Abstract
The transcription factor TWIST-1 is up-regulated in CD34+ cells in myelodysplastic syndrome and is involved in resistance to apoptosis. There is evidence that TWIST-1 affects apoptosis via microRNAs (miRs). Expression of miRs was determined in myeloid cell lines and primary CD34+ marrow cells from patients with myelodysplastic syndrome and healthy donors using NanoString/array and validated by real-time-polymerase chain reaction. Expression levels of miR10a and miR10b were significantly higher in CD34+ marrow cells from 28 patients with myelodysplastic syndrome than in CD34+ cells from healthy donors (P=0.05 and P=0.012, respectively). Levels of miR10a/b correlated with TWIST-1 miR levels in CD34+ myelodysplastic marrow cells (miR10a, R=+0.69, P<0.0001; miR10b, R=+0.56, P=0.0008). Inhibition of miR10a/10b in clonal cells interfered with proliferation and enhanced sensitivity to apoptosis, which involved NF-κB-dependent p53 activation. These data support a role for miR10a/10b in the regulation of apoptosis in myelodysplastic syndrome and suggest the TWIST-1/miR10a/b-axis as a therapeutic target in myelodysplastic syndrome.
Introduction
Myelodysplastic syndromes (MDS) are characterized by abnormal maturation and differentiation of hematopoietic cells and a high risk of progression to leukemia.414 Recent studies on gene mutations in clonal hematopoietic precursors, interactions with the microenvironment, and the regulatory function of microRNAs (miRs) are providing new insights into the pathophysiology of these complex disorders.2–6
MiRs, small non-coding RNAs (19–25 nucleotides), play a key role in post-transcriptional regulation of specific genes.7 MiRs have been shown to be important regulators of hematopoietic stem cell (HSC) function and hematopoiesis.8,9 Deficiency of certain mature miRs decreases HSC numbers and impairs the development of pro-B cells.8,10 Given the key role of miRs in regulating hematopoiesis and HSC function, and their potential role in the development of clonal MDS cells, investigations into the role of miRs in the pathogenesis of MDS are warranted.
