Abstract
Valvular Interstitial Cells (VICs) are a common substrate for congenital and adult heart disease yet the signaling mechanisms governing their formation during early valvulogenesis are incompletely understood. We developed an unbiased strategy to identify genes important in endocardial epithelial-to-mesenchymal transformation (EMT) using a spatial transcriptional profile. Endocardial cells overlaying the cushions of the atrioventricular canal (AVC) and outflow tract (OFT) undergo an EMT to yield VICs. RNA sequencing (RNA-seq) analysis of gene expression between AVC, OFT, and ventricles (VEN) isolated from chick and mouse embryos at comparable stages of development (chick HH18; mouse E11.0) was performed. EMT occurs in the AVC and OFT cushions, but not VEN at this time. 198 genes in the chick (n=1) and 105 genes in the mouse (n=2) were enriched 2-fold in the cushions. Gene regulatory networks (GRN) generated from cushion-enriched gene lists confirmed TGFβ as a nodal point and identified NF-κB as a potential node. To reveal previously unrecognized regulators of EMT four candidate genes, Hapln1, Id1, Foxp2, and Meis2, and a candidate pathway, NF-κB, were selected. In vivo spatial expression of each gene was confirmed by in situ hybridization and a functional role for each in endocardial EMT was determined by siRNA knockdown in a collagen gel assay. Our spatial-transcriptional profiling strategy yielded gene lists which reflected the known biology of the system. Further analysis accurately identified and validated previously unrecognized novel candidate genes and the NF-κB pathway as regulators of endocardial cell EMT in vitro.
1. INTRODUCTION
Valvular heart disease comprises a major portion of cardiovascular disease in children and adults. Congenital Heart Disease (CHD), which includes a spectrum of valvular defects, occurs in 1 out of 100 live births and is responsible for ten percent of infant deaths per year[1, 2]. Ninety percent of children with CHD now live to adulthood due to the surgical and medical advances of the past half century[3]. As a result, adults now represent the largest age group with CHD, but because they are a relatively new class of patient, treatment approaches are as yet unclear[4]. Also, it is now well recognized that there is a developmental basis to adult cardiovascular and valve disease. It is reasonable to assume that the homeostatic and repair functions present in adults would use many of the same mechanisms involved in the original development and remodeling of these tissues[4]. Therefore, the careful examination of genes and signaling pathways governing early valve development is likely to provide novel insight into the mechanisms underlying valve disease and suggest potential new therapies.
Valvular interstitial cells (VICs) are a common substrate of both congenital and adult valvular disease[4]. These cells are derived primarily from specialized endothelium, located in the atrioventricular (AVC) and outflow tract (OFT) cushion regions of the developing heart tube, which undergoes EMT[1]. VICs direct both the remodeling of the ECM during valvulogenesis in the embryo and the maintenance of the structure of the valve leaflets in the adult. Despite a continued interest in the process of endocardial EMT (reviewed [5-7]) our understanding remains incomplete and often progresses at the pace of a single gene study at a time. We directed our analysis to an unbiased approach identifying genes and pathways associated with endocardial EMT. Previous studies have used a variety of gene expression analysis approaches to identify cushion-enriched genes in the AVC[8-11]. Comparisons of normal and aberrant cushion development, a strategy predicated on the availability of models with known heart defects, yielded only genes associated with the perturbed signaling pathway[9]. While this approach has utility, many of the genes and signaling pathways associated with normal cushion development have yet to be determined. A second approach identified genes temporally regulated in the AVC by comparing different stages of development[8, 11]. This strategy identified pathways that change over time in the AVC but pathways specific to endothelial EMT were not directly queried. Both of these approaches focused on the use of a single spatial region of the heart for analyses.
We recognized that the inherent regional specificity observed in the developing heart tube presents a unique opportunity for analysis of endocardial EMT. EMT occurs specifically in the endothelium of the AVC and OFT but not in the adjacent endothelium of the common ventricle (VEN). Thus, a spatial transcriptional profile of these three regions of the heart tube enables a direct comparison between regions that do and do not undergo EMT. Further, the generation of data from these regions allows for genes expressed in the nontransforming ventricle to be subtracted from those genes expressed in the AVC and OFT in order to enrich for genes associated with endothelial EMT. This approach expands on earlier studies which focused on the AVC by comparisons that allow background subtraction of extraneous, non-target cell populations present during EMT. For example, at E10.5 proepicardial cells attach to the AVC[12] and subsequently migrate over the myocardium coincident with a population of PAX-3-positive cardiac neural crest cells entering the OFT[13] . The approach here, which identifies genes with enriched expression specific to both the AVC and OFT, excludes genes expressed by either of these non-target cell populations, which are contained in only a single cushion. Therefore, this focuses our analysis on identifying genes directing endocardial EMT and their associated pathways in early valvulogenesis.
2. METHODS
2.1 Embryo dissection
HH18 chick (n=200, yielding 8μg of RNA) and E11.0 ICR (n=100; n=60; yielding 17μg and 10μg of RNA, respectively) mouse hearts were dissected into AVC, OFT, and VEN (Figure 1A-B) and the samples were pooled. Standard phenol-chloroform extraction of RNA was performed.
