This study was undertaken to test the ability of hiPSCs to give rise to retinal cells under nonxenogeneic conditions. hiPSCs were maintained in traditional, feeder-free, or xeno-free culture conditions, and their ability to differentiate to a retinal fate was tested. This study represents the first demonstration of nonxenogeneic differentiation of hiPSCs into neural retinal cell types such as photoreceptors and retinal ganglion cells, which is likely to have important implications for the treatment of diseases such as age-related macular degeneration and glaucoma. The results of this study also highlight the applicability of nonxenogeneic growth and differentiation of hiPSCs to other cellular lineages for translational applications.
Keywords: Pluripotent stem cells, Retina, Differentiation, Developmental biology
Abstract
Human induced pluripotent stem cells (hiPSCs) possess tremendous potential for the field of regenerative medicine because of their ability to differentiate into any cell type of the body. Such ability has profound implications for translational medicine, because these cells have been implicated for use in cell replacement, disease modeling, and pharmacological screening. However, the translation of established methods for deriving retinal cell types from hiPSCs has been hindered by the use of xenogeneic products for their growth and differentiation. Thus, the ability to derive retinal cell types in the absence of xenogeneic products would represent a significant advancement. The following studies were therefore undertaken to test the ability of hiPSCs to give rise to retinal cells under nonxenogeneic conditions. hiPSCs were maintained in traditional, feeder-free, or xeno-free culture conditions, and their ability to differentiate to a retinal fate was tested. Upon differentiation under all three conditions, cells acquired advancing features of retinal development, eventually yielding cell types of the mature retina. Reverse transcription-polymerase chain reaction and immunocytochemistry confirmed early trends in gene and protein expression patterns in xeno-free derived hiPSCs similar to those in cells derived in mouse embryonic fibroblasts and in feeder-free conditions. Results from this study demonstrate that hiPSCs can be maintained and directed to differentiate into retinal cell types under nonxenogeneic conditions, similar to cells derived using current xenogeneic methodologies. The demonstration of this capability will facilitate future efforts to develop hiPSC-based therapies for retinal disorders and also help to advance in vitro studies of human retinal development.
Introduction
Human induced pluripotent stem cells (hiPSCs) have been implicated to hold great potential for regenerative medicine because of their ability to generate any cell type of the body, as well as their unique ability to generate patient-specific cell populations. These cells maintain the entire unique set of genomic information for each individual patient, representing a great opportunity for the development of personalized treatment profiles for a wide spectrum of diseases [1–7]. However, significant issues still need to be addressed before their full potential is realized, because graft rejection and zoonosis constitute two major issues resulting from the use of animal products or other undefined components during routine culture of these cells. These risks must be minimized or eliminated before effective cell replacement therapy can be realized [8, 9]. Hence, a necessary step in this field of research is the development of nonxenogeneic differentiated progeny derived from hiPSCs, which could then be successfully used for translational research and regenerative medicine.
Traditionally, the use of human pluripotent stem cells (hPSCs), including human embryonic stem cells and hiPSCs, for retinal applications has relied upon their growth on a layer of mitotically inactivated mouse embryonic fibroblasts (MEFs) in the presence of media containing fetal bovine serum or knockout serum replacement using animal components [3, 10–17]. Additional efforts have been made to eliminate the use of undefined growth conditions through the use of feeder-free systems using chemically defined media [18–22]. However, such approaches often rely upon defined or semidefined animal components in the media, as well as the growth of cells upon a Matrigel substrate [23]. More recent efforts have focused on the growth of hPSCs under xeno-free conditions, along with the differentiation to limited cellular lineages [24–28]. However, the successful growth and differentiation of hPSCs to a retinal lineage has been largely unexplored. Thus, for future retinal applications, a need exists to establish conditions for the growth and differentiation of hPSCs in the absence of xenogeneic materials.
The use of MEF and feeder-free systems for the growth of hiPSCs has been reviewed extensively in literature [12, 15, 16, 23, 29, 30], allowing for their role as controls for our experiments. We have previously demonstrated the derivation of a variety of retinal cells from hPSCs including photoreceptors, retinal ganglion cells, and retinal pigmented epithelium (RPE) using a stepwise differentiation protocol [3, 15]. In the current study, we have adapted this procedure to demonstrate the growth and differentiation of hiPSCs to a retinal phenotype in the absence of xenogeneic materials. Care was taken to ensure that all ancillary components used in the culturing of hiPSCs were free of animal origin. Here, we present the first xeno-free approach to the growth and differentiation of hiPSCs into retinal cells that closely resembles previously established methods for derivation of retinal cells. These results represent an important step in the advancement of hiPSCs toward translational purposes.
Methods
Maintenance of Undifferentiated Colonies
The IMR90-4 line of hiPSCs was maintained in the undifferentiated state under three different conditions; MEF, feeder-free (FF), and xeno-free (XF). For the MEF system, hiPSCs were grown on plates of mitotically inactivated MEF, with the supplementation of hiPSC medium (Dulbecco's Modified Eagle's Medium: Nutrient Mixture F-12 [DMEM/F12] with 20% knockout serum replacement [Life Technologies, Rockville, MD, http://www.lifetech.com; catalog no. 18028], 0.1 mM β-mercaptoethanol, 1 mM l-glutamine, minimum essential medium [MEM] nonessential amino acids, and 4 ng/ml fibroblast growth factor 2 [FGF2]) on a daily basis. The feeder-free system consisted of Matrigel coating of plates (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com; catalog no. 354277) and the use of mTESR1 medium (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com; catalog no. 05870) for maintenance of cells in an undifferentiated state. For the xeno-free system, cells were grown on Synthemax plates (Corning Enterprises, Corning, NY, http://www.corning.com; catalog no. 3877XX1) in nonxenogeneic Nutristem medium (Stemgent, Cambridge, MA, http://www.stemgent.com; catalog no. 01-0005).