We showed previously that the transcription factor TWIST-1 is up-regulated in CD34+ MDS marrow cells.11 We also showed that marrow stroma-dependent signals resulted in downregulation of TWIST-1 and sensitivity to tumor-necrosis-factor alpha (TNFα)-induced apoptosis in clonal myeloid cell lines and in CD34+ marrow cells from patients with MDS.11 In the present study, we investigated whether TWIST-1-regulated miRs might be involved. Results show that interactions between TWIST-1-dependent miR-10 family members and p53 play a central role in regulating TNFα-mediated apoptosis in MDS clonal cells.
Design and Methods
Patients
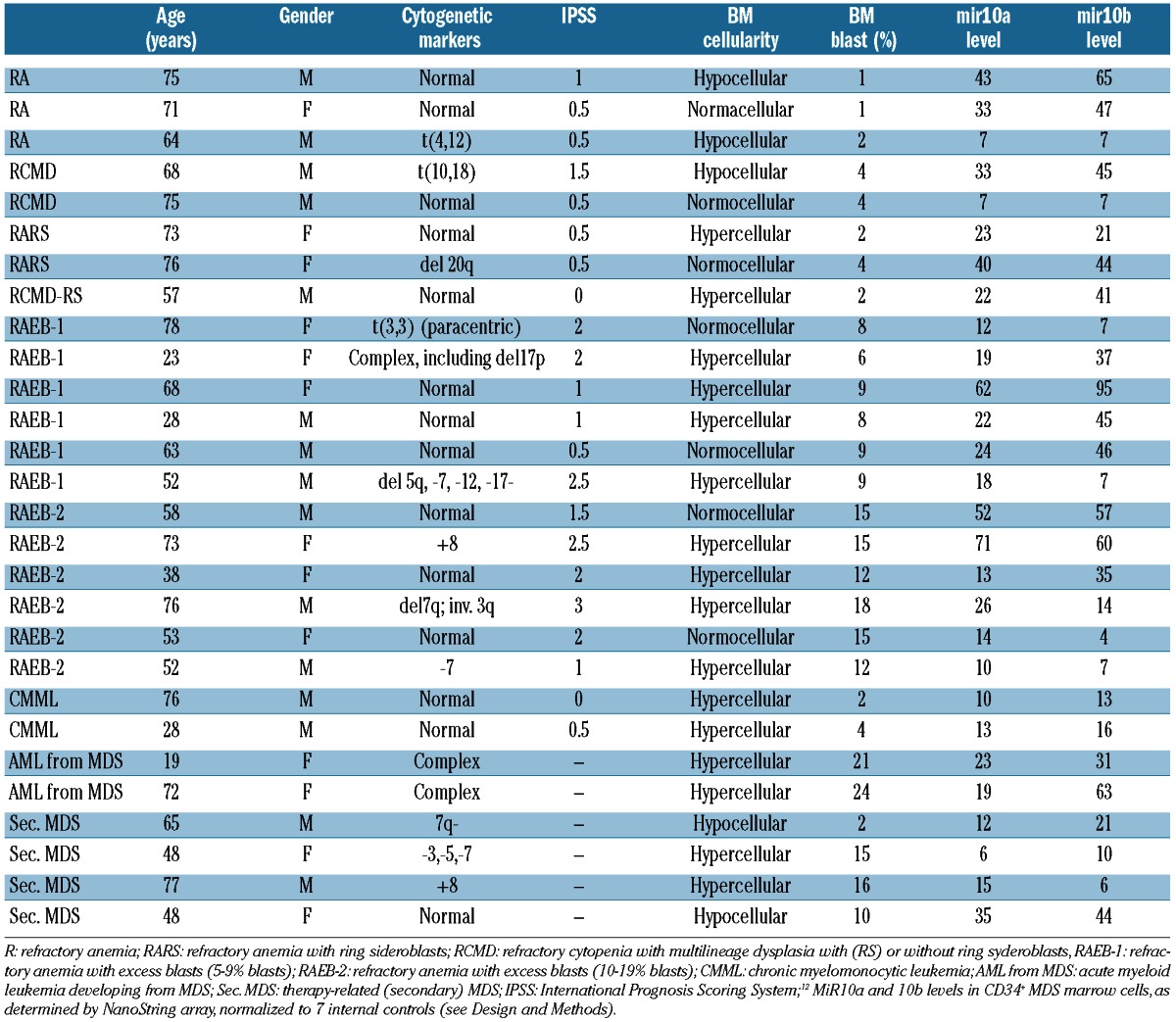
Patients’ characteristics are summarized in Table 1. Thirteen healthy donors, aged between 33 and 70 years (median 45 years), served as controls. All patients and donors had given informed consent as required by the Institutional Review Board of the Fred Hutchinson Cancer Research Center, Seattle, WA, USA (FHCRC) (IRB file n. 5381).
Table 1.
Patients’ and disease characteristics.
Cells and reagents
Bone marrow mononuclear cells were separated on Ficoll-Hypaque gradients, and CD34+ cells were isolated by magnetic-activated cell sorting (MACS; Milteny Biotec, Auburn, CA, USA) yielding a purity of 95–98% as determined by flow cytometry. The leukemia-derived cell lines KG1a and ML-1 were obtained from ATCC (Rockville, MD, USA).13 The MDS-derived cell line MDS-L was a gift from Prof. Tohyama (Hamamatsu, Japan).14 PL-21 cells were a gift from Dr. Stirewalt (FHCRC, Seattle, WA, USA).
Recombinant human TNFα was purchased from PeproTech Inc (London, UK).
NanoString nCounter miR assay
Total cell derived RNA or synthetic miR pools (IDT; 30 pmol per oligonucleotide) were used as input for nCounter miR sample preparation reactions. All sample preparations and hybridization reactions were carried out according to the manufacturer’s instructions, using 5 μL of the 5-fold diluted sample. All hybridization reactions were incubated at 65°C for a minimum of 18 h. Hybridized probes were purified and counted on the nCounter Prep Station and Digital Analyzer (NanoString, Seattle, WA, USA). For each assay, a high-density scan (600 fields of view) was performed.