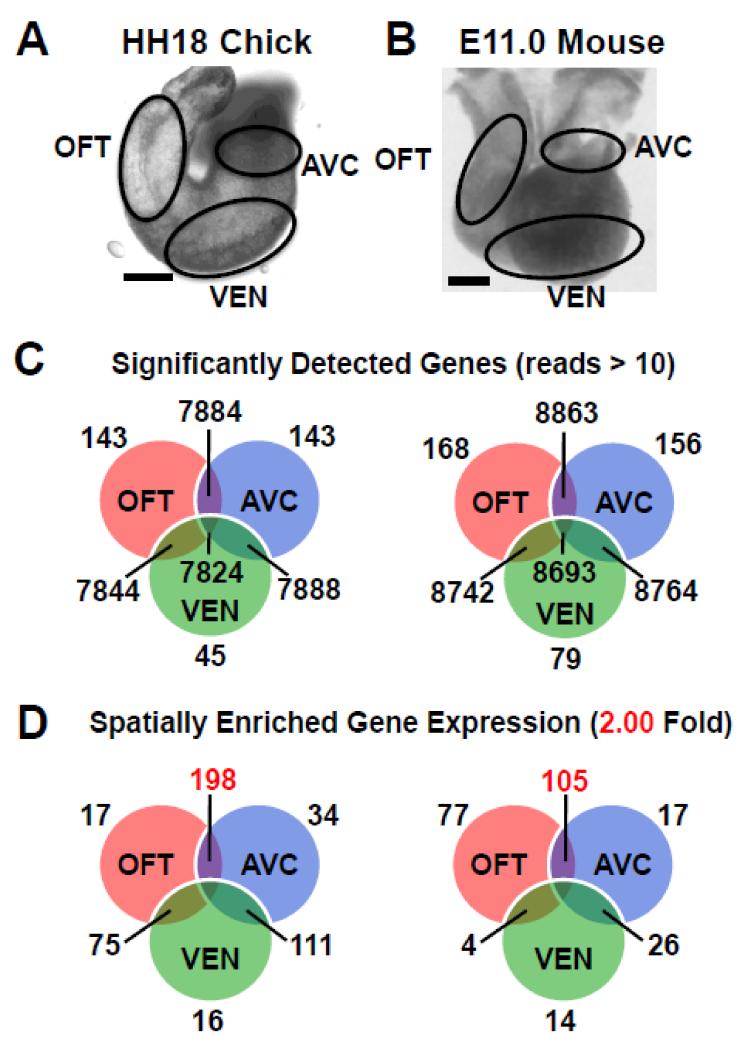
Figure 1. Spatial transcriptional profile of heart identifies cushion-enriched genes.
A-B, AVC, OFT and VEN were dissected from equivalent stages of chick HH18 (A) and mouse E11.0 (B). C, RNA-seq analysis identified genes with significantly expressed (>10 reads) in each spatial region of the developing heart tube. D, Mapping genes with enriched expression (>2-fold) onto each spatial region identifies those genes with cushion-enriched expression (in red), 198 in the chick and 105 in the mouse. Scale bars - 100μM.
2.2 RNA-seq
RNAseq libraries were generated as described without normalizations or RNA/cDNA fragmentation[14]. Libraries were sequenced as 50bp paired end sequences on a single lane of the Illumina HiSeq2000 producing 7 million reads per chick sample and an average of 10 million reads per mouse sample. HiSeq 2000 reads are aligned by TOPHAT[15] (http://tophat.cbcb.umd.edu/) to produce bam files. Gene expression profiles were generated as described[16] using a Bayesian p-value[17]. Reads were normalized to total mRNA (total aligned reads on gene-loci per million). Data files have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed via series record GSE43194. Variability was assessed as described in Figure S1 and S2.
2.3 RT-qPCR
First-strand synthesis was done with SuperScript III kit (Invitrogen). SYBR Fas Green (Biorad) was used for qPCR. Primer sequences are in Table S1.
2.4 Collagen Gel Assay
Collagen gel assays (reviewed[5]) with siRNA or small molecule addition were performed as described[18]. AVC were excised from stage 16 chick embryos, transected, and explanted endocardial side down onto a collagen I gel and incubated at 37°C with siRNA constructs. Control for siRNA toxicity was a randomized GC matched construct that do not correspond to any sequence. The positive control was siRNA targeting Tgfbr3[19]. siRNA sequences are listed in Table S2. EMT was quantified by counting the number cells with mesenchymal morphology that invaded into the collagen gel (Table S3).
2.5 In Situ Hybridization
Dioxygenin labeled probes were generated from cDNA derived from E13.5 whole heart tubes using primers in Table S4. In situ hybridization was performed as described[20] and embryos were cryosectioned.
3. RESULTS
3.1 Defining a spatial transcriptional profile of the developing heart tube
Chick and mouse models were used for their relative strengths as model systems and the ability to use cross species comparison to identify key regulatory genes. Equivalent stages in the chick (HH18) and mouse (E11.0) were chosen when robust EMT occurs in both the OFT and AVC but not the VEN at this time[21]. Spatial transcriptional profiles of chick and mouse heart tubes were generated as described in methods (Figure 1A-B).
The dynamic nature of gene expression profiles generated by RNA-seq identified approximately 8,000 genes expressed throughout either the chick or mouse heart tube. A smaller number of genes were also localized to specific regions such as the AVC, OFT, & VEN (Figure 1C). As a first step to enrich for genes mediating EMT, we subtracted genes found in the VEN sample from the AVC & OFT samples. By identifying genes upregulated in both cushions in the comparison we eliminated the influence of the epicardial cells (AVC) and neural crest cells (OFT) which are found only in a single cushion sample and therefore not in the shared gene list we identified (Figure 2A; see Wt1 (AVC, VEN), Sox10 (OFT),BMP10 (VEN)).
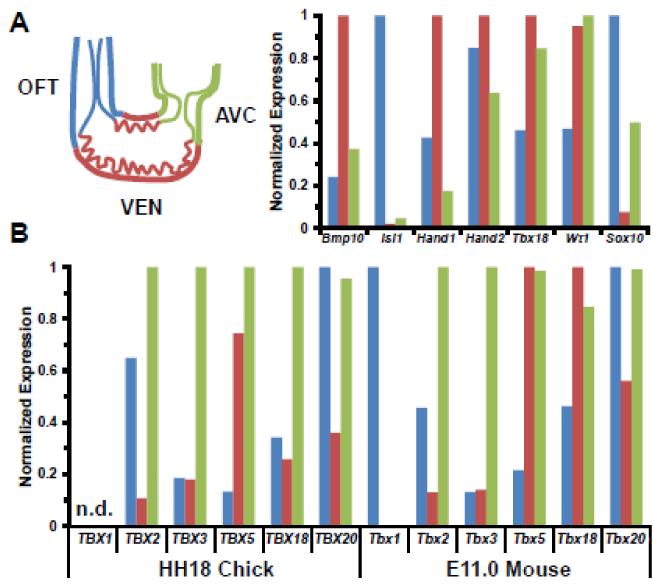
Figure 2. Spatial expression of select genes in the HH18 chick and E11.0 mouse heart tube.