The cells were passaged approximately every 4–5 days, at 70%–80% confluence. Colonies containing clearly visible differentiated cells were manually removed before passaging. The remaining colonies were lifted off the plate enzymatically by treatment with dispase (Life Technologies; catalog no. 17105-041) (2 mg/ml) followed by three washes with DMEM/F12. In the case of the xeno-free culture system, the colonies were mechanically isolated with a cell scraper instead of dispase to maintain xeno-free conditions. The colonies were then broken up into smaller clusters by manual trituration and were replated at a ratio of 1:6.
Differentiation of hiPSCs
Colonies of undifferentiated hiPSCs grown in FF and MEF conditions were directed to differentiate via the formation of embryoid bodies (EBs) through treatment with dispase, as described previously [3, 15]. For XF samples, colonies were mechanically isolated with a cell scraper instead of dispase to gently remove the colonies from the plates. Cells grown in the MEF system were initially grown in EB medium (hiPSC medium minus FGF2) for 4 days and then transferred to neural induction medium (NIM; DMEM/F12 [1:1], 1% N2 supplement, MEM nonessential amino acids, 2 μg/ml heparin). Cells grown under feeder-free and xeno-free conditions were slowly transitioned into NIM by transferring the EBs to a 3:1 ratio of mTESR1/Nutristem:NIM on day 0, 1:1 on day 1, and 1:3 on day 2 followed by transfer to complete NIM at day 3 of differentiation. To ensure nonxenogeneic growth of cells, NIM lacking heparin was used in xeno-free samples.
At day 7 of differentiation, the cells grown under MEF and feeder-free systems were plated on six-well plates coated with laminin (20 μg/ml), whereas Synthemax plates were used for nonxenogeneic cultures. The cells acquired advanced neural rosette morphologies by 17 days of differentiation followed by their transfer to retinal differentiation medium (RDM; DMEM/F12 (3:1), 2% B27 supplement (nonxenogeneic B27 [Life Technologies; catalog no. A11576SA] was used for XF cultures), and 1% penicillin-streptomycin. Retinal neurospheres identified by a bright ring appearance around the periphery were manually isolated as previously described [15]. Neurospheres were fed every 2–3 days and were maintained in suspension up to 60 days of differentiation. For RPE differentiation, the cells remained adherent at day 17 of differentiation and were fed with RDM every 3–4 days until approximately 60 days of differentiation.
Immunocytochemistry
Samples were collected at specified time points of differentiation and were plated onto coverslips coated with polyornithine and laminin (20 μg/ml) for FF and MEF samples and Synthemax substrate (25 μg/ml) for XF samples. The cells were fixed with 4% paraformaldehyde for 30 minutes followed by three washes with phosphate-buffered saline (PBS) for 5 minutes each. The cells were treated with 0.2% Triton X-100 for 10 minutes followed by blocking for an hour in 10% donkey serum. Primary antibody was added at the recommended dilution in 0.1% Triton X-100 and 5% donkey serum and incubated overnight at 4°C. A complete list of primary antibodies used for immunocytochemistry can be found in supplemental online Table 1. Primary antibody was then removed, and the cells were washed three times with PBS followed by blocking with 10% donkey serum. The secondary antibody was diluted along with 4′,6-diamidino-2-phenylindole (DAPI) in 0.1% Triton X-100 and 5% donkey serum for 1 hour. The samples were washed three times with PBS and mounted on slides for fluorescence microscopy.
RPE were cultured as previously described [3]. Briefly, pigmented patches were microdissected manually and replated on polyornithine/laminin or Synthemax-coated coverslips. RPE cells were expanded in RDM supplemented with FGF2 (20 ng/ml) and epidermal growth factor (20 ng/ml) for 1 week. The mitogens were then removed for 2 weeks, and the cells were stained as described above.
Reverse Transcription-Polymerase Chain Reaction
Cells were collected at specified time points of differentiation, and RNA was extracted using the PicoPure RNA isolation kit (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) or the RNeasy RNA isolation kit (Qiagen, Hilden, Germany, http://www.qiagen.com). cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, http://www.bio-rad.com) or the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). Polymerase chain reaction (PCR) amplification was performed using GoTaq qPCR Master Mix (Promega, Madison, WI, http://www.promega.com). PCR experiments were run for 30 cycles followed by analysis of PCR products on 2% agarose gels. A complete list of primers used for reverse transcription (RT)-PCR can be found in supplemental online Table 2.
Data Quantification
The images were quantified using a twofold approach. First, positive aggregates were identified by nuclear colocalization of antibody and DAPI signal, and the percentage of positive cellular aggregates was counted. Additionally, each positive cellular aggregate was assessed to determine the percentage of positive cells as compared with the total number of DAPI-positive nuclei. A minimum of three images were taken of positive aggregates across a minimum of five samples. ImageJ software was used to quantitate the number of antibody-stained nuclei in each of the images. The percentages of positive aggregates, as well as the positive cells stained, were statistically evaluated using unpaired t test or analyses of variance to determine significant differences across experimental growth conditions.