Real-time polymerase chain reaction
RNA was extracted using the RNeasy Mini kit (QIAGEN, Valencia, CA, USA). Primers for miR-10a, miR-10b and U6 were synthesized by Applied Biosystems (Carlsbad, CA, USA). Total RNA (100 ng) was reverse transcribed using the Taqman MiR Reverse Transcriptase kit (Applied Biosystems, Carlsbad, CA, USA). Real-time quantitative (q)RT-PCR was performed and assessed as described.11
Lentivirus-mediated knockdown of miR-10a/b and conditional knockdown of TWIST-1
Lentiviral vectors mZ-CTRL (miRZip-control) and mZ-10a and mZ-10b (miRZip-10a/b) were from System Biosciences (Mountain View, CA, USA). The miRZip delivers short anti-sense RNAs that are stably expressed and competitively bind their endogenous miR targets, thereby inhibiting their function. For conditional inhibition of TWIST-1 a doxycyclin inducible lentiviral construct pTRIPZ (Openbiosystems, Huntsville, AL, USA; RHS4696-99682454) was used.
Apoptosis, proliferation and cell cycle analysis
Lentivirally transfected KG1a or PL-21 cells were plated in triplicates in 24-well plates (2×105 cells/well) containing TNFα at 0.5, 15, or 25 ng/mL. At 48 h apoptosis was assessed by flow cytometry using Annexin V–FITC labeling.11 Proliferation and cell cycle phase were determined using the APC-BrdU Flow Kit (BD Biosciences, Mountainview, CA, USA).
pGL3-p53 luciferase reporter assay
For a dual-luciferase reporter assay, cells were plated in 12-well plates (5×105 cells/well) on the day before transfection. Co-transfections with the pGL3-p53 promoter (2 μg/well) and pRL-SV40 (0.2 μg/well; internal control) were performed using the Nucleofector Kit L (Amaxa Biosystems, Cologne, Germany). Firefly luciferase activity was normalized to that of Renilla luciferase (activity expressed as relative light units, RLU).
Electrophoretic mobility shift assay (EMSA)
One × 107 cells were harvested and washed, and nuclear proteins were extracted with Nuclear Extraction Kit (Panomics, Fremont, CA, USA). Equal amounts of nuclear protein were tested for NF-kB protein/DNA binding using EMSA Gel Shift Kit (Panomics) with a biotin end-labeled NF-κB oligonucleotide (5’-CCACAGTTGGGATTTCCCAA CCTGACCAG-3’).
Western blotting
Western blotting was carried out using standard techniques.11,15
Statistical analysis
Two independent samples were compared using the 2-sample t-test; P<0.05 was considered significant. Pearson’s linear regression was used to evaluate the relationship between miR levels (miR10a and miR10b) and TWIST-1 mRNA.
Results and Discussion
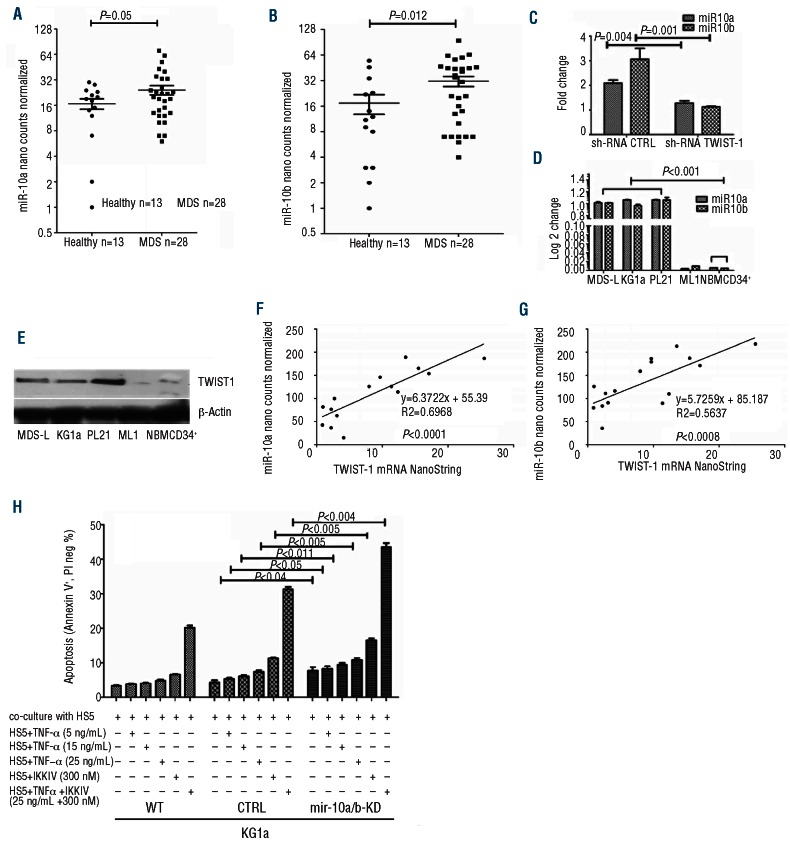
TWIST-1 suppresses the biological activity of p53 by interacting with the DNA-binding domain of p53. The TWIST-1-p53 axis is an important regulator for the control of apoptosis in many disorders.11,16,17 However, the association between p53 and TWIST-1-dependent miRs, an important inhibitory system of transcription/protein translation in the development of MDS, had not been investigated so far. Our data show that miRs-10a/b were expressed in MDS at levels significantly higher than in healthy controls (Figure 1A and B) (mean±SEM 28.2±4.52 vs. 17.2±3.03, P=0.05, and 31.5±4.78 vs. 16.4±4.19, P=0.012, for