For each gene, expression was normalized to the region with the highest read count A, Regional expressed patterns in the mouse heart are consistent with previous observations at E10.5-E11.5. Bmp10 (VEN), Isl1 (OFT), Hand1 (VEN), Hand2 (endocardium throughout heart tube), Tbx18 and Wt1 (Epicardium attaching to AVC and migrating towards VEN), and Sox10 (neural crest in OFT and in OFT and AVC cushions). B, Expression of T-box genes in chick and mouse are consistent with previous reports (reviewed in [50]). n.d.- TBX1 has not been annotated in the chick genome and is not included in the analysis.
This approach yielded a robust list of genes upregulated in the cushions. We mapped genes enriched >2-fold in each spatial region (Figure 1D) Genes associated with distinct regions of the heart tube in chick and mouse cushions, including the T-box family members, had spatial expression patterns consistent with the literature (Figure 2B). We observe a few hundred genes enriched in the cushions, consistent with the hypothesis that the AVC and OFT should share common genes involved in endothelial EMT (Figure 1D). Comparisons of the AVC & VEN or OFT & VEN did not reveal as many enriched genes since they do not share EMT as a common process. Therefore, genes with a significant p-value (p<.001) and >2-fold higher expression in the cushions (AVC & OFT) compared to VEN are considered to be enriched in these compartments and were potential candidates for involvement in EMT.
Overall, 198 genes were identified in the chick (Table S5) and 105 in the mouse (Table S6) that were >2-fold higher expressed in the cushions. A literature search was used to further characterize these gene lists. In total, 15 of the 198 genes in chick and 18 of the 105 genes in mouse have a previously described role in EMT (Table S7). Genes known to be expressed regionally in the myocardium (Bmp4[22], BMP2[22]) were identified as having cushion enriched expression in chick and mouse as expected. Of note, 31 of 105 identified in the mouse were found to be expressed in the endothelium or mesenchyme derived from endothelium. Thus, the RNA-seq methodology was sensitive enough to detect changes in gene expression in the endothelium and mesenchyme when the majority of the sample is derived from the myocardium.
Relaxation of the stringency of gene enrichment to a level of >1.25-fold (p<0.001) (Figure S3) revealed 574 genes enriched in the chick (Table S5) and 250 enriched in the mouse (Table S6). Of these, 54 genes in the chick and 41 genes in the mouse were associated with abnormal heart development and function. Several genes known to play important roles in endocardial EMT were revealed in the >1.25-fold gene lists. Though there is an increased chance of including false positive genes by lowering the stringency, there may also be novel candidate genes in the 1.25 fold gene lists that would otherwise be missed with a 2 fold cutoff. In particular, small changes (<2-fold) in the expression levels of transcription factors may have a significant effect on cell behavior[23].
3.2 Gene ontology (GO) analysis of cushion-enriched genes
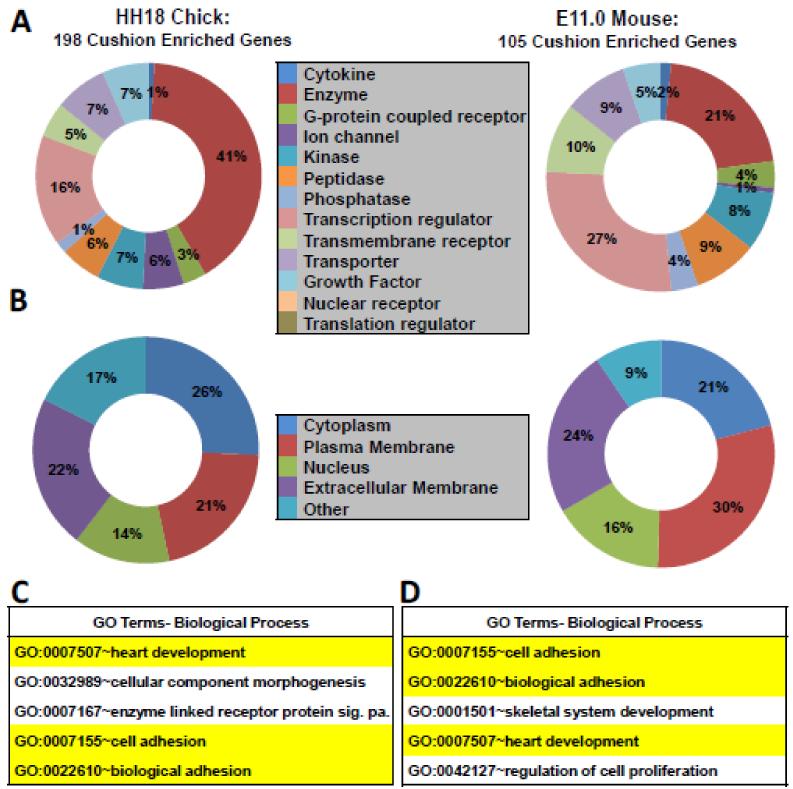
To gain a better understanding of the genes in the >2-fold cushion enriched gene lists we examined the associated predicted protein location, predicted protein function and biological processes (Figure 3). Enriched genes encoded proteins with various predicted functions and cell locations, including proteins predicted to be transcriptional regulators (Figure 3A-B). To examine which biological processes were associated with the cushion-enriched gene lists in chick and mouse we performed functional annotation using Database for Annotation, Visualization, & Integrated Discovery (DAVID) software[24]. Several biological processes were identified in chick (Table S8) and mouse (Table S9) genes lists. Consistent with the shared underlying morphological processes occurring in the cushions of each, we identified biological processes enriched in both chick and mouse associated with the process of EMT (Figure 3C-D). Biological processes related to bone development (skeletal system organization) were observed and are consistent with the prior observation that there is a high degree of conservation in signaling pathways which regulate bone and valve development[8]. GO analysis of the >1.25-fold gene lists yielded results similar to those obtained with the >2-fold gene lists (Figure S4). Thus, GO analysis of cushion-enriched gene lists in chick and mouse identified shared processes expected for a population of endothelial cells undergoing EMT.
Figure 3. Gene ontology, predicted protein location, and protein function of cushion-enriched genes.
