Abstract
We analyzed TP53 mutations in 483 chronic lymphocytic leukemia patients at different phases of the disease and found a higher incidence of mutations at the later phases and a distinctive mutation profile in each phase. p53 function evaluated by immunoblotting and flow cytometry after cell irradiation was impaired in 28 of 109 cases. Three phenotypically different dysfunctions were observed: type I, associated with heterozygous missense TP53 mutations (typically present at diagnosis) and partially resistant to radiation-induced killing; types II and III, with a higher incidence of microdeletions, nonsense mutations and bi-allelic TP53 defects (common in progressive and chemoresistant cases) and a complete radioresistance. Furthermore, in 4 of 28 patients, all chemoresistant, we found p53 dysfunctions without TP53 mutations. In chronic lymphocytic leukemia patients, a disease phase-specific variability in the p53 mutation profile and function takes place, and both analyses could be useful to guide treatment choices.
Introduction
Mutations of the TP53 gene on chromosome 17p have been found in 4–37% of chronic lymphocytic leukemia (CLL) patients, with the highest incidence in patients with fludara-bine-refractory disease.1,2 The presence of TP53 mutations (alone and with del17p) are usually associated with an adverse prognosis characterized by advanced clinical stage, rapid disease progression, chemoresistance, and shorter overall survival.3–6 However, a considerable prognostic heterogeneity has been documented even in the subgroup of CLL patients harboring TP53 abnormalities.7–9 This heterogeneity may reflect the size of the mutated leukemic clone, the position and type of mutation, and the functional properties of the mutated protein.
In the present study, we analyzed a large cohort of CLL patients followed at a single center for TP53 mutations by sequencing with the aim of identifying the incidence and profile of TP53 mutations in different phases of the disease, i.e. diagnosis, progression and chemoresistance. Furthermore, CLL cells were examined for impaired p53 response to ionizing radiation (IR) and p53 functional defects were compared to the status and type of TP53 mutation. Using this approach, we could document: i) the existence of a molecular and functional pattern of p53 alterations specific for each phase of the disease; ii) the relevance of the p53 functional assay to assess the consequences of TP53 mutations at the protein level and to recognize cases characterized by TP53-independent mechanisms of chemoresistance.
Design and Methods
Detailed methodologies are fully described in the Online Supplementary Appendix. Briefly, peripheral blood mononuclear cells from 483 CLL patients consecutively observed at our center were analyzed after obtaining informed consent: 182 at the time of diagnosis, 240 with progressive disease prior to treatment, and 61 resistant to first- or second-line treatment. The study was approved by the Institutional review board of La “Sapienza” University of Rome on November 18, 2010.
Screening for TP53 mutations (exons 4–9) was carried out by polymerase chain reaction (PCR) amplification and direct sequencing on an ABI PRISM 3100 automated DNA sequence analyzer (Applied Biosystems, Foster City, CA, USA). To determine the p53 functional status, CLL cells were exposed to IR, cultured for 8 h, and examined for upregulation of p53 by Western blot. IR-induced apoptosis was measured by flow cytometry after 24-h culture using the Annexin-V technique, and the data analyzed with the CellQuest software (Becton Dickinson).
Results and Discussion
TP53 mutation profile in CLL at different phases of the disease
The screening of the TP53 gene revealed the highest incidence of mutations in chemoresistant CLL (13 of 61, 21.3%) (2 patients showed 2 mutations each), an intermediate incidence (27 of 240, 11.2%) in progressive CLL, while only a small minority (4 of 182, 2.2%) of CLL patients at presentation harbored TP53 mutations. We could observe a peculiar pattern of mutations in each phase of the disease (Online Supplementary Table S1 and Figure S1A-B). CLL at diagnosis was uniformly characterized by the classic pattern of TP53 inactivation, consisting of heterozygous mis-sense mutations generating transitions (Online Supplementary Table S2). Moreover, we did not find any of the most common hot spot mutations reported in cancer, suggesting that hot spot mutations may not be so prevalent in CLL at diagnosis. In contrast, progressive and chemoresistant CLL showed non-missense (nonsense mutations and microdeletions) as well as homozygous mutations (Online Supplementary Tables S3–S4). In particular, we found a higher incidence of microdeletions in patients with progressive disease (14.8%) and a higher incidence of nonsense mutations (26.7%) in chemoresistant patients. The prevalence of nonsense mutations in chemoresistant CLL may thus suggest that loss of function is a strong selection factor in the acquisition of chemoresistance. Finally, we observed a trend towards more hot spot mutations in progressive (22.2%) compared to chemoresistant (6.7%) CLL, suggesting that there is a biological link between rapid cell proliferation/advanced DNA damage and a selective mutational pressure towards the acquisition of TP53 alterations.