miR10a and 10b, respectively). These data are in agreement with reports by others6,19 and suggest a pattern of dysregulation of miRs10a/b in MDS.19 MDS disease stage (WHO classification) was correlated with miR10b expression (early stage MDS vs. healthy controls P=0.05, and late stage MDS vs. healthy controls P=0.08) (Online Supplementary Figure S1A and B). The observed increase in miR10a/b levels was presumably related to an expansion of the myeloblast pool (Figure 1F and G). Since we restricted our analysis to CD34+ cells (CD34 is expressed in most cases of MDS) and controlled for cell numbers, results are thought to reflect a true increase in miR10a/b levels, although a contribution by small numbers of non-clonal CD34+ cells cannot be excluded. Patients with early stage/low grade MDS typically show increased spontaneous apoptosis20 both in clonal and non-clonal cells, and inhibition of miR10a/b would presumably not be of therapeutic interest. Since TWIST-1 has been shown to induce expression of miRs10a/b by binding directly to the promoters of miRs10a/b,19,21 we determined miR10a/b levels in KG1a cells with TWIST-1 knockdown (Figure 1C and Online Supplementary Figure S1C). Results indicated that miR10a/b expression was dependent upon the expression of TWIST-1.
Figure 1.
TWIST-1 and miR10a/b levels and their effects on apoptosis and proliferation. (A) and (B) MiR10a and 10b levels in CD34+ MDS marrow cells, as determined by NanoString array, normalized to 7 internal controls (see Design and Methods). Both miR10a and 10b levels were higher in patients with MDS than in healthy controls (mean±SEM 28.2±4.52 vs. 17.2±3.03, P=0.05 and 31.5±4.78 vs. 16.4±4.19, P=0.012, respectively; Student’s t-test for comparison of continuous variables). (C) Expression of miR10 a/b is decreased following knockdown of TWIST-1; shown are the fold changes in message as determined by RT-PCR in KG1a and PL-21 cells transfected with a control construct (shRNA control), and with a TWIST-1-specific construct (shRNA TWIST-1) (mean±SEM of 3 experiments, fold change, normalized to shRNA control; Student’s t-test for comparison of continuous variables). (D) TWIST-1-dependent expression of miR10a/b in myeloid cell lines as determined by RT-PCR (mean±SEM, fold changes normalized for the expression in MDS-cells). (E) TWIST-1 protein expression in the myeloid cell lines MDS-L, KG1a,PL-21 and ML1, and in primary CD34+ cells from healthy donors (NBMCD34+). (F) and (G) correlation of TWIST-1 mRNA and miR10a and miR10b levels in primary CD34+ MDS marrow cells. miR10a/b progressively increased with TWIST-1 mRNA levels in BMCD34+ MDS cells (miR10a R=+0.69, P<0.0001;miR10b, R=+0.56, P=0.0008, using Pearson’s linear regression). (H) Apoptosis rate in KG1a cells with miR10a/b knockdown (KG1a miR10a/b KD) in comparison to unmodified cells (wt) and cells transfected with a control vector (KG1a miRCTRL). Cells were co-cultured with HS5 stroma as described18 and treated with TNFα (5–25 ng/mL), IKK inhibitor (300 nM) or both. Shown is the rate of early stage apoptosis (Annexin V+ PI−) (mean±SEM of 3 experiments; Student’s t-test for comparison of continuous variables).