Results
Maintenance of Pluripotency Under MEF, Feeder-Free, and Xeno-Free Conditions
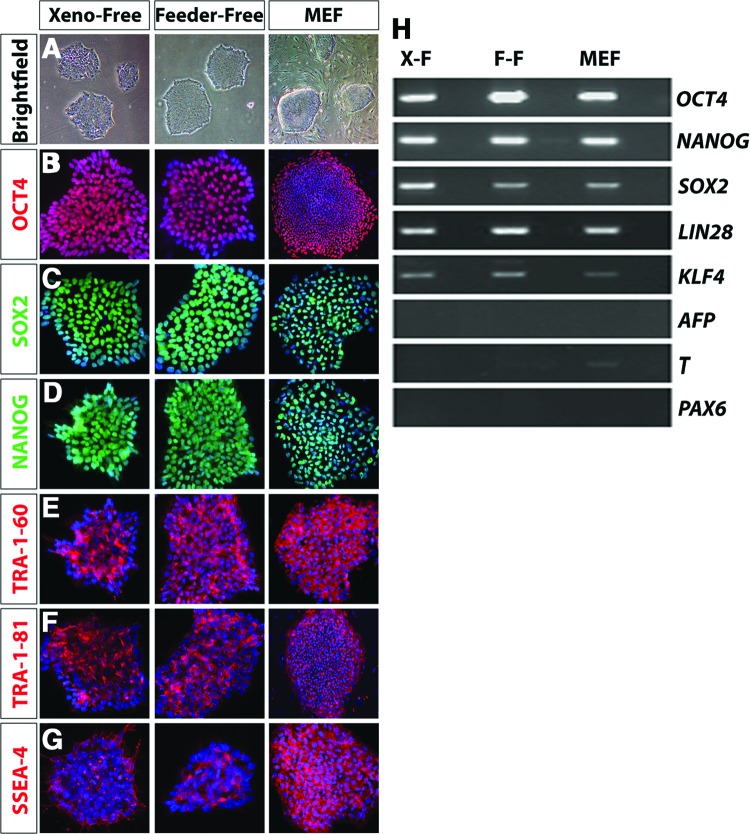
As a prerequisite to xeno-free differentiation of hiPSCs, the ability to effectively expand hiPSCs and maintain pluripotency must be established. Thus, the first experiments were designed to analyze and compare pluripotency characteristics in hiPSCs cultures under these three conditions. After a minimum of five passages in either MEF, feeder-free, or xeno-free systems, the colonies of hiPSCs exhibited a uniform appearance without marked differences in the morphologies of the colonies under different culture conditions (Fig. 1A). Under all three conditions, immunocytochemistry results confirmed the expression of key pluripotency associated factors in colonies of hiPSCs including OCT4, SOX2, NANOG, TRA-1-60, TRA-1-81, and SSEA-4 (Fig. 1B–1G). Maintenance of the pluripotent state was further confirmed through RT-PCR analysis, in which key pluripotency genes were found to be expressed under all conditions (Fig. 1H). In addition to the expression of characteristic pluripotency genes, colonies of hiPSCs grown under each culture condition also largely lacked the expression of differentiation markers including α-FETOPROTEIN, PAX6, and BRACHYURY, further confirming their undifferentiated state.
Figure 1.
Maintenance of pluripotency under xeno-free, feeder-free, and MEF growth conditions. (A): Colonies grown under xeno-free, feeder-free, and MEF conditions exhibited similar morphological features when viewed under bright-field microscopy. Magnification, ×4. (B–D): Uniform and homogeneous expression of pluripotency associated factors such as OCT-4, SOX2, and NANOG was observed in the undifferentiated colonies, irrespective of the system they were grown in. Magnification, ×20. (E–G): Similar expression of cell surface markers including TRA-1-60, TRA-1-81, and SSEA-4 was observed in the xeno-free cells when compared with feeder-free and MEF systems, further confirming the pluripotency of these cells. Magnification, ×20. (H): Reverse transcription-polymerase chain reaction analysis confirmed the expression of pluripotency genes in hiPSCs maintained in all conditions, as well as the relative absence of markers of differentiation. Abbreviations: F-F, feeder-free; MEF, mouse embryonic fibroblast; X-F, xeno-free.
Specification of Neural and Retinal Progenitor Cells
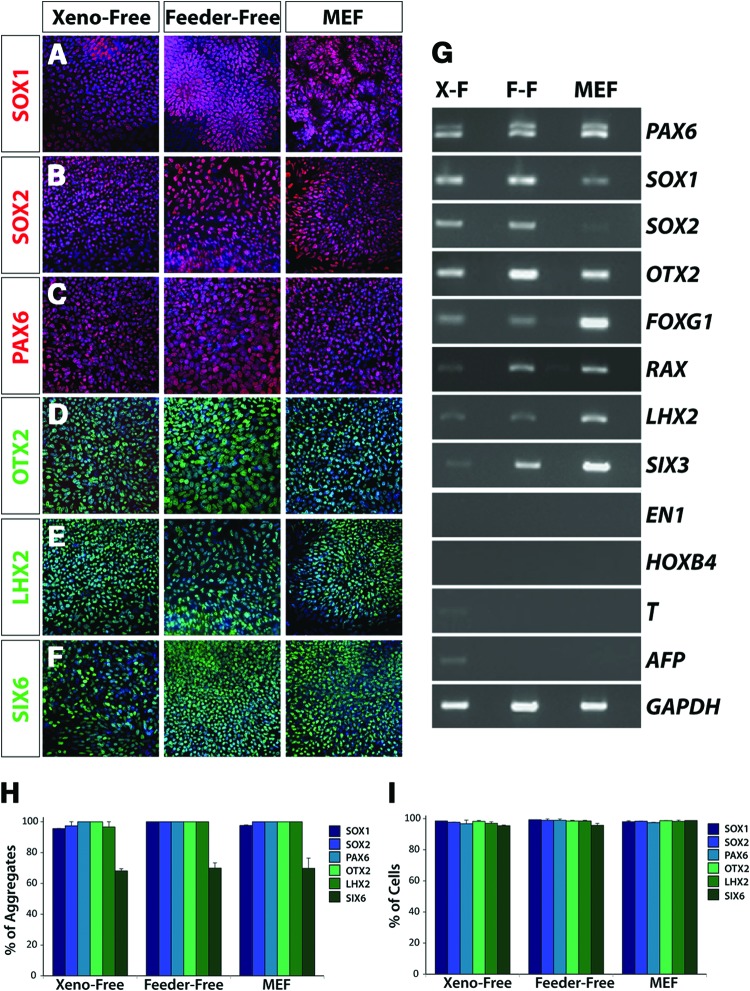
Prior to the specification of mature retinal cell types, hiPSCs undergo a series of differentiation events analogous to the major stages of retinal development including the primitive eye field, optic vesicle, and optic cup [3, 15]. Following modifications to previously established protocols, hiPSCs initially acquired features of the primitive anterior neuroepithelium, including the eye field. Immunocytochemistry experiments illustrated the expression of transcription factors SOX1, SOX2, PAX6, OTX2, LHX2, and SIX6 (Fig. 2A–2F), which were collectively indicative of the acquisition of an anterior neural phenotype. Importantly, inappropriate regional and temporal gene expression was generally not observed within these cells, confirmed by the lack of expression of markers associated with other germ layers (BRACHYURY and AFP; Fig. 2G) and posterior markers of the midbrain and hindbrain (EN-1 and HOXB4; Fig. 2G). RT-PCR analysis also confirmed the establishment of a retinal identity within the anterior neural population, demonstrating the expression of eye field transcription factors [31], including PAX6, RAX, SIX3, SIX6, and LHX2 (Fig. 2G). To ensure that possible differences among growth conditions were not due to the lack of heparin in the nonxenogeneic culture condition, a comparative analysis was performed that demonstrated a potentially superfluous inclusion of heparin in traditional (MEF and FF) neural differentiation protocols [3, 15, 32, 33] (supplemental online Fig. 1). Importantly, the expression patterns of all transcription factors at this stage were consistent under all three conditions tested, as confirmed by quantification of immunocytochemistry results (Fig. 2H, 2I), illustrating the potential to derive retinal cell types under nonxenogeneic conditions (Fig. 2G).
