Abstract
We examined whether lenalidomide exposure up-regulates miRNAs and mRNAs, previously shown to play a role in the disease phenotype of del(5q) myelodysplastic syndrome, in pre-treatment CD34+ marrow cells. We hypothesized that increased expression would predict for clinical response. Changes in miR-143, miR-145, miR-146a, miR-146b, miR-378, miR-584, SPARC and RPS14 were examined in del(5q) (n=10) and non-del(5q) (n=18) myelodysplastic syndrome patient samples. Significantly increased expression of miR-143 (1.8-fold and 1.5-fold in del(5q) and non-del(5q), respectively), and miR-145 (1.9-fold and 1.6-fold in del(5q) and non-del(5q), respectively) was observed. In the del(5q) myelodysplastic syndrome cohort, transfusion independence correlated with a 1.3-fold or more increase in miR-145 expression and response over 12 months correlated with a 1.5-fold or more increase. Knockdown of miR-143 and miR-145 in cord blood CD34+ cells resulted in increased erythroid progenitor activity. Lenalidomide selectively abrogated progenitor activity in cells depleted of miR-143 and miR-145 supporting a key role for miR-143/145 in the sensitivity to lenalidomide of del(5q) myelodysplastic syndrome patients.
Introduction
Lenalidomide is an effective agent in the treatment of myelodysplastic syndromes (MDS) especially in cases with interstitial deletions of the long arm of chromosome 5 (del(5q)).1–4 The disease phenotype in patients harboring this cytogenetic anomaly is related to haploinsufficiency of genes found in the commonly deleted region (CDR) on chromosome arm 5q.5–9 In non-del(5q) MDS patients, lenalidomide appears to function by a distinct mechanism to induce erythropoiesis although the clinical response is much less robust.10–12 In patients harboring the del(5q) anomaly, the activity of lenalidomide may correlate with effects on genes found on the remaining sister chromosome thereby recapitulating the normal diploid state. Reinduction of the tumor suppressor gene SPARC has been documented following lenalidomide exposure.13 Loss of key phosphatases (Cdc25c and PP2a) centromeric to the CDR has also been shown to play a role in lenalidomide-induced apoptosis of clonal hematopoietic precursors.5
A number of microRNAs (miR) have been implicated in the pathophysiology of MDS. In MDS with del(5q), haploinsufficiency of miR-143, miR-145 and miR-146a has been shown to play a causative role in the development of the characteristic megakaryocytic dysplasia, thrombocytosis and dominance of the del(5q) malignant clone.6,8 Both miR-145 and miR-146a regulate a number of anti-apoptotic and proliferation pathways.14–17 In MDS, dysregulation of such miRNAs has been proposed as one mechanism providing a growth advantage to affected clones within the marrow niche.6,18 That said, only miR-145, along with miR-143, are found in the chromosome 5q CDR.
We examined whether lenalidomide could re-induce the expression of these key miRNAs and mRNAs found in the CDR that are retained on the remaining normal sister chromosome. We hypothesized that this would selectively remove the pro-survival signal in affected clones and would be predictive of response. By using pre-treatment patient samples and short in vitro exposure to the drug, we expected to identify direct targets of lenalidomide in the malignant CD34+ clone. We also modeled the selective sensitivity of del(5q) clones to lenalidomide by examining clonogenic progenitor potential of human CD34+ cells depleted of miR-143 and miR-145.
Design and Methods
Cell selection and culture
Initial optimization studies were performed using 14 cord blood (CB) samples from healthy donors. Lineage-negative cells from mouse marrow were also examined. Clinical samples were from 29 patients with MDS (10 del(5q) and 19 non-del(5q)). The cohort consisted of patient samples from British Columbia, Washington, Florida and Germany. Sample use was approved by the institutional review board at each site. Karyotype was determined as per local practice.
