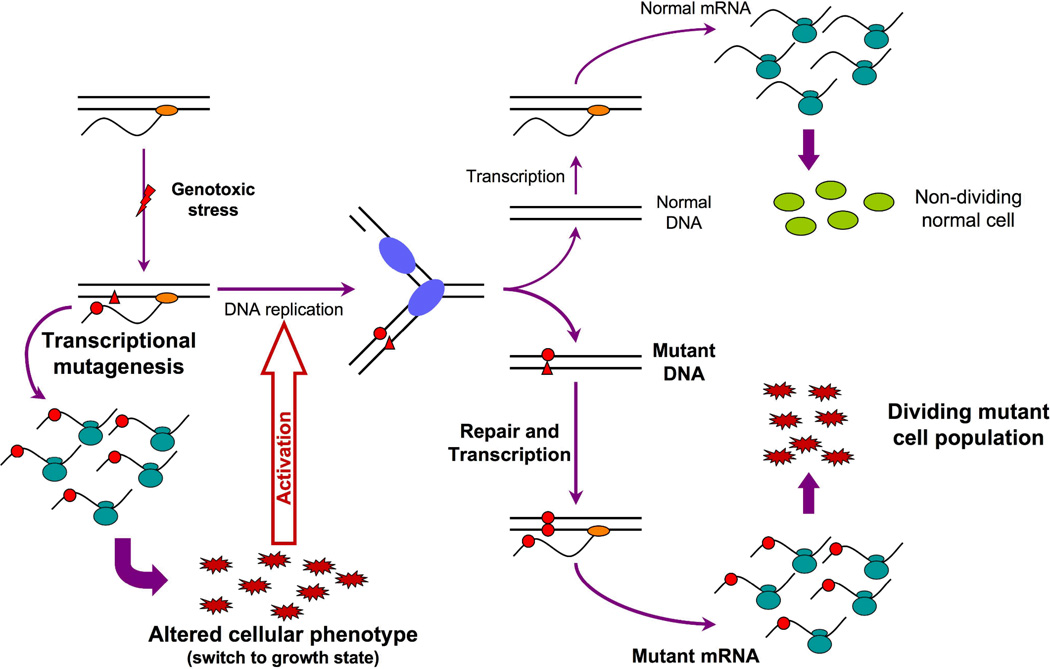
Figure 2. Potential role of transcriptional mutagenesis in tumor development.
Following genotoxic stress (red thunderbolt), a DNA lesion (red triangle) can appear on the transcribed strand of a gene resulting in the production of high levels of erroneous protein via transcriptional mutagenesis as shown in Figure 1. The resulting mutant proteins (red jagged ovals) may have the ability to alter the phenotype of the cell in such a way as to confer a growth advantage leading to initiation of DNA replication. If left unrepaired, the DNA lesion will subsequently be encountered by the replication machinery (blue ovals, DNA synthesizing lagging and leading strands) and will likely cause similar miscoding during DNA synthesis resulting in the fixation of the mutation into the genome of one progeny cell (bottom right). Subsequent rounds of replication in this progeny will lead to a dividing cell population harboring the mutation that conferred the growth advantage and thus could lead to tumor development. Double blue ovals, ribosomes; orange ovals, RNA polymerase ; red dots, misincorporated nucleotides; green ovals, normal proteins.
Interestingly, within the database of tumor-associated p53 base substitutions, about 5% of tumors are reported to contain two or more base substitutions. A retrospective analysis of this database indicates that most base substitution multiplets arise from a transient, hyper-mutagenic event in one cell that subsequently proliferated into a clonal tumor76. Additionally, this p53 transient hyper-mutagenesis involves a base substitution, the driver mutation, that arises along the transcribed strand of the p53 gene and changes the amino acid sequence of the p53 protein. This strongly indicates that selection acted while the driver mutation was asymmetrical, i.e. before it was resolved into a double-stranded substitution. Accordingly, the best scenario for the generation of such multiple base substitutions is a transient switch to growth state induced by transcriptional mutagenesis and subsequent retromutagenesis in the dividing cell77.
Conclusions and future perspectives
As data are accumulating, it is becoming clearer that transcriptional mutagenesis could play a significant role in tumor development and other biological outcomes (Boxs 2 and 3). Although vector based analyses of transcriptional mutagenesis are very informative, the validation for the role of transcriptional mutagenesis in tumor development remains to be precisely established. The next challenge will be to express a reporter cellular gene containing a DNA lesion in its transcribed strand present in one copy per cell and to follow the subsequent phenotypic change that should result from the production of erroneous proteins. One will then have to follow the progeny of a single cell to determine if cancerous growth can be initiated by the transient expression of oncogenic proteins or the disruption of signaling pathways.