Figure 2.
Primitive retinal specification of human induced pluripotent stem cells (hiPSCs) grown under xeno-free conditions. Within the first 10 days of differentiation, hiPSCs acquired features of the primitive anterior neuroepithelium under all growth conditions. (A-C): The near uniform expression of SOX1, SOX2, and PAX6 indicated the acquisition of a primitive neural fate from hiPSCs. Magnification, ×20. (D–F): hiPSCs also expressed retinal-associated genes including OTX2, LHX2, and SIX6. Magnification, ×20. (G): Reverse transcription-polymerase chain reaction analysis highlighted similar patterns of gene expression across all growth conditions at this time point. Importantly, in addition to the expression of early neuroretinal genes, the regional and temporal specificity of gene expression was confirmed through the absence of genes including EN-1, HOXB4, BRACHYURY, and AFP. (H, I): Quantification of immunocytochemistry experiments revealed comparable percentages of cell aggregates that expressed the indicated transcription factors (H), as well as equivalent percentages of cells within immunopositive aggregates expressing each transcription factor (I). Magnification, ×20. Abbreviations: F-F, feeder-free; MEF, mouse embryonic fibroblast; X-F, xeno-free.
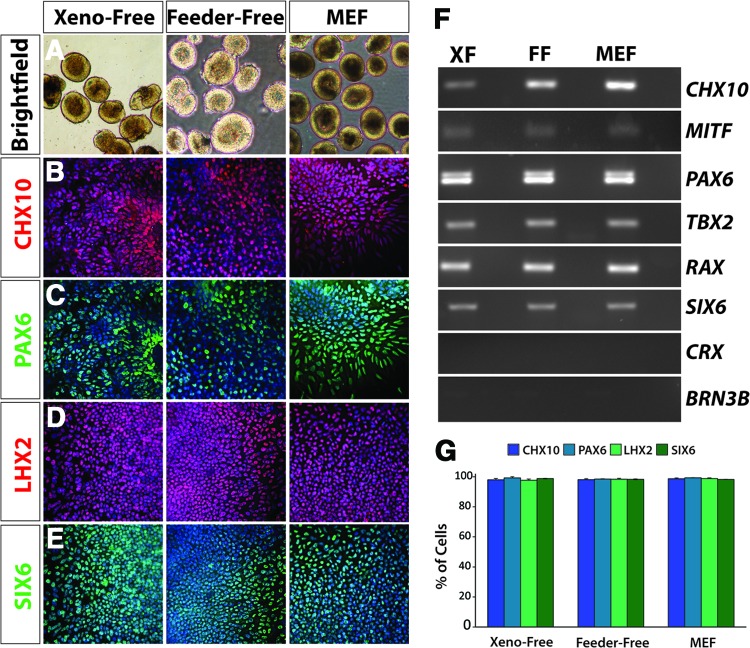
Following the acquisition of a primitive eye-field phenotype, hiPSCs were directed to differentiate through subsequent stages of retinal development, including those analogous to the optic vesicle and optic cup. We have previously demonstrated the ability to identify and isolate two morphologically distinct populations of cells in cultures of differentiating hiPSCs within the first 20 days of differentiation with characteristics analogous to retinal and forebrain progenitor cells, respectively [3]. Neurospheres previously demonstrated to acquire a retinal fate possessed a phase-bright ring around the periphery (Fig. 3A), and this characteristic was manually isolated and analyzed for the expression of retinal progenitor markers. Immunocytochemistry and RT-PCR analyses demonstrated the expression of these transcription factors in xeno-free, feeder-free, and MEF conditions (Fig. 3). Global analyses revealed the expression of a full complement of retinal progenitor cell-associated transcription factors including CHX10 (Fig. 3B), MITF, and TBX2 (Fig. 3F). The expression of retinal and neural transcription factors such as PAX6, LHX2, SIX6 (Fig. 3C–3E), and RAX (Fig. 3F) were also maintained in these neurospheres. Additionally, the coexpression of many of these transcription factors is noteworthy, such as CHX10 and PAX6 (Fig. 3B, 3C), as well as LHX2 and SIX6 (Fig. 3D, 3E) within the same field of cells, further supporting the retinal progenitor nature of these cells. The absence of expression of mature retinal genes such as CRX and BRN3, along with expression of transcription factors such as CHX10 underscored the retinal progenitor nature of these populations as expected, with the expression of these transcription factors expressed similarly across all three systems (Fig. 3F). No significant differences in gene expression patterns across the three growth conditions were observed, confirming the similarity of the XF system to traditional MEF and FF systems (Fig. 3G). Furthermore, the retinal progenitor marker CHX10 remained highly expressed in the presence or absence of heparin in neural induction medium, suggesting a dispensable role for this traditional media supplement (supplemental online Fig. 2).
Figure 3.