A, Predicted protein function of genes identified in the mouse and chick cushion-enriched gene lists generated using IPA software. Percent of total genes with defined function is depicted. Genes with unknown function in chick (89) and mouse (44) are not shown. B, Predicted protein location of genes identified in mouse and chick cushion using IPA software. C-D, Analysis of genes found >2-fold enriched in chick and mouse AVC and OFT using DAVID software. Selected significantly enriched biological processes (from DAVID BP-FAT) are depicted for chick and mouse (p<0.0001 for all terms). Significance was based on p-values generated by DAVID software[24]. Biological processes highlighted in yellow are shared between chick and mouse.
3.3 Gene regulatory network (GRN) analysis of cushion-enriched genes
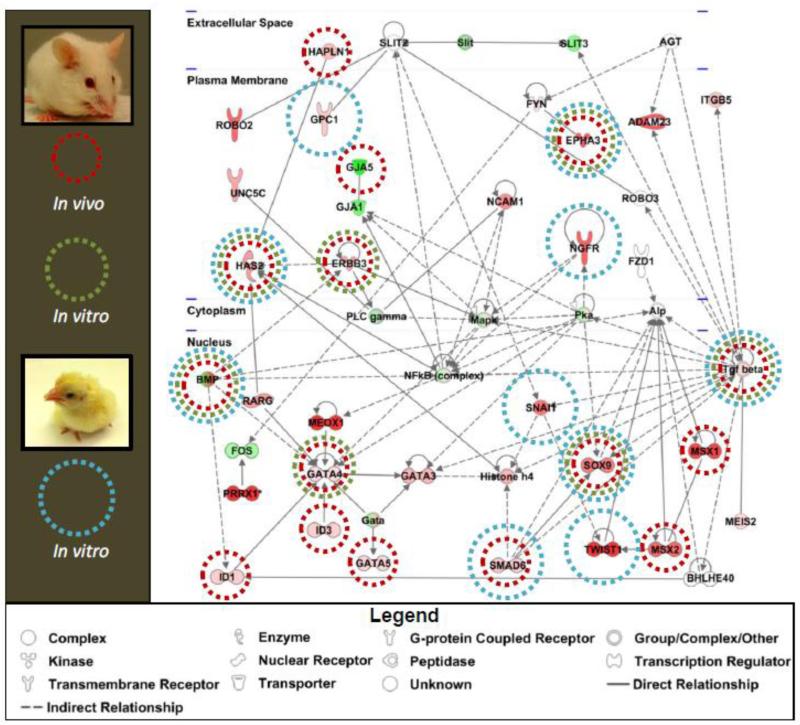
To gain a better understanding of how the genes enriched in the cushion interact with each other, GRN analysis was undertaken of the chick and mouse >2-fold differentially expressed gene lists using Ingenuity Pathway Analysis software (IPA) (www.ingenuity.com). Figure 4 depicts an example mouse gene network. Consistent with the known importance of endocardial EMT in valve development several genes in this network have been previously demonstrated to regulate EMT in vitro in mice (green circles) or in chick (blue circles). We observed several genes in the network that when targeted in mice resulted in phenotypes associated with abnormal cushion or valve development (red circles). This high number of genes known to be important in cushion or valve development provides confidence that this gene network reflects the biological processes occurring in the cushions. Our analysis confirms TGFβ as a signaling node in regulating endocardial EMT and valvulogenesis (reviewed[6]). This network also identified NF-κB as a node and the role of NF-κB in endocardial EMT is undescribed. Thus, GRN analysis of the cushion-enriched gene lists in chick and mouse provided insight into the relationships between genes known to regulate endocardial EMT and identified candidate signaling pathways for functional analysis.
Figure 4. Gene regulatory networks generated from cushion-enriched-gene lists identifies known regulators of endocardial EMT.
Gene regulatory networks were generated with IPA software using >2-fold cushion-enriched genes in chick and mouse. An example mouse network is depicted. Lines denote interactions between genes observed in the literature across all systems, with solid lines reflecting a direct interaction and doted lines denoting an indirect interaction. Gene names over shapes denote nodes. Green nodes have decreased expression and red have increased expression. Additional genes were annotated based on known interactions in the literature (Bmp). Hapln1 and Meis2 nodes were added to identify potential interactions of novel candidate genes in this network. Genes were marked that are known to regulate EMT in mice in vivo (green circles), in mice in vitro (red circles), and in chick in vitro (blue circles).
3.4 Identification of gene candidates for functional analysis
We selected candidate genes for further analysis using our knowledge of the biology of the system to target genes involved in critical signaling pathways. Hapln1, Id1, Meis2, and Foxp2 comprised our slate of candidate genes which were selected from both the >2-fold (Hapln1 (chick) and Foxp2 (mouse)) and >1.25 fold (Id1 (mouse) and Meis2 (mouse)) cushion enriched gene lists. For each candidate, whole mount in situ hybridization in E11.0 mouse embryos confirmed the expression pattern predicted by RNA-seq analysis (Figure 5). HAPLN1, or CRTL1, interacts with Hyaluronic Acid (HA) and additional ECM proteins in the extra-cellular space to affect ECM organization[25, 26]. The deletion of Hapln1 in mice results in hypoplastic valves[10]. Inhibitor of DNA binding 1, ID1, is a helix-loop-helix (HLH) protein that acts as a dominant negative antagonist of basic HLH transcription factors and can regulate both TGFβ and BMP signaling. MEIS2 has recently been reported to be required for cardiac looping in zebra fish but has no described function in valve development[27]. FOXP2 is a forkhead/winged-helix transcription factor that has been extensively studied in the context of neural development. It is required for birds to learn song and mutations in FOXP2 have been linked to human speech pathologies[28]. Although some candidates have been studied in valvulogenesis, none have a described role in endocardial EMT.