Box 2. Other biological consequences of transcriptional mutagenesis.
In addition to its potential involvement in tumor development, transcriptional mutagenesis may also have other consequences for eukaryotic cells. Transcritpional mutagenesis also provides a mechanism for the generation of prions in neuronal cells. If the mutant protein generated by transcriptional mutagenesis is more stable in the β-sheet conformation, this event could allow the generation of adequate levels of protein to promote prion fiber nucleation, subsequently leading to the conversion of normal protein to the prion conformation and causing fibril formation and finally neuronal death89. This mechanism of nucleation induced by the transient production of mutant protein can also be used to explain the etiology of other neurodegenerative syndromes, characterized by aggregates of misfolded proteins, including Alzheimer's and Parkinson's diseases. Indeed, mutant forms of amyloid precursor protein and ubiquitin B protein are detected in protein aggregates found in the dystrophic neurites that contribute to the characteristic pathology (neuritic plaques, neurophil threads and neurofibrillary tangles) of Alzheimer's disease but not in brains of individuals without dementia90. Mutant proteins were found to originate from mutant mRNAs that were produced in the affected cells, but examination of the genomic DNA failed to reveal any evidence of mutation, so they are presumed to have arisen as errors of transcription90. Whether these transcription errors are due to transcriptional mutagenesis or lapses in RNA polymerase fidelity remain to be established. However, it is well documented that oxidative DNA damage accumulates in aging brains and that increased levels of oxidized guanine are found in DNA from ventricular cerebrospinal fluid of patients with Alzheimer's disease compared with controls91. These increased levels of DNA damage are also correlated with decreased DNA repair capacity92. Additionally, when a nucleation-prone mutant protein is produced in a specific neuronal cell, the protein aggregation phenomena could be detrimental to neighboring cells. Indeed, addition of aggregated α-synuclein forms, found in neuronal cells from patients with Alzheimer's or Parkinson's disease, to the culture medium of neuroblastoma cells causes apoptotic death89. These observations strongly suggest that transcriptional mutagenesis could also play a significant role in the etiology of neurodegenerative diseases.
Box 3. Biological consequences of transcriptional mutagenesis for prokaryotes.
The retromutagenesis mechanism can also be viewed as an environmental adaptation pathway in prokaryotes as such retromutations would have been tested and selected for conferring a growth advantage as mutant mRNAs before they became heritable mutations. The fitness increase brought by this environmental adaptation is cell-selfish in that it is limited to an immediate growth or replication advantage for that host cell in that environment. This cell-selfish mode of evolution can help to confer an immediate growth advantage while minimizing DNA mutational load76. Such a process has been proposed to explain adaptive mutagenesis induced by starvation in Escherichia coli, a setting in which mutations arise rapidly and are confined to those that allow the cells to grow93, 94. In non-proliferating cells, the contribution of transcriptional mutagenesis to the mutant protein pool, and thus the cellular phenotype, is likely to be much more apparent, especially because the capacities of certain DNA repair pathways are diminished in those conditions94, 95. A similar role for transcriptional mutagenesis in bacterial and other haploid unicellular organisms has been proposed for the acquisition of antibiotic resistance in microbial pathogens34.
Additionally, a more global approach to investigations of transcriptional mutagenesis is now possible with the emergence of new technologies. For example, the employment of massively parallel sequencing of cellular mRNA populations78 to determine the extent erroneous transcription can be applied to a wide variety of genotoxic agents and cell types with different DNA repair capacities. Future studies addressing these issues will provide additional insights into the mechanisms and consequences of transcriptional mutagenesis and further establish the role of this process in tumor development.
Acknowledgments
We would like to thank past and present members of the Doetsch lab for their helpful discussions and enthusiasm for the concept of transcriptional mutagenesis. DB is supported by Univeristé Paris-Sud 11 and CNRS. For transcriptional mutagenesis studies, PWD is supported by National Institutes of Health Grant CA120288.
Biography
Damien Brégeon:
Damien Brégeon completed is Ph. D. in the laboratory of Miroslav Radman at the Université Pierre et Marie Curie. He began postdoctoral research in 2001 to study transcriptional mutagenesis in bacterial (laboratory of P.W. Doetsch) and mammalian cells (laboratory of A. Sarasin, Institut Gustave Roussy, France). In 2006, he was appointed as Maître de Conférence at the Université Paris-Sud 11 where he is studying translational error mechanisms.