Derivation of definitive retinal progenitors from human induced pluripotent stem cells (hiPSCs) under xeno-free conditions. (A): Under each of the three growth conditions, retinal progenitor spheres can be isolated under bright-field microscopy using morphological cues. The retinal spheres were identified by a light outer ring around the periphery, a morphological feature absent in nonretinal spheres. Magnification, ×4. (B–E): Retinal progenitor cells expressed characteristic markers such as CHX10, PAX6, LHX2, and SIX6. Magnification, ×20. (F): Reverse transcription-polymerase chain reaction analysis confirmed the expression of these retinal progenitor markers and indicated similar expression levels of cells grown in a nonxenogeneic environment when compared with the cells grown using traditional systems. Markers of more mature cells were not expressed at this stage, including CRX and BRN3, indicating the temporal specificity of these retinal progenitor cells. (G): After manual isolation of retinal neurospheres based on morphological cues, similar levels of expression were observed between samples for each of the indicated transcription factors. Abbreviations: FF, feeder-free; MEF, mouse embryonic fibroblast; XF, xeno-free.
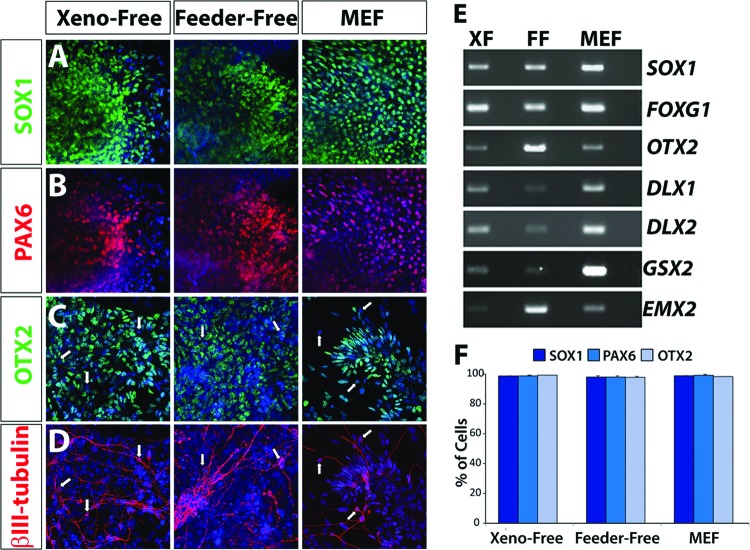
Nonretinal neurospheres were previously demonstrated to possess a forebrain progenitor fate [3]. In the current study, these neurospheres were similarly comprised of neural progenitors possessing an anterior identity (SOX1/PAX6/OTX2-positive), as well as βIII-tubulin-positive neurons (Fig. 4A–4D). As demonstrated by RT-PCR analysis, these neurospheres expressed a full complement of anterior neural transcription factors (Fig. 4E). These results demonstrated the ability to enrich for retinal progenitor cells apart from other neural cell types maintained under xeno-free conditions, as previously established for traditional systems. Additionally, the levels and patterns of expression of SOX1, PAX6, and OTX2 were highly similar across the XF, FF, and MEF systems of differentiation (Fig. 4F).
Figure 4.
Nonretinal neurospheres retain an anterior neural identity. (A, B): Nonretinal neurospheres expressed markers indicative of neural progenitor fate such as SOX1 and PAX6 after 25 days of differentiation. Magnification, ×20. (C): Further analysis of these cells demonstrated their anterior neural phenotype based on the expression of OTX2. Magnification, ×20. (D): At this stage of differentiation, the first expression of neuronal-specific markers such as βIII-tubulin was observed. The arrows in (C, D) indicated that βIII-tubulin-positive neurons had lost the expression of progenitor-associated transcription factors such as OTX2. Magnification, ×20. (E): Reverse transcription-polymerase chain reaction analysis demonstrated that these cells expressed a variety of anterior neural transcription factors. (F): Quantification of immunocytochemistry experiments performed on nonretinal neurospheres revealed similar percentages of cells within aggregates expressing each indicated transcription factor. Abbreviations: FF, feeder-free; MEF, mouse embryonic fibroblast; XF, xeno-free.
Differentiation of Mature Retinal Cell Types
For translational relevance leading to clinical applications, it will be necessary to derive retinal cells under nonxenogeneic conditions, including cells of both the RPE and neural retina. In the current study, hiPSCs were therefore directed to become neural retinal and RPE cells under xeno-free conditions, similar to those previously documented in the MEF and feeder-free systems [3]. Initially, RPE differentiation was induced through modifications to previously established methods [3, 15], and pigmented, hexagonal RPE-like cells were first apparent approximately 1 month following the start of differentiation. The number of pigmented cells increased in abundance over next few weeks (Fig. 5A). Immunocytochemistry at 50 days of differentiation revealed the expression of RPE-characteristic tight junction proteins such as ZO-1, as well as RPE-associated transcription factors, including OTX2 (Fig. 5B). RPE from both xeno-free and traditional cultures expressed a full complement of RPE-associated genes such as MITF, OTX2, RPE65, BEST1, EZRIN, CRALBP, ZO-1, and PEDF, as confirmed by RT-PCR (Fig. 5C). In addition, the percentage of OTX2/ZO-1-positive cells remained unchanged irrespective of the culture system used (Fig. 5D).
Figure 5.