Notably, transitions at the CpG sites were more frequently involved in the mutational process occurring in CLL at diagnosis (2 of 4, 50%) and progression (9 of 27, 33.3%), while being less common in chemoresistant CLL (3 of 15, 20%). Since transitions at CpG dinucleotides are believed to be caused by endogenous processes,10,11 our data further support the theory of the prevalence of spontaneous TP53 mutagenic mechanisms in untreated CLL, while highlighting the role of therapy as an exogenous mutational driving force in treated CLL.
Finally, by combining the results of FISH and TP53 mutation analysis, we also observed that the association between TP53 mutations and percentage of del17p increased in relation to the disease phase, confirming the existence of a selection towards a more complex TP53 alteration profile at disease progression and chemoresistance. The main biological characteristics of TP53 mutated patients at the time of the study are shown in the Online Supplementary Table S1.
Detection of p53 dysfunction by Western blotting
The results of TP53 gene analysis prompted us to assess whether the heterogeneity in TP53 mutations could translate into different functional properties of the p53 mutants.
We identified a normal p53 response, characterized by p53 not detectable in non-irradiated cells but strongly up-regulated after IR, in 81 of 109 patients (74.3%) analyzed. On the contrary, the remaining 28 (25.7%) patients showed an impaired p53 response to IR. In line with the results obtained from TP53 mutation analysis, the highest incidence of p53 dysfunctions was found at the later stages of the disease, i.e. 10 of 20 chemoresistant CLL (50%), 17 of 72 progressive CLL (23.6%), and only 1 of 17 CLL at diagnosis (5.9%).
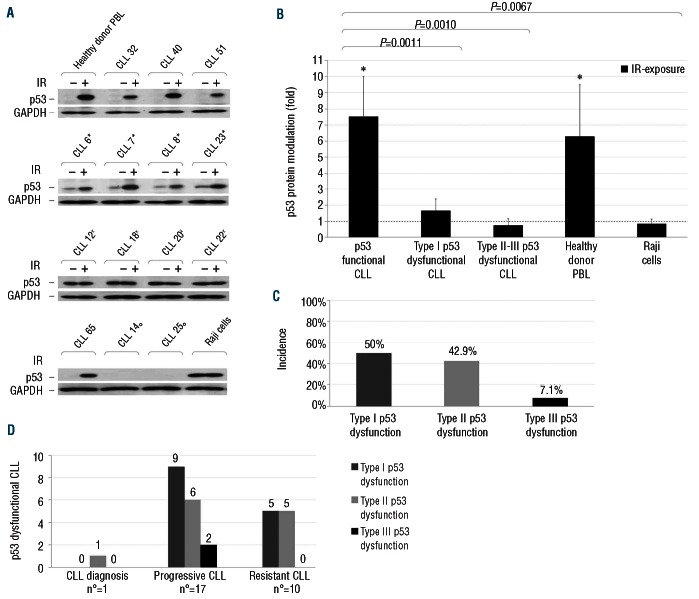
Three types of p53 dysfunctions were identified, each characterized by a distinct mode of protein accumulation pre- and post-exposure to IR: 54.5% of cases were characterized by a high constitutive p53 expression followed by further upregulation after irradiation (type I dysfunction); 40.9% showed the same levels of p53 stabilization pre- and post-irradiation (type II dysfunction); in 4.5%, p53 proved undetectable before and after IR exposure (type III dysfunction) (Figure 1A). The different patterns of p53 response observed were also confirmed by densitometric analysis that showed the most evident protein modulation after IR exposure in p53 functional samples (Figure 1B). Type I dysfunctions were prevalent in the totality of p53 dysfunctional cases (50%), as well as in progressive CLL (52.9%). Resistant CLL were equally associated to type I (50%) and type II (50%) dysfunctions, while type III dysfunctions were only detected in progressive CLL (Figure 1C and D). It was not possible to characterize the type of p53 dysfunction among CLL patients at diagnosis because of the few p53 dysfunctional samples at that phase of the disease.
Figure 1.