Footnotes
Web pages and databases
p53 mutations database : http://www-p53.iarc.fr/
Paul W. Doetsch's lab webpages : http://www.biochem.emory.edu/labs/medpwd/
Damien Brégeon' lab webpages : http://www.igmors.u-psud.fr/spip.php?article358
Contributor Information
Damien Brégeon, Université Paris Sud-11, Institut de Génétique et Microbiologie, CNRS UMR 8621, Bât 400, F-91405 Orsay Cedex, France, Tel : +33 1 69 15 35 61, Fax : +33 1 69 15 46 29, damien.bregeon@igmors.u-psud.fr.
Paul W. Doetsch, Departments of Biochemistry and Radiation Oncology, Winship Cancer Institute, Emory University School of Medicine, 1510 Clifton Rd NE, Atlanta, Georgia 30322, USA, Tel : +1 (404) 727-0409, Fax : +1 (404) 727-2618, medpwd@emory.edu
References
- 1.Weinberg RA. The Biology of Cancer. New York: Garland Science; 2006. [Google Scholar]
- 2.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, D.C: ASM Press; 2006. [Google Scholar]
- 5.Loeb LA, Monnat RJ., Jr DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 6.Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair (Amst) 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- 7.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc. Natl. Acad. Sci. U S A. 2009;106:1766–1771. doi: 10.1073/pnas.0813128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell. Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 10.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 11.Taddei F, et al. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 12.Bellacosa A, Moss EG. RNA repair: damage control. Curr. Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 13.Brégeon D, Sarasin A. Hypothetical role of RNA damage avoidance in preventing human disease. Mutat. Res. 2005;577:293–302. doi: 10.1016/j.mrfmmm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U S A. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kireeva ML, et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remington KM, Bennett SE, Harris CM, Harris TM, Bebenek K. Highly mutagenic bypass synthesis by T7 RNA polymerase of site-specific benzo[a]pyrene diol epoxide-adducted template DNA. J. Biol. Chem. 1998;273:13170–13176. doi: 10.1074/jbc.273.21.13170. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RJ, Bonawitz ND, Reines D. Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J. Biol. Chem. 2002;277:24420–24426. doi: 10.1074/jbc.M202059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 19.Brakier-Gingras L, Phoenix P. The control of accuracy during protein synthesis in Escherichia coli and perturbations of this control by streptomycin, neomycin, or ribosomal mutations. Can. J. Biochem. Cell. Biol. 1984;62:231–244. doi: 10.1139/o84-033. [DOI] [PubMed] [Google Scholar]
- 20.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Bacher JM, de Crecy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl. Acad. Sci. U S A. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl. Acad. Sci. U S A. 2009;106:6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Zhou W, Doetsch PW. RNA polymerase bypass at sites of dihydrouracil: implications for transcriptional mutagenesis. Mol. Cell. Biol. 1995;15:6729–6735. doi: 10.1128/mcb.15.12.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 25.Tornaletti S. DNA repair in mammalian cells: Transcription-coupled DNA repair: directing your effort where it's most needed. Cell. Mol. Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doetsch PW. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat. Res. 2002;510:131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 27.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 28.You HJ, Viswanathan A, Doetsch PW. In vivo technique for determining transcriptional mutagenesis. Methods. 2000;22:120–126. doi: 10.1006/meth.2000.1052. [DOI] [PubMed] [Google Scholar]
- 29.Zhou W, Doetsch PW. In: Microbial Genome Methods. Adolph KW, editor. CRC Press: Boca Raton; 1996. pp. 151–165. [Google Scholar]
- 30.Brégeon D, Doetsch PW. Reliable method for generating double-stranded DNA vectors containing site-specific base modifications. Biotechniques. 2004;37:760–762. 764, 766. doi: 10.2144/04375ST01. [DOI] [PubMed] [Google Scholar]
- 31.Brégeon D, Doetsch PW. Assays for transcriptional mutagenesis in active genes. Methods Enzymol. 2006;409:345–357. doi: 10.1016/S0076-6879(05)09020-8. [DOI] [PubMed] [Google Scholar]
- 32.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 33.Brégeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli . Mol. Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 34.Clauson CL, Oestreich KJ, Austin JW, Doetsch PW. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli . Proc. Natl. Acad. Sci. U S A. 2010;107:3657–3662. doi: 10.1073/pnas.0913191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brégeon D, Peignon PA, Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8:388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuraoka I, et al. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 2003;278:7294–7299. doi: 10.1074/jbc.M208102200. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Doetsch PW. Escherichia coli RNA and DNA polymerase bypass of dihydrouracil: mutagenic potential via transcription and replication. Nucleic Acids Res. 1998;26:1707–1712. doi: 10.1093/nar/26.7.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan A, Doetsch PW. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 1998;273:21276–21281. doi: 10.1074/jbc.273.33.21276. [DOI] [PubMed] [Google Scholar]