Cryopreserved cells were separated based on CD34+ status by positive selection (EasySep Human CD34 Positive Selection Kit, StemCell Technologies Inc., Vancouver, Canada). Lineage-negative murine cells were similarly selected (EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit, StemCell Technologies Inc., Vancouver, Canada). Cells were cultured in vitro for 48 h with either lenalidomide (10 μM) or DMSO. The small and large RNA fractions were isolated (mirVana PARIS kit, Ambion, Austin, TX, USA). Expression was determined by RT-qPCR (Invitrogen, Carlsbad, CA, USA). miRNAs assayed included miR-143, miR-145, miR-146a and miR-146b, as well as miR-378 and miR-584 which are also found within the CDR. Expression was normalized to 5S miRNA. mRNAs examined included RPS14 and SPARC. mRNA expression was normalized to GAPDH mRNA. Fold-change was compared to baseline defined by expression in DMSO-exposed control samples. Further details are described in the Online Supplementary Appendix and Online Supplementary Table S1.
Correlation with clinical outcome
We attempted to correlate the in vitro miRNA and mRNA expression changes with clinical outcome. A range of fold-changes (1.3-, 1.5- and 2-fold) were examined in relation to response.19,20 Lenalidomide administration and dose modification was at the discretion of the treating physicians at each center. Complete clinical data were available for 28 of 29 patients. Transfusion independence (TI) was examined as the primary clinical end point. A durable response was defined as one lasting for more than 12 months.
Knockdown experiments
To model both the survival advantage and the selective inhibition of del(5q) hematopoietic progenitors induced by the lack of key miRNAs, CB CD34+ progenitors were depleted of miR-143 and miR-145 using lentiviral miRNA decoy constructs as described previously.6,21 Decreased expression of miRNAs was confirmed by RT-qPCR. In addition, changes in miR-143, miR-145 and SPARC expression were examined in transduced cells in the presence of lenalidomide and compared to controls with DMSO alone. Clonogenic progenitor assays were performed following treatment of transduced cells in vitro with lenalidomide (10 μM) or vehicle (DMSO). Colony forming cells (CFCs) were scored after 14 days. Further details are described in the Online Supplementary Appendix.
Results and Discussion
Initial optimization studies with CB samples were performed to identify the cell population most sensitive to lenalidomide-induced changes in miRNA and mRNA expression. Exposure to lenalidomide led to the upregulation of miR-143, miR-145 and SPARC. No significant increase in miR-146a, miR-146b or RPS14 was observed. Importantly, the CD34+ fraction was found to be most sensitive to changes in miRNA and mRNA expression (Online Supplementary Figure S1A and B). Mouse marrow hematopoietic progenitor cells (lineage-negative) were also examined but did not respond to lenalidomide exposure with induction of miR-143 or miR-145, suggesting that mouse cells cannot be used to study the effect of this immunomodulatory drug (Online Supplementary Figure S1C).
We next examined the effect of lenalidomide on marrow samples from 29 patients with MDS (10 del(5q) and 19 non-del(5q)). Of particular importance was the focus on CD34+ cells from pre-treatment marrow samples as this fraction is enriched for the diseased clone. As such, any changes would be attributed to effects on the dysplastic population rather than re-expansion of normal CD34+ cells post-therapy as might be observed if comparisons are made between pre- and post-treatment marrow samples.