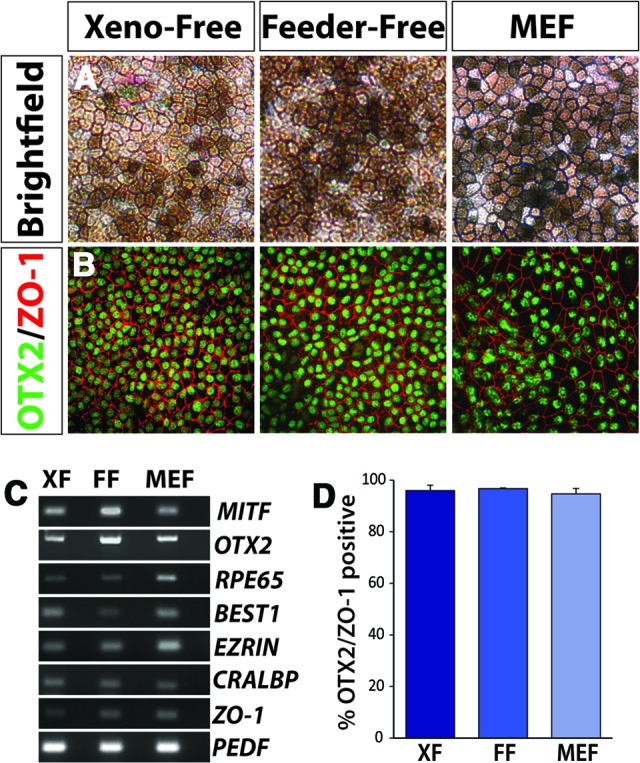
Retinal pigmented epithelium (RPE) derived from human induced pluripotent stem cells (hiPSCs) under xeno-free and traditional growth conditions. (A): Bright-field microscopy demonstrated the characteristic pigmented, hexagonal morphology associated with RPE specification. This phenotype was first apparent approximately 1 month following the start of differentiation and increased over the next few weeks. Magnification, ×20. (B): hiPSC-derived RPE grown under all three conditions expressed characteristics such as the transcription factor OTX2 and the tight junction protein ZO-1. Magnification, ×20. (C): Reverse transcription-polymerase chain reaction from xeno-free, feeder-free, and MEF cultures similarly expressed a number of RPE-associated genes. (D): Following manual isolation and expansion of RPE derived under each growth condition, comparable percentages of cells coexpressing OTX2 and ZO-1 were observed. Abbreviations: FF, feeder-free; MEF, mouse embryonic fibroblast; XF, xeno-free.
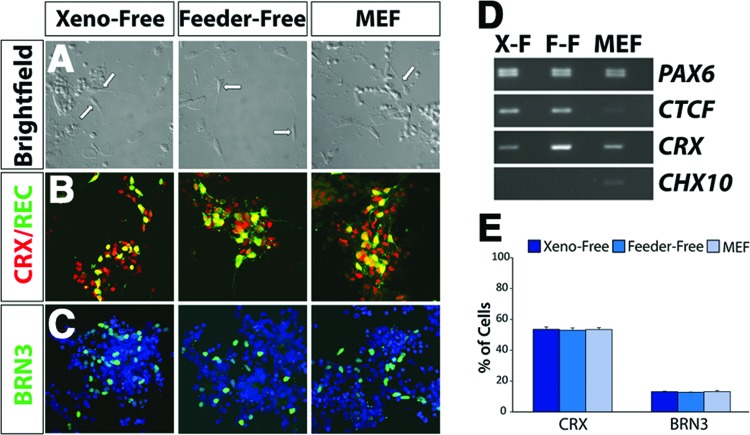
Beyond the ability to derive RPE cells, cells were observed with morphologies of primitive photoreceptor-like cells in vitro after 60 days of differentiation (Fig. 6A). The cells expressed genes associated with varied neuroretinal phenotypes, with numerous cells expressing the photoreceptor precursor-specific transcription factor CRX, as well as RECOVERIN, indicative of a photoreceptor-specific fate (Fig. 6B). Additionally other cells expressed BRN3, indicative of a retinal ganglion cell fate (Fig. 6C). RT-PCR demonstrated the acquisition of advancing features of retinal cells over 50 days of differentiation in a xeno-free environment (Fig. 6D). The percentages of cells expressing retinal ganglion cell (BRN3-positive) and photoreceptor (CRX-positive) phenotypes were found to be highly comparable across all three systems of growth (Fig. 6E).
Figure 6.
Neuroretinal cell types derived under xeno-free conditions. (A): After 60 days of differentiation, a subset of cells acquired morphologies of primitive photoreceptor-like cells in vitro, including a unipolar appearance with one short process, as indicated by arrows. Magnification, ×20. (B): At this time point, a subset of cells expressed genes associated with photoreceptor cells, including CRX and RECOVERIN. Magnification, ×40. (C): Other cells expressed markers specific to retinal ganglion cells such as the transcription factor BRN3. Magnification, ×40. (D): Reverse transcription-polymerase chain reaction analysis indicated the expression of genes specific to differentiated retinal subtypes including CRX, whereas the retinal progenitor marker CHX10 is no longer expressed in these cells. (E): Aggregates of cells after 60 days of differentiation exhibited comparable percentages of cells within immunopositive colonies expressing CRX and BRN3, indicative of photoreceptors and retinal ganglion cells, respectively. Abbreviations: F-F, feeder-free; MEF, mouse embryonic fibroblast; X-F, xeno-free.
Discussion
The data presented in the current study are the first demonstration of the derivation of multiple retinal cell types from hiPSCs in the absence of xenogeneic components. We established the generation of retinal cell types, including RPE, photoreceptors, and retinal ganglion cells, from hiPSCs under xeno-free conditions similar to those cells maintained and derived by traditional methods of differentiation. Such an ability is a necessary step as hiPSC research is further developed for translational purposes.
Several reports have focused upon the ability to derive retinal cell types from hiPSCs, with prospects for future therapeutic approaches for patients with devastating blinding disorders. However, most of these studies have relied upon xenogeneic culture systems using media containing animal products and/or mouse feeder cells to support the growth of hiPSCs [3, 11, 13, 15–17, 34–37]. The ability to derive retinal cells from hiPSCs under nonxenogeneic conditions has profound implications for future approaches to the treatment of retinal degenerative disorders, including age-related macular degeneration, retinitis pigmentosa, and glaucoma. Recent reports have focused on the ability to derive RPE cells in a less xenogeneic culture environment [38]. This is of particular importance because the first clinical trials for human embryonic stem cell-based products are under way for the potential treatment of age-related macular degeneration [39]. Although such studies have successfully generated RPE cells, these systems have remained somewhat xenogeneic through the maintained use of serum and other animal components, raising the risk of zoonosis. Furthermore, studies to date have excluded the ability to derive cells of the neuroretina, including photoreceptors and retinal ganglion cells [38, 40].