Detection of p53 dysfunctions in CLL patients. Irradiated and non-irradiated CLL cells were lysed after 8-h culture. Protein extracts were subjected to SDS-PAGE and Western blot analysis, as described in the Online Supplementary Design and Methods. (A) Representative results of: CLL showing a normal p53 function; CLL showing type I (asterisk), type II (dagger) and III (empty circle) p53 dysfunctions. Healthy donor PBL: negative control; Raji cells: positive control. GAPDH staining is shown as loading control. p53 and GAPDH images derive from different parts of the same gel. (B) Protein bands were quantified by densitometry and p53 protein levels were expressed as folds of protein modulation by IR-exposure, with respect to the control culture set to 1 (hatched line) after normalization to GAPDH. Asterisks indicate P<0.05 (Student’s t-test) against control. Results for 31 p53 functional CLL samples, 14 type I, 12 type II, 2 type III p53 dysfunctional CLL samples, 10 healthy donor PBL samples and Raji B-cell line samples from 10 independent experiments. (C) Distribution of the different types of p53 dysfunctions in the totality of CLL dysfunctional patients (n=28). (D) Distribution of the different types of p53 dysfunctions in CLL dysfunctional patients according to their disease phase.
Comparison between p53 functionality and the TP53 gene status
To determine the correlation between p53 functionality and the presence of TP53 mutations, we next compared the presence of a normal or impaired p53 response to IR with the TP53 status. While all p53 functional cases had a wild-type TP53 gene sequence, 24 of 28 dysfunctional CLL were characterized by the presence of TP53 mutations (Online Supplementary Table S5).
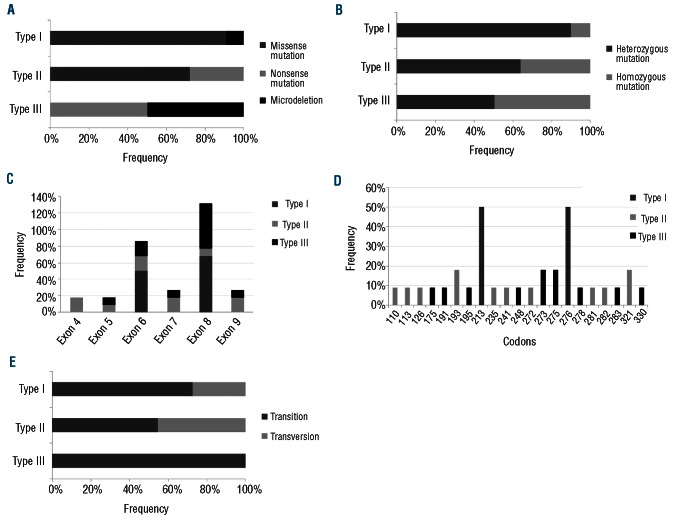
Each p53 dysfunction exhibited a peculiar association with the TP53 mutation profile. Type I dysfunctions were associated with the more classic pattern of TP53 inactivation; i.e. missense and heterozygous mutations (10 of 11), which explains their prevalence in CLL cells. In contrast, type II and III dysfunctions were related to a more complex pattern of TP53 mutations, characterized by a higher incidence of homozygous (4 of 11 and 1 of 2, respectively) and nonsense (3 of 11 and 1 of 2, respectively) mutation as well as by the prevalence of microdeletions in type III (1 of 2) and transversions in type II (5 of 11) dysfunctions (Figure 2). We also observed that each type of p53 dysfunction was associated with mutations affecting TP53 sequence at different positions (Figure 2); at the same time, some mutational variants at the same codon induced equal functional consequences. Overall, these data suggest that each type of TP53 mutation may differently affect the function of the mutated protein observed in vitro.
Figure 2.
Correlation between p53 dysfunctions and CLL TP53 mutation profile. (A) Mutation type, (B) allelic status, (C) exon distribution, (D) codon distribution and ( ) type of mutational event. Type I p53 dysfunctions were found to be associated with the most recurrent pattern of TP53 mutations, i.e. heterozygous and missense mutations generating transitions, mainly located in the DNA binding domain. In contrast, type II and III p53 dysfunctions were related to a more complex pattern of TP53 mutations, highlighted by a higher incidence of homozygous and nonsense mutation as well as microdeletions and transversions.
Four of 28 CLL harbored p53 functional defects in the absence of TP53 mutations detected by Sanger sequencing. Del(17p) was detected in 1 of 3 cases with available FISH analysis. To exclude the presence of ‘false-negatives’, we performed the mutational analysis of TP53 using the AmpliChip p53 Test array,12 a methodology that is more sensitive in identifying mutations in small clones. The AmpliChip detected TP53 mutations in 1 of 4 dysfunctional CLL cases, the one presenting del(17p). The p53 functionality defects observed in the remaining 3 cases could instead be due to the existence of TP53-independent mechanisms of protein stabilization. In our cases, ATM mutations, as well as other potential causes of p53 dysfunction, such as alterations in the ubiquitin pathway and proteasomal proteolysis, are under investigation.13–15
Interestingly, all p53 dysfunctional/TP53 wild-type cases were characterized by resistant disease. The association between this phase of the disease and the occurrence of a p53 dysfunction in the absence of mutation further supports the view that cytotoxic drugs may utilize the redundancy of the p53 pathway to induce chemorefractoriness. Therefore, as p53 dysfunctions due to TP53-independent mechanisms could contribute to the acquisition of refractoriness to conventional DNA-damaging chemotherapy agents, similarly to TP53 disruption, our study suggests the importance of detecting them in order to improve CLL clinical management. The frequency of TP53-independent p53 dysfunctions should be taken into consideration as they account for 10.7% of CLL cases carrying p53 functional defects.