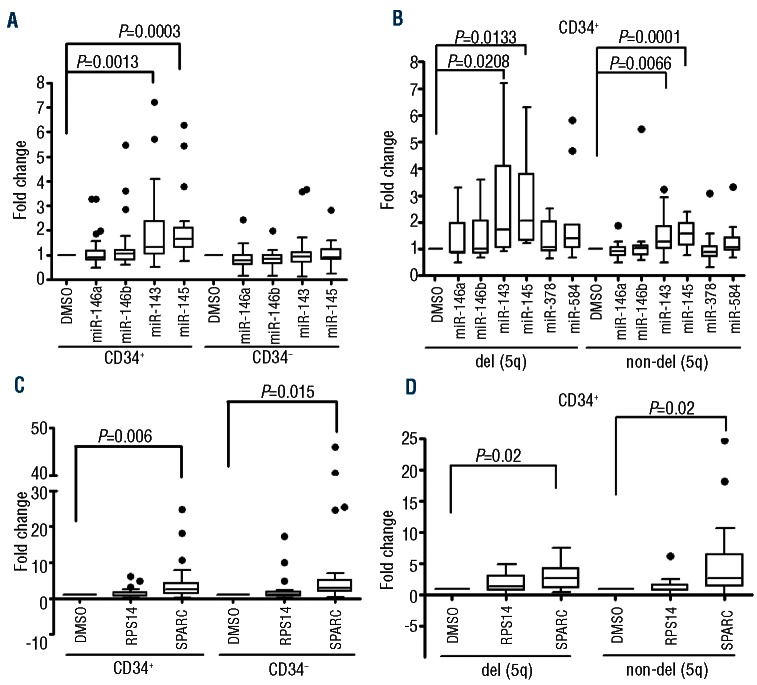
Examining the entire cohort, only the CD34+ fraction showed evidence of a significant change in miRNA expression (Figure 1A). In the del(5q) cohort, a statistically significant change in miR-143 (1.8-fold) and miR-145 (1.9fold) expression was seen (Figure 1B). In the non-del(5q) cohort a similar but less pronounced miRNA upregulation was noted (1.5− and 1.6−fold increase in miR-143 and miR-145, respectively; Figure 1B). No statistically significant changes in miR-146a, miR-146b, miR-378 or miR-584 expression were observed. The effect of lenalidomide on mRNA expression was also examined. RPS14 levels did not change significantly after exposure to lenalidomide irrespective of cell fraction or del(5q) status. SPARC levels did however increase significantly (Figure 1C). This was irrespective of del(5q) status (2.8− and 5.2−fold in del(5q) and non-del(5q), respectively; Figure 1D). Overall, the miRNA and mRNA changes observed in patient samples were similar to those seen in normal CD34+ cells from cord blood suggesting a direct effect on gene expression rather than a secondary effect of eliminating the malignant cell population.
Figure 1.
miRNA and mRNA changes in marrow samples from patients with MDS after in vitro exposure to lenalidomide. (A) Expression of miR-143, miR-145, miR-146a and miR-146b was examined. A statistically significant increase in expression of only miR-143 and miR-145 was noted and was restricted to the CD34+ cell fraction. (B) The miRNA fold-change in CD34+ cells was reanalyzed based on del(5q) status. Significant fold-change was seen in both miR-143 and miR-145 irrespective of del(5q) status. Expression of miR-378 and miR-584 were also examined in this cell fraction and significant differential expression was not seen. (C) Expression of SPARC and RPS14 levels were also examined based on CD34 status. mRNA expression was normalized to 18s rRNA. A statistically significant increase in SPARC expression was noted irrespective of CD34 status or (D) del(5q) status.
Given the important role these gene products play in hematopoiesis we postulated that induction after in vitro exposure to lenalidomide could act as a marker for therapeutic responsiveness. As this finding has yet to be described, no thresholds correlating gene expression and response have been established. As such, we examined a range of fold-changes in relation to clinical outcome. As expected, a high rate of TI was seen in del(5q) patients (90%) compared with non-del(5q) patients (22%). In del(5q) patients a 1.3−fold or more increase in miR-145 expression correlated with TI (Table 1). Seven of 9 responding del(5q) patients maintained their TI for more than 12 months and this correlated significantly with a 1.5−fold or more increase in miR-145 expression. No statistically significant correlation between response or response duration and miR-143 expression was seen. A trend to significance was also noted for the correlation between a 1.3−fold or more increase expression of SPARC mRNA and TI but not with response duration. Interestingly, the single non-responding patient in the del(5q) cohort did not show increases in either miR-145 or SPARC expression, suggesting microdeletion or epigenetic regulation in the CDR of the retained allele. In the non-del(5q) cohort there was no statistically significant correlation with response or response duration and miRNA or mRNA expression at any threshold.
Table 1.
Correlation between both clinical response and response duration with miRNA/mRNA expression; TI (transfusion independence).