- 41.Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc. Natl. Acad. Sci. U S A. 1993;90:6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Doetsch PW. Transcription bypass or blockage at single-strand breaks on the DNA template strand: effect of different 3' and 5' flanking groups on the T7 RNA polymerase elongation complex. Biochemistry. 1994;33:14926–14934. doi: 10.1021/bi00253a032. [DOI] [PubMed] [Google Scholar]
- 43.Zhou W, Doetsch PW. Efficient bypass and base misinsertions at abasic sites by prokaryotic RNA polymerases. Ann. N. Y. Acad. Sci. 1994;726:351–354. doi: 10.1111/j.1749-6632.1994.tb52849.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuraoka I, et al. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. U S A. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galli F, et al. Oxidative stress and reactive oxygen species. Contrib. Nephrol. 2005;149:240–260. doi: 10.1159/000085686. [DOI] [PubMed] [Google Scholar]
- 46.Sedelnikova OA, et al. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat. Res. 704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 48.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. Embo J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Htun H, Johnston BH. Mapping adducts of DNA structural probes using transcription and primer extension approaches. Methods Enzymol. 1992;212:272–294. doi: 10.1016/0076-6879(92)12017-k. [DOI] [PubMed] [Google Scholar]
- 50.Tornaletti S, Maeda LS, Lloyd DR, Reines D, Hanawalt PC. Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2001;276:45367–45371. doi: 10.1074/jbc.M105282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimitri A, et al. Transcription of DNA containing the 5-guanidino-4-nitroimidazole lesion by human RNA polymerase II and bacteriophage T7 RNA polymerase. DNA Repair (Amst) 2008;7:1276–1288. doi: 10.1016/j.dnarep.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YH, Bogenhagen DF. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J. Biol. Chem. 1993;268:5849–5855. [PubMed] [Google Scholar]
- 53.Damsma GE, Cramer P. Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J. Biol. Chem. 2009;284:31658–31663. doi: 10.1074/jbc.M109.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-->T and A-->C substitutions. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 55.Moriya, et al. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli . Mutat. Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 56.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 57.Singer B, Dosanjh MK. Site-directed mutagenesis for quantitation of base-base interactions at defined sites. Mutat. Res. 1990;233:45–51. doi: 10.1016/0027-5107(90)90150-3. [DOI] [PubMed] [Google Scholar]
- 58.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 59.Wood ML, Esteve A, Morningstar ML, Kuziemko GM, Essigmann JM. Genetic effects of oxidative DNA damage: comparative mutagenesis of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 61.Kouchakdjian M, et al. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 62.Brooks PJ, et al. The oxidative DNA lesion 8,5'-(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 63.Damsma GE, Alt A, Brueckner F, Carell T, Cramer P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 2007;14:1127–1133. doi: 10.1038/nsmb1314. [DOI] [PubMed] [Google Scholar]
- 64.Dimitri A, Burns JA, Broyde S, Scicchitano DA. Transcription elongation past O6-methylguanine by human RNA polymerase II and bacteriophage T7 RNA polymerase. Nucleic Acids Res. 2008;36:6459–6471. doi: 10.1093/nar/gkn657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dosanjh MK, Loechler EL, Singer B. Evidence from in vitro replication that O6-methylguanine can adopt multiple conformations. Proc. Natl. Acad. Sci. U S A. 1993;90:3983–3987. doi: 10.1073/pnas.90.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 67.Choi DJ, Roth RB, Liu T, Geacintov NE, Scicchitano DA. Incorrect base insertion and prematurely terminated transcripts during T7 RNA polymerase transcription elongation past benzo[a]pyrenediol epoxide-modified DNA. J. Mol. Biol. 1996;264:213–219. doi: 10.1006/jmbi.1996.0635. [DOI] [PubMed] [Google Scholar]
- 68.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Doetsch PW. Template strand gap bypass is a general property of prokaryotic RNA polymerases: implications for elongation mechanisms. Biochemistry. 1996;35:14999–5008. doi: 10.1021/bi961455x. [DOI] [PubMed] [Google Scholar]
- 70.Zhou W, Reines D, Doetsch PW. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995;82:577–585. doi: 10.1016/0092-8674(95)90030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tudek B. Base excision repair modulation as a risk factor for human cancers. Mol. Aspects Med. 2007;28:258–275. doi: 10.1016/j.mam.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 73.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 74.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat. Rev. Mol. Cell. Biol. 11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 75.Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–259. doi: 10.4161/cc.5.3.2414. [DOI] [PubMed] [Google Scholar]