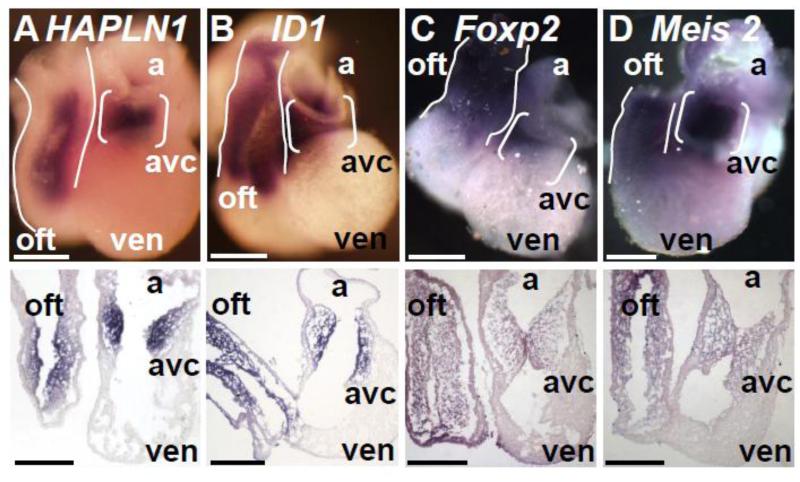
Figure 5. Confirmation of cushion-enriched gene expression in E11.0 hearts.
A-D, Whole mount in situ hybridization with probes specific for (A) Hapln1, (B) Id1, (C) Foxp2, and (D) Meis2 in E11.0 embryos in whole hearts (top) and 10 μM cryosections (bottom) revealed enriched expression in the AVC (brackets) and OFT (outlines) compared to VEN. avc- atrioventricular canal, oft- outflow tract, a- atria, ven- ventricle. Scale bars: 100μM.
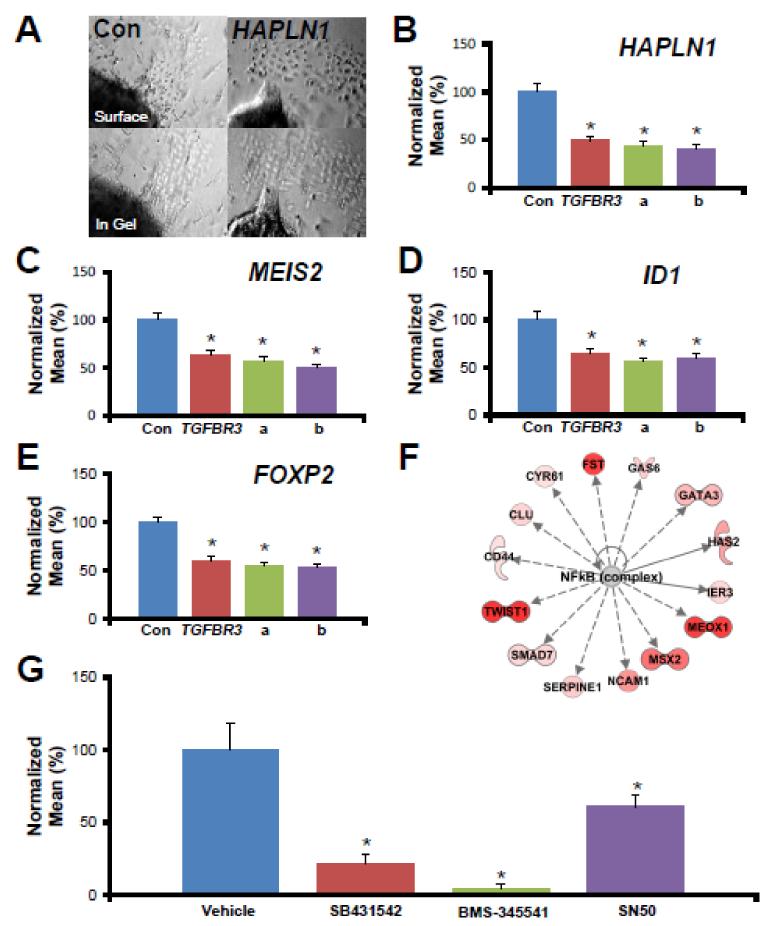
A functional analysis using siRNA to target candidate genes in the well-defined chick AVC explant collagen gel assay in vitro was used to assess any role in endocardial EMT (reviewed[5, 6]). Each of two independent siRNA constructs targeting candidates resulted in a decrease of endocardial EMT (Figure 6 A-E). qPCR analysis of explants confirmed both the expression and successful targeting of these genes in the chick AVC explants (Figure S5). These results establish a role for each candidate in the regulation of endocardial EMT in vitro and confirm our analysis was successful in identifying genes that regulate endocardial EMT. The lack of effect of control siRNA on EMT and the reported successful targeting of genes and pathways in this model that display no effect on EMT[18, 19] supports the validity and specificity of these results.
Figure 6. Candidate genes are required for endocardial EMT in vitro.
A, Representative photomicrographs of AVC explants incubated with HAPLN1-targeted siRNA. B-E, Candidate gene expression in HH16 chick embryos was knocked down with siRNA in an in vitro collagen gel assay. The mean number of cells in the collagen gel was determined and normalized to the number of cells in controls (100%). Con - GC-content matched, randomized siRNA constructs with no homology to any known chick gene, TGFβR3 - siRNA targeting a gene known to be required for EMT in vitro. a or b - independent constructs targeting indicated gene. * - p<0.01. F, Transcriptional network depicting genes known to be downstream of NF-κB which had increased expression in the AVC and VEN. See legend in Figure 4. G, NF-κB inhibition by two independent small molecule inhibitors (10 μM BMS 345541 and 2 μM SN50) resulted in a loss of function an in vitro collagen gel assay. SB431542, an ALK5 kinase inhibitor, was used as a positive control. Vehicle - 0.01% DMSO. * - p<0.01.
3.5 Selection and validation of signaling pathway identified by GRN analysis
An advantage of GRN analysis lies in the ability to generate networks from a list of differentially expressed genes to predict the involvement of genes and signaling pathways where expression is unchanged. This approach can identify potential regulatory nodes that may escape notice if only gene expression is examined. One such example from our GRN analysis is the NF-κB pathway (Figure 4) where signaling activity is regulated on the protein level (reviewed[29]) making it unlikely to appear in a screen of gene expression. Indeed, NF-κB pathway signaling components were not found in our cushion-enriched gene lists yet our strategy of using GRN analysis allowed us to identify NF-κB as a potential regulator of endocardial EMT. Many upstream regulators of NF-κB were observed and more edges were associated with NF-κB than other nodes, such as Pka (Figure 4). We also examined those genes regulated by NF-κB in the cushion-enriched gene list. Figure 6F depicts genes enriched in the AVC and OFT which are known to be directly or indirectly regulated by NF-κB signaling, including several genes known to be important in endocardial EMT (Has2, Msx2, Twist1). NF-κB signaling has long been associated with adult heart disease (reviewed[30]) and has recently been linked to FGF-2 mediated EMT in corneal endothelial cells[31]. However, the role of NF-κB signaling in endocardial EMT has not been investigated. Together, these data and the known role of NF-κB in EMT in other systems led us to select NF-κB for further study.