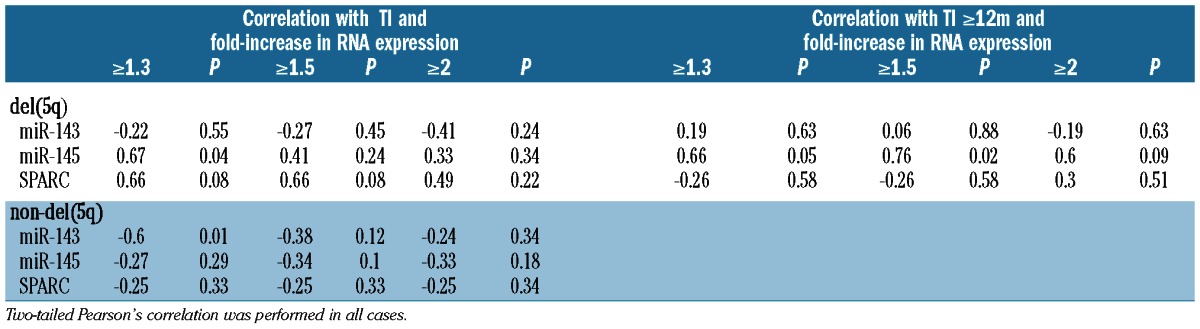
These data support the hypothesis that re-induction of key gene products found within the CDR is required for the selective drug activity in del(5q) patients. In addition, induction of miR-145 may act as marker for improved therapeutic outcomes. While a specific threshold of expression may exist, small sample size limits conclusions in this regard. Indeed, it is difficult to obtain large numbers of samples with del(5q) with sufficient numbers of viable cells to perform these experiments.
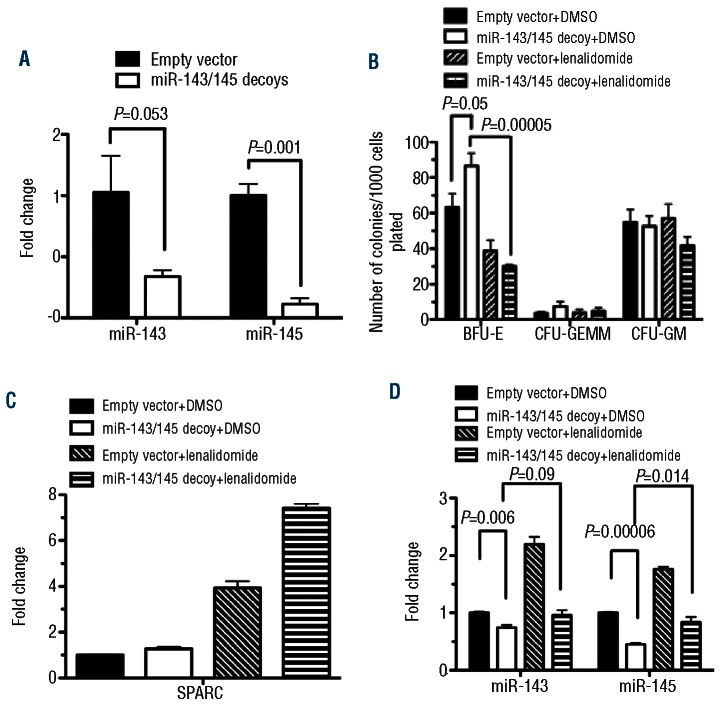
In order to further support our hypothesis, we attempted to model the survival advantage and the selective inhibition of del(5q) hematopoietic progenitors by lenalidomide. Viral transduction with an miRNA decoy successfully decreased intracellular miR-143 and miR-145 levels (Figure 2A). The depletion of target miRNAs led to increased erythroid progenitor activity as measured by BFU-E numbers, compared to empty vector transduced cells (Figure 2B). These data support the hypothesis that depletion of these key gene products imparts a survival advantage on affected clones. We then examined the effect of lenalidomide exposure on these cells. Importantly, there was no significant decrease in BFU-E, CFU-GEMM or CFU-GM formation in control cells transduced with the empty vector (Figure 2B). However, in cells transduced with the miRNA decoy, a statistically significant decrease in BFU-E was seen in the presence of lenalidomide (Figure 2B). This finding implies that, in addition to gaining a survival advantage over their normal CD34+ counterparts, cells lacking miR-143 and miR-145 are also selectively sensitive to lenalidomide. It is important to note that in miR-143/145-decoy cells there was no effect on baseline SPARC expression or the expected increase after lenalidomide exposure (Figure 2C). In addition, miR-143 and miR-145 expression was markedly diminished in transduced cells indicating that expression was effectively inhibited by the decoy (Figure 2D). These functional data corroborate the observations correlating miR-145 induction and therapeutic response.