- 76.Holmquist GP. Cell-selfish modes of evolution and mutations directed after transcriptional bypass. Mutat. Res. 2002;510:141–152. doi: 10.1016/s0027-5107(02)00259-2. [DOI] [PubMed] [Google Scholar]
- 77.Rodin SN, Rodin AS, Juhasz A, Holmquist GP. Cancerous hyper-mutagenesis in p53 genes is possibly associated with transcriptional bypass of DNA lesions. Mutat. Res. 2002;510:153–168. doi: 10.1016/s0027-5107(02)00260-9. [DOI] [PubMed] [Google Scholar]
- 78.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Derheimer FA, et al. RPA and ATR link transcriptional stress to p53. Proc. Natl. Acad. Sci. U S A. 2007;104:12778–12783. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 81.Yamaizumi M, Sugano T. U.v.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene. 1994;9:2775–2784. [PubMed] [Google Scholar]
- 82.Frosina G. The current evidence for defective repair of oxidatively damaged DNA in Cockayne syndrome. Free Radic. Biol. Med. 2007;43:165–177. doi: 10.1016/j.freeradbiomed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 83.Cooper PK, Nouspikel T, Clarkson SG. Retraction. Science. 2005;308:1740. doi: 10.1126/science.308.5729.1740b. [DOI] [PubMed] [Google Scholar]
- 84.Cozzarelli NR. Editorial expression of concern. Proc. Natl. Acad. Sci. U S A. 2003;100:11816. doi: 10.1073/pnas.2034938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Le Page F, et al. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2005;123:711. doi: 10.1016/j.cell.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 86.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 87.Pastoriza-Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb(−)/Ogg1(−) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 88.Larsen E, Kwon K, Coin F, Egly JM, Klungland A. Transcription activities at 8-oxoG lesions in DNA. DNA Repair (Amst) 2004;3:1457–1468. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 89.El-Agnaf OM, et al. Aggregates from mutant and wild-type alpha-synuclein proteins and NAC peptide induce apoptotic cell death in human neuroblastoma cells by formation of beta-sheet and amyloid-like filaments. FEBS Lett. 1998;440:71–75. doi: 10.1016/s0014-5793(98)01418-5. [DOI] [PubMed] [Google Scholar]
- 90.van Leeuwen FW, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 91.Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer's disease ventricular CSF. J. Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- 92.Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech. Ageing Dev. 2008;129:366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bridges BA. Starvation-associated mutation in Escherichia coli: a spontaneous lesion hypothesis for "directed" mutation. Mutat. Res. 1994;307:149–156. doi: 10.1016/0027-5107(94)90287-9. [DOI] [PubMed] [Google Scholar]
- 94.Bridges BA. Mutation in resting cells: the role of endogenous DNA damage. Cancer Surv. 1996;28:155–167. [PubMed] [Google Scholar]
- 95.Brégeon D, Matic I, Radman M, Taddei F. Inefficient mismatch repair: genetic defects and down regulation. J. Genet. 1999;78:21–28. [Google Scholar]
- 96.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. Embo J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C --> T transitions in vivo. Proc. Natl. Acad. Sci. U S A. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belousova EA, et al. DNA polymerases beta and lambda bypass thymine glycol in gapped DNA structures. Biochemistry. 2010;49:4695–4704. doi: 10.1021/bi901792c. [DOI] [PubMed] [Google Scholar]
- 99.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu F, et al. Peroxynitrite-induced reactions of synthetic oligo 2'-deoxynucleotides and DNA containing guanine: formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- 101.Neeley WL, et al. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 102.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 103.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U S A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuraoka I, et al. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase eta and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J. Biol. Chem. 2001;276:49283–49288. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 105.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 106.Chary P, Lloyd RS. In vitro replication by prokaryotic and eukaryotic polymerases on DNA templates containing site-specific and stereospecific benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide adducts. Nucleic Acids Res. 1995;23:1398–1405. doi: 10.1093/nar/23.8.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mah MC, Boldt J, Culp SJ, Maher VM, McCormick JJ. Replication of acetylaminofluorene-adducted plasmids in human cells: spectrum of base substitutions and evidence of excision repair. Proc. Natl. Acad. Sci. U S A. 1991;88:10193–10197. doi: 10.1073/pnas.88.22.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta PK, et al. Mutagenesis by single site-specific arylamine-DNA adducts. Induction of mutations at multiple sites. J. Biol. Chem. 1989;264:20120–20130. [PubMed] [Google Scholar]
- 109.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the "A rule". J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]