To test for a role of NF-κB in endocardial EMT we took advantage of the availability of small molecule inhibitors of NF-κB signaling. We chose 2 different inhibitors (10 μM BMS 345541 and 2 μM SN50) with differing mechanisms of action to block NF-κB function in the collagen gel assay. BMS 345541 specifically inhibits the phosphorylation of IκB kinase by inhibition of IKK[32] which prevents IκB targeting for degradation and the subsequent release of NF-κB into the nucleus. SN50 is a synthetic peptide which blocks the nuclear import of the NF-κB complex itself[33]. Some explants were also incubated with 2.5 μM SB431542, an ALK5 inhibitor previously characterized in this system[18], as a positive control for small molecule inhibition of EMT, or vehicle as a negative control. Incubation with BMS 345541 or SN50 resulted in a 31% or 79% reduction in the number of cells invading into the collagen gel compared to vehicle-incubated explants, respectively (Figure 6 G). Together, these data indicate that NF-κB signaling can regulate endocardial EMT in vitro.
4. DISCUSSION
In this study we used an unbiased approach to identify important regulators of cardiac valvular EMT. Previous studies have used a similar spatial analysis in E10.5 mouse hearts in an effort to identify genes associated with endocardial EMT and valvulogenesis. Analysis of the AVC and VEN by microarray[10] and, more recently, examination of micro-dissected atria, AVC, OFT, and VEN by TAG-seq[34], generated lists of genes enriched in the regions of the heart tube undergoing EMT. This information was used to identify and target a single gene, Hapln1, in vivo in mice. However, our approach was unique in that it coupled the power of spatial analysis with a rapid in vitro bioassay to confirm a role for candidate genes and pathways in endocardial EMT. To that end we took advantage of the longstanding collagen gel chick AVC explant assay and expanded our spatial analysis to include equivalent stages of both mouse and chick heart tubes. The inclusion of the chick model allowed for cross species comparison and provided primary data obtained from the species upon which our in vitro assay is based.
The genes identified as enriched in these two models were subjected to GO and GRN analysis which revealed biological processes, signaling pathways and gene interactions known to be important in endocardial EMT. Among others, key components of TGFβ (TGFβ2 chick/mouse), BMP (Bmp2 mouse), retinoic acid (CYP26B1 chick/mouse), and WNT (WNT4 chick, Wnt9b mouse) signaling pathways were revealed. Several genes common to both lists were known to play a role in valve formation (Table S10). Thus, the cushion-enriched gene lists accurately reflect the biological processes associated with endocardial EMT and suggest candidate genes and networks for functional confirmation.
We identified and confirmed the involvement of novel genes and a novel pathway in endocardial EMT in vitro. HAPLN1 is expressed throughout the heart tube at E10.0 and becomes restricted to the endocardium and mesenchyme of the AVC and OFT by E11.0 (Figure 5A)[10]. Deletion of Hapln1 in mice resulted in hypoplastic valves similar to, but less severe than, the null phenotypes of HA and Versican, both of which HAPLN1 binds[10, 26, 35]. The authors noted that it was unclear whether the lack of cellularization of the cushions in these animals was due to a defect in post-EMT cell proliferation or in endothelial EMT itself[10]. Our data suggests that the hypoplastic valves observed in Hapln1 null mice may be at least partially due to aberrant endocardial EMT. Similarly, Id1 is expressed in the endothelium and cushion mesenchyme at E10.5/E11.0 (Figure 5B) [36, 37]. Deletion of both Id1 and Id3 leads to hypoplastic cushions associated with discontinuous endothelium in the context of disrupted heart development[37]. Targeting of Id1 in the endothelium of Id3 null mice yields a less severe phenotype that displays signs of aberrant cushion mesenchyme development associated with valvular and septal defects[38]. These studies point to a potential role for Id1 in VIC development, yet Id1 had not been shown to regulate endothelial EMT. Our results suggest that compromised endothelial EMT may contribute to the phenotypes seen in Id1 null mice.
Members of the FOXP family of transcription factors have been implicated in heart development including FOXP1[39] and FOXP4[40]. At E11.0, Foxp2 is expressed abundantly in the endocardium and mesenchyme of the OFT and AVC when compared to the VEN as well as in the myocardium of the OFT (Figure 5C). Both FOXP2 and FOXP4 have been shown to promote the delamination of neuroepithelial cells in the mouse[41] demonstrating a role for each in regulating EMT in vivo. Foxp4 was also enriched in our RNA-seq data sets and can form heterodimers with FOXP2[42]. The presence of Foxp4 in the cushions suggests that FOXP4 may compensate for the loss of FOXP2, explaining the lack of a reported cushion or valve phenotype in Foxp2 null mice[43].
A second transcription factor, MEIS2, is expressed in the endocardium and mesenchyme of the AVC and OFT at E11.0 (Figure 5D). Recent studies in zebrafish identified a role for meis2 in cardiac looping[27], yet no data concerning the role of MEIS2 in chick or mouse heart development has been reported. MEIS2 induces proximal gene expression while BMP signaling inhibits MEIS2 expression to promote distal limb gene expression and support proper limb patterning[44]. The well described role of BMP in regulating endocardial EMT[45] provides a potential role for MEIS2 as a modulator of this process and subsequent valve maturation.
Although the expression of components of the NF-κB pathway were not enriched in the AVC or OFT, GRN analysis revealed that several genes regulated by NF-κB had enriched expression in the AVC and OFT. Perturbation of the NF-κB signaling pathway with small molecule inhibitors revealed a requirement for NF-κB signaling in endocardial EMT. As the role of NF-κB in driving EMT in cancer cells via regulation of Snail/Slug/Twist is well described (reviewed[46]), the NF-κB pathway may represent a mechanism by which transcription factors that repress the epithelial phenotype in endocardial EMT are regulated. Mouse embryos lacking p50 survive to adulthood with no apparent developmental defects [47] while mice lacking p65[48] die at E15.5 from liver degeneration by apoptosis. No valvular phenotypes were reported. Our findings suggest a targeted re-examination of NF-κB signaling in valve development may reveal previously unappreciated defects. It is also possible that disruption of NF-κB signaling in VICs may result in adult valvular disease or altered response to injury.