We had previously11 shown that expression of TWIST-1 is dysregulated in CD34+ MDS cells and showed increased levels in KG1a, PL-21 and MDS-L cell lines. The present data in KG1a (mutant p53), PL-21 (wild-type p53), MDS-L (mutant p53) and ML1 cells (wild-type p53) show that expressions of miR10a/b correlated with TWIST-1 expression and was independent from the p53 mutational status. ML-1 cells, which do not express TWIST-1 protein, also showed very low levels of miRs10a/b (Figure 1D and E). Bone marrow CD34+ cells from healthy controls showed miR10a/b and TWIST-1 protein levels that were statistically significantly lower than in clonal myeloid cells. TWIST-1 mRNA levels in selected CD34+ MDS marrow cells (available for only part of the samples) showed a statistically highly significant association with miR10a/b levels (miR10a, R=+0.69, P<0.0001; miR10b, R=+0.56, P=0.0008) (Figure 1F and G).
To determine the role of miRs10a/b for TNFα-dependent apoptosis, miRs10a/b were stably knocked down (KD) in KG1a (KG1a miR10a/b KD) and PL-21 cells (PL-21 miR10a/b KD); KD cells were co-cultured with HS5 stroma and exposed to TNFα or I-κB Kinase (IKK) inhibitors (to prevent NF-κB translocation). Apoptosis was significantly enhanced in KG1a KD and PL-21 KD cells (Figure 1H, and Online Supplementary Figure S1D) and analysis of BrdU incorporation showed a G1 arrest in cells with miR10a/b KD (Online Supplementary Figure S1E). These data support the concept that miRs10a/b, which are transcriptionally regulated by TWIST-1, facilitated apoptotic responses in association with decreased cell proliferation.21,22
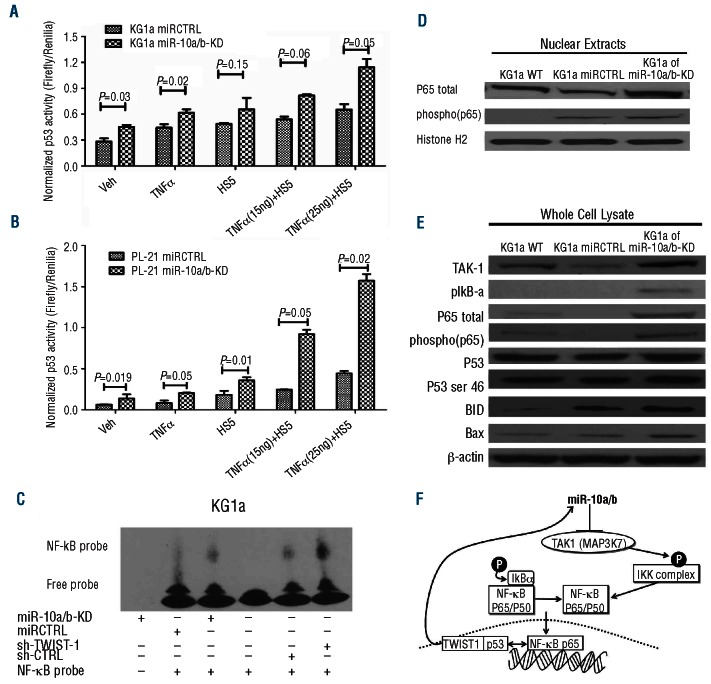
TWIST-1/NF-κB/p53 interactions are central to the control of apoptosis and proliferation, as shown in other dis-orders.11,16,17 A possible role for miRs in modulating this axis has not yet been investigated. To assess p53 function in our TWIST-1-dependent miR10a/b working model we used a luciferase reporter assay in unmodified and KD modified KG1a and PL-21 cells. The data suggested increased p53 promoter activity in KG1a and PL-21 cells with miR10a/b KD when cultured alone or co-cultured with HS5 stroma in the presence of TNFα (Figure 2A and B). EMSA assays and Western blotting of nuclear extracts showed that inhibition of either TWIST-1 or miRs10a/b increased the activity of NF-κB compared to scrambled controls (Figure 2C and D and Online Supplementary Figure S2A and B). Others have previously shown that p53 and NF-B inhibit each other’s ability to enhance gene expression and that this process is controlled by the relative levels of p53 and NF-κB.23 Our model confirms these data: whole cell lysates from KG1a and PL-21 cells with miR10a/b KD showed that TAK-1, which phosphorylates and activates I-κB,24 was subject to post-transcriptional regulation by miRs-10a/b22 (Figure 2E and Online Supplementary Figure