To build upon previous studies and establish a truly nonxenogeneic system with which to derive retinal cells from hiPSCs, we sought to maintain a xeno-free environment in our system with a focus on both media and substrate components. First, synthetically coated culture plates (a defined, feeder-free alternative for the growth of hiPSCs) were used. Second, medium devoid of xenogeneic components was used to specify neural and retinal fates. The ability to derive retinal phenotypes under nonxenogeneic conditions also has important implications for the development of stem cell-based approaches to a variety of disorders, because many groups have demonstrated the ability to derive a variety of retinal cell types following similar differentiation paradigms [12, 17].
The results presented in this study offer numerous advantages over previously described approaches to derive retinal cells from hiPSCs. First, the nonxenogeneic system established within this report is completely chemically defined, whereas previous efforts to establish a nonxenogeneic culture system often relied upon the use of serum or knockout serum-containing media at some stage of the differentiation process [38, 41]. The lack of animal sera throughout this protocol helps to create a more reproducible culture system, because significant variability often exists between lots of animal-derived products. Additionally, beyond the use of commercially available media supplements, this method does not require the use of additional exogenous growth factors that may complicate efforts to establish a nonxenogeneic, more reproducible culture system. Furthermore, as previously demonstrated under traditional culture systems [3], we establish the ability to identify and enrich for retinal progenitor cell populations based upon morphological characteristics and demonstrate this capability under nonxenogeneic conditions. Such an ability to derive highly enriched populations of retinal progenitor cells under nonxenogeneic culture conditions is an essential step for the development of translational applications of hiPSCs.
Previous studies using similar neural and retinal differentiation protocols have often used heparin as a component of the differentiation medium. However, the necessity of heparin in these media has not been thoroughly tested. For the establishment of a nonxenogeneic culture system, proteins of animal origin must be either removed or replaced with suitable alternatives. The experiments described in this study demonstrate that although heparin has been widely used for neural and retinal specification media, the presence of heparin is perhaps superfluous and may likely be eliminated as needed in future studies. Thus, the studies described within this report not only demonstrate the ability to derive retinal cell types under nonxenogeneic culture conditions, but the results also underscore the ability to use such an approach to the development of refinements to existing protocols.
Beyond the implications for advanced therapeutic approaches to retinal disease, the establishment of a nonxenogeneic system with which to maintain and differentiate hiPSCs also has significance for the use of these cells as an in vitro model of human development or for disease modeling. Previous studies have demonstrated the ability of hiPSCs to recapitulate each of the major stages of retinogenesis, yielding each of the major cell types of the retina [3, 15, 17]. However, questions remained about how well such systems faithfully recapitulate in vivo development, because these systems have relied upon factors that would not be found in a human in vivo system, including mouse cells, as well as animal serum and other proteins. With the establishment of a nonxenogeneic system with which to direct the differentiation of hiPSCs to a retinal fate, future studies of human retinogenesis using hiPSCs will likely be more easily translatable to an in vivo environment and may more faithfully recapitulate disease-associated phenotypes associated with patient-specific samples. Furthermore, such an ability to derive retinal cell types under nonxenogeneic conditions will also simplify efforts to use hiPSCs for pharmacological screening, because these cells are more likely to faithfully recapitulate the in vivo environment because of the lack of xenogeneic components. Normalizing culture conditions will also make subtle phenotypes more easily attributable to underlying factors that may otherwise be overlooked because of inherent variability associated with xenogeneic conditions. Thus, investigators will be able to more reliably draw comparisons between patient-derived cell lines and models.
These results establish that hiPSCs can be specified to differentiate into mature neural cell types, such as retinal neurons, under nonxenogeneic conditions. Such results have important implications as new stem cell-based approaches are developed for the treatment of human disease, including those proceeding to clinical trials for the potential treatment of age-related macular degeneration [39]. Before cells grown under nonxenogeneic conditions can be used in therapeutic applications, other precautions will be necessary. Existing cell lines need to be assayed for xenogeneic material that had been previously acquired. It may therefore be advantageous to derive new lines of hiPSCs under nonxenogeneic conditions, as described previously for hiPSCs [28]. Additionally, routine culturing of these cells in a research lab does not afford the same level of protection for a patient as those cells grown under good manufacturing practices (GMP) [42]. Before the translation of this research to therapeutic applications, it will be advantageous and perhaps necessary to expand the results presented here to include the derivation and differentiation of these cells under GMP-compliant environmental conditions. The results presented here describe for the first time a chemically defined, nonxenogeneic culture system for the derivation of retinal cell types, with profound implications for future approaches to regenerative medicine.
Conclusion
In summary, the results of this study demonstrate the ability to differentiate hiPSCs into a variety of retinal cell types under nonxenogeneic culture conditions. This study represents the first demonstration of nonxenogeneic differentiation of hiPSCs into neural retinal cell types such as photoreceptors and retinal ganglion cells, which is likely to have important implications for the treatment of diseases such as age-related macular degeneration and glaucoma. Of importance, no marked differences in the maintenance and differentiation of hiPSCs into retinal cells were observed between each of the three culture conditions. The results of this study also highlight the applicability of nonxenogeneic growth and differentiation of hiPSCs to other cellular lineages for translational applications. Although additional studies are still necessary before widespread application of hiPSCs for translational applications, the current study serves to establish the feasibility of nonxenogeneic growth and differentiation of hiPSCs for applications related to retinal development and disease.
Acknowledgments
We thank Corning Life Sciences for supplying reagents and supplies. This work was supported by a grant from the Indiana University Collaborative Research Grant fund of the Office of the Vice President for Research, American Health Assistance Foundation Grant G2012027, and startup funds from the School of Science at Indiana University Purdue University Indianapolis.