It is clear that the in vitro bioassay does not always recapitulate in vivo phenotypes (reviewed[5]). The more complex ECM and signaling environment present in vivo may allow endocardial cells to compensate in ways cells exposed to the simpler collagen gel system cannot. For example, targeting of Tgfbr2 in vivo does not inhibit endocardial EMT, but AVC explants from these mice fail undergo EMT in vitro[49]. However, later stages of cushion development are abnormal in Tgfbr2 conditionally null mice, suggesting that the mesenchyme resulting from EMT is abnormal. Therefore, this system provides a robust bioassay to identify potential targets for gene perturbation in vivo.
Other candidate genes identified by our analysis were left untested, but represent interesting avenues for future work. For example, ECM remodeling is fundamentally important in valve development. We identified several genes encoding ECM related proteins that were enriched in mouse cushions including collagens (Col2a1, Col6a3, Col9a1, Col9a3), hyaluronic acid (Has2, Cd44), and Eln. Knowledge of the ECM, which the mesenchymal cells and VICs must interact with and remodel subsequent to EMT, may provide important clues on how to better design in vitro systems that promote VIC differentiation and maintenance. Future studies combining spatial and temporal transcriptional profiles will likely provide further insight into endocardial EMT.
Supplementary Material
Highlights.
We generated spatial transcriptional profiles of E11.0 mouse and HH18 chick hearts
Genes enriched in the valve forming regions of the heart were indentified
Gene network analysis identified candidate genes and pathways
Hapln1, Id1, Meis2, and Foxp2 were required for endocardial EMT in vitro
NF-κB signaling pathway was validated as a regulator of endocardial EMT in vitro
ACKNOWLEDGEMENTS
The authors thank Tyson Foods, Inc. for chicken eggs and Drs. Chris Brown, Todd Townsend, Tom Doetschman, and Mr. Josh Gorham for helpful discussions, advice, and technical assistance.
SOURCES OF FUNDING
This study was funded by an initiative of the Roadmap for Medical Research/Common Fund, Systems-based Consortium for Organ Design and Engineering, NIH U54 092551 (DMD, HSB, JCS, JVB) and NHLBI Cardiac Development Consortium 1U01HL098166 (JGS).
Abbreviations
- AVC
Atrioventricular canal
- CHD
Congenital heart disease
- EMT
Epithelial-to-mesenchymal transformation
- GRN
Gene regulatory networks
- OFT
Outflow tract
- RNA-seq
RNA sequencing
- VEN
Ventricle
- VIC
Valvular interstitial cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
None disclosed.
REFERENCES
- [1].Barnett JV, Desgrosellier JS. Early events in valvulogenesis: a signaling perspective. Birth Defects Res C Embryo Today. 2003 Feb;69(1):58–72. doi: 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- [2].Loffredo CA. Epidemiology of cardiovascular malformations: prevalence and risk factors. Am J Med Genet. 2000;97(4):319–25. doi: 10.1002/1096-8628(200024)97:4<319::aid-ajmg1283>3.0.co;2-e. Winter. [DOI] [PubMed] [Google Scholar]
- [3].Khairy P, Hosn JA, Broberg C, Cook S, Earing M, Gersony D, et al. Multicenter research in adult congenital heart disease. Int J Cardiol. 2008 Sep 26;129(2):155–9. doi: 10.1016/j.ijcard.2008.03.014. [DOI] [PubMed] [Google Scholar]
- [4].Doetschman TBJ, Runyan R, Camenisch TD, Heimark RL, Granzier HL, Conway SJ, Azhar A. Transforming growth factor beta signaling in adult cardiovascular diseases and repair. Cell and tissue research. 2012;347(2):539–48. doi: 10.1007/s00441-011-1241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lencinas A, Tavares AL, Barnett JV, Runyan RB. Collagen gel analysis of epithelial-mesenchymal transition in the embryo heart: an in vitro model system for the analysis of tissue interaction, signal transduction, and environmental effects. Birth Defects Res C Embryo Today. 2012 Dec;93(4):298–311. doi: 10.1002/bdrc.20222. [DOI] [PubMed] [Google Scholar]
- [6].DeLaughter DM, Saint-Jean L, Baldwin HS, Barnett JV. What chick and mouse models have taught us about the role of the endocardium in congenital heart disease. Birth Defects Res A Clin Mol Teratol. 2011 Jun;91(6):511–25. doi: 10.1002/bdra.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res. 2012 Jun 8;110(12):1628–45. doi: 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chakraborty S, Cheek J, Sakthivel B, Aronow BJ, Yutzey KE. Shared gene expression profiles in developing heart valves and osteoblast progenitor cells. Physiol Genomics. 2008 Sep 17;35(1):75–85. doi: 10.1152/physiolgenomics.90212.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006 Sep;133(18):3607–18. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wirrig EE, Snarr BS, Chintalapudi MR, O’Neal JL, Phelps AL, Barth JL, et al. Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev Biol. 2007 Oct 15;310(2):291–303. doi: 10.1016/j.ydbio.2007.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vrljicak P, Chang AC, Morozova O, Wederell ED, Niessen K, Marra MA, et al. Genomic analysis distinguishes phases of early development of the mouse atrio-ventricular canal. Physiol Genomics. 2010 Feb 4;40(3):150–7. doi: 10.1152/physiolgenomics.00142.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981 Sep;201(1):157–68. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- [13].Conway SJ, Henderson DJ, Copp AJ. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development. 1997 Jan;124(2):505–14. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- [14].Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol. 2011 Apr; doi: 10.1002/0471142727.mb0412s94. Chapter 4: Unit4 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Christodoulou DC, Gorham JM, Kawana M, DePalma SR, Herman DS, Wakimoto H. Ausubel Frederick M, et al., editors. Quantification of gene transcripts with deep sequencing analysis of gene expression (DSAGE) using 1 to 2 microg total RNA. Current protocols in molecular biology. 2011 Jan; doi: 10.1002/0471142727.mb25b09s93. Chapter 25: Unit25B 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997 Oct;7(10):986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- [18].Townsend TA, Wrana JL, Davis GE, Barnett JV. Transforming growth factor-beta-stimulated endocardial cell transformation is dependent on Par6c regulation of RhoA. J Biol Chem. 2008 May 16;283(20):13834–41. doi: 10.1074/jbc.M710607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Townsend TA, Robinson JY, Deig CR, Hill CR, Misfeldt A, Blobe GC, et al. BMP-2 and TGFbeta2 shared pathways regulate endocardial cell transformation. Cells Tissues Organs. 