S2C), consistent with the report by Fang et al.,22 who observed that miR10a/b levels affected expression of TAK-1, thereby altering phosphorylation of I-κB and nuclear translocation of NF-κB.24 Furthermore, miR10a KD in KG1a and PL-21 cells resulted in increased levels of the pro-apoptotic proteins, BID and BAX (downstream of p53), supporting the role of miR10a in TNFα–induced apoptosis via p53 activation. Taken together, these findings suggest a working model in which TWIST-1-dependent miR10a/b expression directly modulates the p53 promoter and affects expression of downstream targets of NF-κB (Figure 2F).
Figure 2.
TWIST-1-dependent miR10a/b expression impacts p53 activity and regulates downstream targets of NF-κB. (A) and (B) p53 transcriptional activity in KG1a miR10a/b KD KG1a miRCTRL cells, PL-21 miR10a/b KD and PL-21 miRCTRL cells; cell were cultured by themselves (none) or with the addition of TNFα (TNFα), in contact with stroma (HS5) or both (TNFα + HS5). Luciferase assays showed increased p53 transcriptional activity in KG1a (p53 mutant) and PL-21 cells (p53 wild type) with knockdown of mir10a/b both in the presence and absence of HS5 stroma cells and TNFα (15 ng/mL and 25 ng/mL) exposure, in comparison to scrambled controls (miRCTRL). (Results show the mean±SEM of 3 experiments; Student’s t-test). (C) Activation of NF- κB in KG1a miR10a/b KD, KG1a and sh-TWIST-1 as shown by Electrophoretic Mobility Shift Assay (EMSA). (D) Nuclear protein lysates from KG1a, miRCTRL and miR10a/b KD cells, separated on 4%–12% Bis-Tris gels and immunoblotted with antibodies against p65 total and phospho-p65. Histone H2 served as loading control (one of 2 similar experiments). (E) Protein lysates from KG1a wt, miRCTRL and miR10a/b KD cells, were separated on 4%–12% Bis-Tris gels and immunoblotted with antibodies against TAK-1, pIKB-α, p65 total, phosphor- p65, p53, p53-ser46, BID and Bax, respectively; β-actin served as loading control (one of 2 similar experiments). (F) Proposed working model summarizing the regulatory network involving TWIST-1, miR10a/b and TAK-1 in myeloid cell lines.
In summary, TWIST-1 and miR10a/b expression are dysregulated in MDS. Expression of miRs10a/b is controlled by TWIST-1 and, via NF-κB and p53, appears to control sensitivity to TNFα–induced (and stroma-dependent) apoptosis in clonal myeloid cells. These findings suggest a role for miR10 family members in the pathophysiology of MDS. This further supports the concept of onco-genic function of the miR10 members, as recently suggested in a mouse tumor model.25 We propose that the TWIST-1/miR10/p53 axis could serve as a potential new target for therapeutic interventions in advanced MDS.
Supplementary Material
Acknowledgments
We thank Helen Crawford and Bonnie Larson for help with manuscript preparation and Nathan Elliot from NanoString Technologies for technical support.