The majority (20 of 25, 80%) of p53 dysfunctional patients with available cytogenetics harbored a 17p deletion. This chromosomal abnormality was characteristic of all type II and III p53 dysfunctional CLL, which were also characterized by the highest percentage of deleted cells. On the contrary, monoallelic TP53 mutations were exclusively found in type I dysfunctional cells, underlining the possibility that p53 stabilization may occur even in the presence of the second wild-type allele. The main biological characteristics of p53 dysfunctional patients are shown in Online Supplementary Table S6.
p53 cellular response to radiation-induced killing
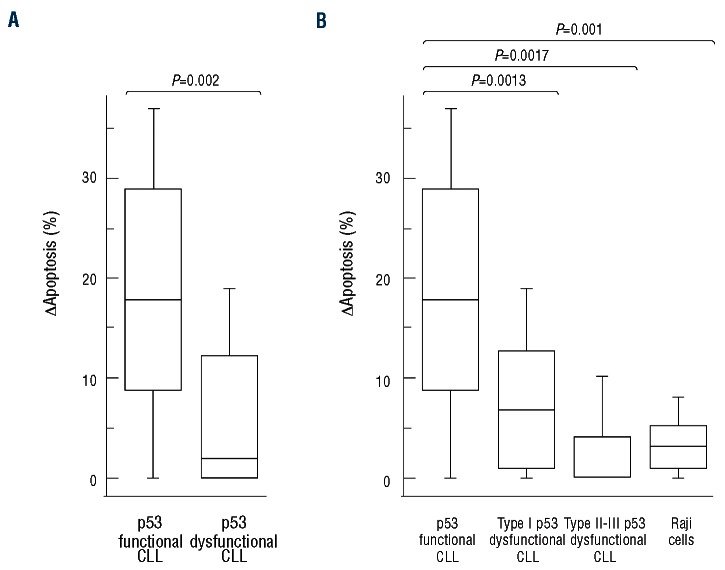
The effect of both p53 dysfunction and TP53 mutation on DNA damage-induced apoptosis was assessed in 31 p53 functional and 15 p53 dysfunctional cases (13 TP53 mutated and 2 TP53 wild-type) at 24 h after DNA damage. As expected, CLL cases with no functional or molecular p53 abnormalities significantly increased cell death after exposure to IR (1.40-fold increase, ∆apoptosis 18.1%±10.7; P=0.0002) (Figure 3A). In contrast, 14 of 15 (93.3%) p53 dysfunctional CLL were radioresistant, independently of the presence of TP53 mutations (1.08-fold increase, ∆apoptosis 5.6%±6.9) (Figure 3A). One p53 dys-functional/TP53 mutant case showed a normal activity of the p53 pathway, consistent with either the possibility that the loss of p53 activity occurred in a subclone of the total CLL tumor population or with the induction of an alternative apoptotic pathway (data not shown). When analyzed comparatively, p53 functional CLL changes in apoptosis appeared significantly greater compared to p53 dysfunctional samples, considered either totally (P=0.0002) or divided according to the type of dysfunction, and to the leukemic control (P=0.001) (Figure 3A and B).
Figure 3.
IR-induced cytotoxicity in CLL cells. Comparison of the changes in apoptosis [∆apoptosis = (IR-induced apoptosis) – (spontaneous apoptosis)] between p53 functional and p53 dysfunctional CLL samples considered either totally (A) or divided according to the type of dysfunction (B). Lymphoid cells were exposed to 5Gy IR and the induction of apoptosis was calculated as percentage of AnnV+/PI- and AnnV+/PI+ cells after 24-h culture. Results for 31 p53 functional CLL samples, 14 p53 dysfunctional CLL samples (8 type I, 4 type II and 2 type III p53 dysfunctions) and Raji B-cell line samples from 10 independent experiments. Box plots define the median values, 25% to 75% of values around the median and the range of values. The corresponding P values (Student’s t-test) are reported.