2011;194(1):1–12. doi: 10.1159/000322035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei Q, Manley NR, Condie BG. Whole mount in situ hybridization of E8.5 to E11.5 mouse embryos. J Vis Exp. (56):2011. doi: 10.3791/2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Camenisch TD, Molin DG, Person A, Runyan RB, Gittenberger-de Groot AC, McDonald JA, et al. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev Biol. 2002 Aug 1;248(1):170–81. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- [22].Keyes WM, Logan C, Parker E, Sanders EJ. Expression and function of bone morphogenetic proteins in the development of the embryonic endocardial cushions. Anat Embryol (Berl) 2003 Sep;207(2):135–47. doi: 10.1007/s00429-003-0337-2. [DOI] [PubMed] [Google Scholar]
- [23].Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000 Apr;24(4):372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- [24].Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- [25].Morgelin M, Paulsson M, Heinegard D, Aebi U, Engel J. Evidence of a defined spatial arrangement of hyaluronate in the central filament of cartilage proteoglycan aggregates. Biochem J. 1995 Apr 15;307(Pt 2):595–601. doi: 10.1042/bj3070595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Matsumoto K, Shionyu M, Go M, Shimizu K, Shinomura T, Kimata K, et al. Distinct interaction of versican/PG-M with hyaluronan and link protein. J Biol Chem. 2003 Oct 17;278(42):41205–12. doi: 10.1074/jbc.M305060200. [DOI] [PubMed] [Google Scholar]
- [27].Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012 Sep 28;151(1):221–32. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Enard W. FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr Opin Neurobiol. 2011 Jun;21(3):415–24. doi: 10.1016/j.conb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- [29].Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- [30].Gordon JW, Shaw JA, Kirshenbaum LA. Multiple facets of NF-kappaB in the heart: to be or not to NF-kappaB. Circ Res. 2011 Apr 29;108(9):1122–32. doi: 10.1161/CIRCRESAHA.110.226928. [DOI] [PubMed] [Google Scholar]
- [31].Lee JG, Kay EP. NF-kappaB is the transcription factor for FGF-2 that causes endothelial mesenchymal transformation in cornea. Invest Ophthalmol Vis Sci. 2012 Mar;53(3):1530–8. doi: 10.1167/iovs.11-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Burke JR, Pattoli MA, Gregor KR, Brassil PJ, MacMaster JF, McIntyre KW, et al. BMS-345541 is a highly selective inhibitor of I kappa B kinase that binds at an allosteric site of the enzyme and blocks NF-kappa B-dependent transcription in mice. J Biol Chem. 2003 Jan 17;278(3):1450–6. doi: 10.1074/jbc.M209677200. [DOI] [PubMed] [Google Scholar]
- [33].Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995 Jun 16;270(24):14255–8. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- [34].Vrljicak P, Cullum R, Xu E, Chang AC, Wederell ED, Bilenky M, et al. Twist1 transcriptional targets in the developing atrio-ventricular canal of the mouse. PLoS One. 2012;7(7):e40815. doi: 10.1371/journal.pone.0040815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Perkins SJ, Nealis AS, Dudhia J, Hardingham TE. Immunoglobulin fold and tandem repeat structures in proteoglycan N-terminal domains and link protein. J Mol Biol. 1989 Apr 20;206(4):737–53. doi: 10.1016/0022-2836(89)90580-9. [DOI] [PubMed] [Google Scholar]
- [36].Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev Dyn. 1996 Nov;207(3):235–52. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- [37].Fraidenraich D, Stillwell E, Romero E, Wilkes D, Manova K, Basson CT, et al. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science. 2004 Oct 8;306(5694):247–52. doi: 10.1126/science.1102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao Q, Beck AJ, Vitale JM, Schneider JS, Gao S, Chang C, et al. Developmental ablation of Id1 and Id3 genes in the vasculature leads to postnatal cardiac phenotypes. Dev Biol. 2011 Jan 1;349(1):53–64. doi: 10.1016/j.ydbio.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004 Sep;131(18):4477–87. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- [40].Li S, Zhou D, Lu MM, Morrisey EE. Advanced cardiac morphogenesis does not require heart tube fusion. Science. 2004 Sep 10;305(5690):1619–22. doi: 10.1126/science.1098674. [DOI] [PubMed] [Google Scholar]
- [41].Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, et al. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012 Apr 26;74(2):314–30. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004 Jan;24(2):809–22. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].French CA, Groszer M, Preece C, Coupe AM, Rajewsky K, Fisher SE. Generation of mice with a conditional Foxp2 null allele. Genesis. 2007 Jul;45(7):440–6. doi: 10.1002/dvg.20305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Capdevila J, Tsukui T, Rodriquez Esteban C, Zappavigna V, Izpisua Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999 Nov;4(5):839–49. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- [45].Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005 Dec;132(24):5601–11. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- [46].Min C, Eddy SF, Sherr DH, Sonenshein GE. NF-kappaB and epithelial to mesenchymal transition of cancer. J Cell Biochem. 2008 Jun 1;104(3):733–44. doi: 10.1002/jcb.21695. [DOI] [PubMed] [Google Scholar]
- [47].Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995 Jan 27;80(2):321–30. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- [48].Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995 Jul 13;376(6536):167–70. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- [49].Azhar M, Runyan RB, Gard C, Sanford LP, Miller ML, Andringa A, et al. Ligand-specific function of transforming growth factor beta in epithelial-mesenchymal transition in heart development. Dev Dyn. 2009 Feb;238(2):431–42. doi: 10.1002/dvdy.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. Jul 15;91(2):212–22. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.