Footnotes
Funding
This work was supported by research funding from the National Institutes of Health, Bethesda, MD, USA, grant ns. K08DK085156, P01HL036444, R01HL095999. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health nor its subsidiary Institutes and Centers.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases (Review). Nat. Rev. Cancer. 2007;7(2):118–29 [DOI] [PubMed] [Google Scholar]
- 2.Hussein K, Theophile K, Busche G, Schlegelberger B, Gohring G, Kreipe H, et al. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk Res. 2010;34(9):1169–74 [DOI] [PubMed] [Google Scholar]
- 3.Dostalova MM, Krejcik Z, Votavova H, Belickova M, Vasikova A, Cermak J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. European Journal of Human Genetics. 2011;19(3):313–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigna E, Recchia AG, Madeo A, Gentile M, Bossio S, Mazzone C, et al. Epigenetic regulation in myelodysplastic syndromes: implications for therapy. [Review]. Expert Opinion on Investigational Drugs. 2011;20(4):465–93 [DOI] [PubMed] [Google Scholar]
- 5.Sokol L, Caceres G, Volinia S, Alder H, Nuovo GJ, Liu CG, et al. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br J Haematol. 2011;153(1):24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B, Diaz T, et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50(11):1854–9 [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function (Review). Cell. 2004;116(2):281–97 [DOI] [PubMed] [Google Scholar]
- 8.Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, et al. MicroRNA miR-125a controls hematopoietic stem cell number. PNAS. 2010;107(32):14229–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs (Review). Science. 2011;331(6017):550–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–74 [DOI] [PubMed] [Google Scholar]
- 11.Li X, Marcondes AM, Gooley TA, Deeg HJ. The helix-loop-helix transcription factor TWIST is dysregulated in myelodysplastic syndromes. Blood. 2010;116(13):2304–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seal S, Hockenbery DM, Spaulding EY, Kiem H-P, Abbassi N, Deeg HJ. Differential responses of FLIP(long) and FLIP(short)-over-expressing human myeloid leukemia cells to TNF-α and TRAIL-initiated apoptotic signals. Exp Hematol. 2008;36(12):1660–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tohyama K, Tohyama Y, Nakayama T, Ueda T, Nakamura T, Yoshida Y. A novel factor-dependent human myelodysplastic cell line, MDS92, contains haemopoietic cells of several lineages. Br J Haematol. 1995;91(4):795–9 [DOI] [PubMed] [Google Scholar]
- 14.Marcondes AM, Li X, Gooley TA, Milless B, Deeg HJ. Identification of DJ-1/Park-7 as a determinant of stroma-dependent and TNFα-induced apoptosis in MDS by using mass spectrometry and phosphopeptide analysis. Blood. 2010;115(10):1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, et al. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 2008;27(42):5543–53 [DOI] [PubMed] [Google Scholar]
- 16.Shiota M, Izumi H, Tanimoto A, Takahashi M, Miyamoto N, Kashiwagi E, et al. Programmed cell death protein 4 down-regulates Y-box binding protein-1 expression via a direct interaction with Twist1 to suppress cancer cell growth. Cancer Res. 2009;69(7):3148–56 [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40(5):1553–60 [DOI] [PubMed] [Google Scholar]
- 18.Li X, Bryant E, Deeg HJ. Simultaneous demonstration of clonal chromosome abnormalities and apoptosis in individual marrow cells in myelodysplastic syndrome. Int J Hematol. 2004;80(2):140–5 [DOI] [PubMed] [Google Scholar]
- 19.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer [Erratum appears in Nature. 2008 Sep 11;455(7210):256]. Nature. 2007;449(7163):682–8 [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. PNAS. 2010;107(30):13450–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404(6780):892–7 [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51 [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes [Erratum appears in Blood 1998 Feb 1;91(3):1100]. Blood. 1997;89(6):2079–88 [PubMed] [Google Scholar]
- 25.Mhyre A, Marcondes AM, Spaulding EY, Deeg HJ. Stroma-dependent apoptosis in clonal hematopoietic precursors correlates with expression of PYCARD. Blood. 2009;113(3):649–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.