Interestingly, among the 14 p53 dysfunctional cases resistant to IR-induced apoptosis, CLL cells with type I dysfunction were partially resistant (∆apoptosis 7.7%±6.7), while CLL cells with type II and III dysfunctions were completely radioresistant (∆apoptosis 2.7%±6.7) (Figure 3B). The different levels of CLL cell radioresistance are consistent with the idea that each type of p53 dysfunction may have a less or more profound impact on p53-dependent cellular mechanisms.
A slightly heterogeneous response to IR was found among type I p53 dysfunctional cases (data not shown), with a variable IR-induced killing that almost certainly reflects the residual activity of the normal protein, which is strictly dependent on the properties of the mutated protein. This may also explain why type II and III dysfunctions, characterized by the virtual absence of a normal protein, show a complete radioresistance.
In conclusion, our findings highlight the existence of a disease phase-specific ‘p53 fingerprint’. p53 response to IR was found to be an effective way of screening CLL patients for the different functional consequences of each TP53 mutation, as well as for the presence of TP53-inde-pendent mechanisms of p53 inactivation. Furthermore, our data suggest that the analysis of the p53 function after IR is, in addition to gene sequencing analysis, a useful test to help decision-making in the clinical management of CLL patients.
Supplementary Material
Footnotes
Funding
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), Special Program Molecular Clinical Oncology, 5 x 1000, n. 10007, Milan, Italy; Ministero dell’Università e della Ricerca Scientifica, Progetto FIRB 2010 (Program “Futuro in Ricerca”), Rome, Italy; Compagnia di San Paolo, Turin, Italy.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Zenz T, Häbe S, Denzel T, Mohr J, Winkler D, Bühler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114(13):2589–97 [DOI] [PubMed] [Google Scholar]
- 2.Zenz T, Kröber A, Scherer K, Häbe S, Bühler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112(8):3322–9 [DOI] [PubMed] [Google Scholar]
- 3.Cordone I, Masi S, Mauro FR, Soddu S, Morsilli O, Valentini T, et al. p53 expression in B-cell chronic lymphocytic leukemia: a marker of disease progression and poor prognosis. Blood. 1998;91(11):4342–9 [PubMed] [Google Scholar]
- 4.Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, et al. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23(1):117–24 [DOI] [PubMed] [Google Scholar]
- 5.Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15(3):995–1004 [DOI] [PubMed] [Google Scholar]
- 6.Pospisilova S, Gonzalez D, Malcikova J, Trbusek M, Rossi D, Kater AP, et al. ERIC recommendations on TP53 mutation analysis in Chronic Lymphocytic Leukemia. Leukemia. 2012;26 (7):1458–61 [DOI] [PubMed] [Google Scholar]
- 7.Best OG, Gardiner AC, Davis ZA, Tracy I, Ibbotson RE, Majid A, et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia. 2009;23 (1):212–4 [DOI] [PubMed] [Google Scholar]
- 8.Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O'Brien S, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114(5):957–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trbusek M, Smardova J, Malcikova J, Sebejova L, Dobes P, Svitakova M, et al. Missense Mutations Located in Structural p53 DNA-Binding Motifs Are Associated With Extremely Poor Survival in Chronic Lymphocytic Leukemia. J Clin Oncol. 2011;29(19):2703–8 [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich M, Zhang XY, Inamdar N. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutat Res. 1990;238(3):277–86 [DOI] [PubMed] [Google Scholar]
- 11.Ames BN, Gold LS. Too many rodent carcinogens: mitogenesis increases mutagenesis. Science. 1990;249(4972):970–1 [DOI] [PubMed] [Google Scholar]
- 12.Chiaretti S, Tavolaro S, Marinelli M, Messina M, Del Giudice I, Mauro FR, et al. Evaluation of TP53 mutations with the AmpliChip p53 research test in chronic lymphocytic leukemia: Correlation with clinical outcome and gene expression profiling. Genes Chromosomes Cancer. 2011;50(4):263–74 [DOI] [PubMed] [Google Scholar]
- 13.Guarini A, Marinelli M, Tavolaro S, Bellacchio E, Magliozzi M, Chiaretti S, et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica. 2012;97(1):47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi D, Gaidano G. ATM and chronic lymphocytic leukemia: mutations, and not only deletions, matter. Haematologica. 2012;97(1):5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettitt AR, Sherrington PD, Stewart G, Cawley JC, Taylor AM, Stankovic T. p53 dysfunction in B-cell chronic lymphocytic leukemia: inactivation of ATM as an alternative to TP53 mutation. Blood. 2001;98(3):814–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.