Author Contributions
A.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; M.M.S.: data analysis and interpretation, manuscript writing; J.S.M.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Jin ZB, Okamoto S, Osakada F, et al. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One. 2011;6:e17084. doi: 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comyn O, Lee E, MacLaren RE. Induced pluripotent stem cell therapies for retinal disease. Curr Opin Neurol. 2010;23:4–9. doi: 10.1097/WCO.0b013e3283352f96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert AD, Yu J, Rose FF, Jr., et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itzhaki I, Maizels L, Huber I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 7.Marchetto MC, Carromeu C, Acab A, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin ZB, Okamoto S, Mandai M, et al. Induced pluripotent stem cells for retinal degenerative diseases: A new perspective on the challenges. J Genet. 2009;88:417–424. doi: 10.1007/s12041-009-0063-5. [DOI] [PubMed] [Google Scholar]
- 9.Sun N, Longaker MT, Wu JC. Human iPS cell-based therapy: Considerations before clinical applications. Cell Cycle. 2010;9:880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucherie C, Sowden JC, Ali RR. Induced pluripotent stem cell technology for generating photoreceptors. Regen Med. 2011;6:469–479. doi: 10.2217/rme.11.37. [DOI] [PubMed] [Google Scholar]
- 11.Buchholz DE, Hikita ST, Rowland TJ, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 12.Hirami Y, Osakada F, Takahashi K, et al. Generation of retinal cells from mouse and human induced pluripotent stem cells. Neurosci Lett. 2009;458:126–131. doi: 10.1016/j.neulet.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 13.Lamba DA, Karl MO, Ware CB, et al. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reh TA, Lamba D, Gust J. Directing human embryonic stem cells to a retinal fate. Methods Mol Biol. 2010;636:139–153. doi: 10.1007/978-1-60761-691-7_9. [DOI] [PubMed] [Google Scholar]
- 15.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26:215–224. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 17.Mellough CB, Sernagor E, Moreno-Gimeno I, et al. Efficient stage specific differentiation of human pluripotent stem cells towards retinal photoreceptor cells. Stem Cells. 2012;30:673–686. doi: 10.1002/stem.1037. [DOI] [PubMed] [Google Scholar]
- 18.Chen G, Gulbranson DR, Hou Z, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Powell S, Brunette E, et al. Expansion of human embryonic stem cells in defined serum-free medium devoid of animal-derived products. Biotechnol Bioeng. 2005;91:688–698. doi: 10.1002/bit.20536. [DOI] [PubMed] [Google Scholar]
- 20.Luo S, Lin G, Sun Z, et al. Maintenance of undifferentiated state of human embryonic stem cells in chemical defined medium at high clone density without exogenous cell factors. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35:1123–1128. doi: 10.3969/j.issn.1672-7347.2010.11.. [DOI] [PubMed] [Google Scholar]
- 21.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig TE, Levenstein ME, Jones JM, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig T, Thomson J. Defined, feeder-independent medium for human embryonic stem cell culture. Curr Protoc Stem Cell Biol. 2007 doi: 10.1002/9780470151808.sc01c02s2. Chapter 1:Unit 1C 2. [DOI] [PubMed] [Google Scholar]
- 24.Bergström R, Strom S, Holm F, et al. Xeno-free culture of human pluripotent stem cells. Methods Mol Biol. 2011;767:125–136. doi: 10.1007/978-1-61779-201-4_9. [DOI] [PubMed] [Google Scholar]
- 25.Ellerstrom C, Strehl R, Moya K, et al. Derivation of a xeno-free human embryonic stem cell line. Stem Cells. 2006;24:2170–2176. doi: 10.1634/stemcells.2006-0130. [DOI] [PubMed] [Google Scholar]
- 26.Mallon BS, Park KY, Chen KG, et al. Toward xeno-free culture of human embryonic stem cells. Int J Biochem Cell Biol. 2006;38:1063–1075. doi: 10.1016/j.biocel.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swistowski A, Peng J, Han Y, et al. Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS One. 2009;4:e6233. doi: 10.1371/journal.pone.0006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker BA, Anfinson KR, Mullins RF, et al. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Translational Medicine. 2012;1:16–24. doi: 10.5966/sctm.2012-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung HC, Lin RC, Logan GJ, et al. Human induced pluripotent stem cells derived under feeder-free conditions display unique cell cycle and DNA replication gene profiles. Stem Cells Dev. 2012;21:206–216. doi: 10.1089/scd.2010.0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idelson M, Alper R, Obolensky A, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Zuber ME, Gestri G, Viczian AS, et al. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- 32.Pankratz MT, Li XJ, Lavaute TM, et al. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaVaute TM, Yoo YD, Pankratz MT, et al. Regulation of neural specification from human embryonic stem cells by BMP and FGF. Stem Cells. 2009;27:1741–1749. doi: 10.1002/stem.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bharti K, Miller SS, Arnheiter H. The new paradigm: Retinal pigment epithelium cells generated from embryonic or induced pluripotent stem cells. Pigment Cell Melanoma Res. 2011;24:21–34. doi: 10.1111/j.1755-148X.2010.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamba DA, McUsic A, Hirata RK, et al. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLoS One. 2010;5:e8763. doi: 10.1371/journal.pone.0008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osakada F, Jin ZB, Hirami Y, et al. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 38.Vaajasaari H, Ilmarinen T, Juuti-Uusitalo K, et al. Toward the defined and xeno-free differentiation of functional human pluripotent stem cell-derived retinal pigment epithelial cells. Mol Vis. 2011;17:558–575. [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz SD, Hubschman JP, Heilwell G, et al. Embryonic stem cell trials for macular degeneration: A preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 40.Rowland TJ, Blaschke AJ, Buchholz DE, et al. Differentiation of human pluripotent stem cells to retinal pigmented epithelium in defined conditions using purified extracellular matrix proteins. J Tissue Eng Regen Med. 2012 doi: 10.1002/term.1458. doi: 10.1002/term.1458. [DOI] [PubMed] [Google Scholar]
- 41.Rajala K, Lindroos B, Hussein SM, et al. A defined and xeno-free culture method enabling the establishment of clinical-grade human embryonic, induced pluripotent and adipose stem cells. PLoS One. 2010;5:e10246. doi: 10.1371/journal.pone.0010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crook JM, Peura TT, Kravets L, et al. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell. 2007;1:490–494. doi: 10.1016/j.stem.2007.10.004. [DOI] [PubMed] [Google Scholar]