Figure 2.
miR-143/miR-145 knockdown increases ery-throid progenitor activity of human CD34+ cord blood cells, which is selectively inhibited by lenalidomide. (A) Decreased expression of miRNAs was confirmed by RT-qPCR. Bars show the means±SE of 2 experiments. Clonogenic progenitor assays were performed following treatment of transduced cells in vitro with lenalidomide. (B) Increased BFU-E was demonstrated after transduction with the miRNA decoy. Lenalidomide exposure abrogated the increased clonogenic progenitor potential in transduced CD34+ cells. (C) SPARC mRNA expression was not affected by the decoy vector. (D) miR-143 and miR-145 was suppressed by the decoy vector. Bars show the means±SE of 3 experiments, each performed in duplicate.
In this study, we demonstrate that lenalidomide induces changes in key miRNA and mRNAs found within the CDR that characterizes del(5q) MDS. It confirms previous observations of lenalidomide-induced SPARC gene expression in dysplastic progenitor cells, and is the first to show induction of miR-143 and miR-145. Importantly, it is the first study attempting to correlate expression changes in these 3 genes with outcome. Despite the small sample size, our findings suggest that miR-145 induction may be an important predictor of response to lenalidomide in patients with MDS with del(5q). Recent work provides a mechanistic basis for the clinical effect of lenalidomide via the miR-145 pathway. Loss of miR-145 has been shown to lead to upregulation of Fli-1 and TIRAP, the downstream effects of which result in characteristic megakaryocytic dysplasia and thrombocytosis, respectively.6,8 Based on the data presented here, down-regulation of miR-143 and miR-145 also imparts increased clonogenic progenitor activity and selective sensitivity to the action of lenalidomide. Importantly, it appears that the drug has limited effect on the hematopoietic differentiation potential of normal CD34+ cells. Rather, lenalidomide selectively inhibits cells with depleted levels of miR-143 and miR-145, as would be found in del(5q) clones.
In MDS with del(5q), lenalidomide treatment likely does not reverse the potential for ineffective erythropoesis; this trait is caused by the loss of genetic material from chromosome 5. Rather, the drug alters the mechanisms required for these abnormal cells to flourish allowing normal progenitors to re-establish themselves within the marrow niche. Importantly, this effect seems to be most pronounced in altering faulty erythroid differentiation as evidenced by the suppression of BFU-E formation in cells lacking miR-143 and miR-145 in vitro. This is in line with the clinical observation that lenalidomide primarily functions to improve the anemia caused by ineffective erythropoiesis; a key feature of MDS with del(5q). However, it is important to recognize that lenalidomide likely does not completely eliminate the malignant clone.22 This corroborates the clinical observation that ongoing treatment with the drug is often required and that its removal will again uncover the clonal advantage leading to a return of the disease phenotype with proliferation of the affected clone.
It is interesting that, despite similar patterns of gene induction, poor correlation of miRNA or mRNA expression with clinical response in the non-del(5q) cohort indicates that lenalidomide is likely acting on alternate pathways in these genetically diverse cells. Given that these patients did not harbor deletions of chromosome 5q, it would not be expected that these genes play a role in the disease phenotype. As such, upregulation by the drug would have little impact on clinical outcomes.
While the study cohort is small, the statistically significant correlation seen between miR-145 expression, TI and response duration, combined with supportive functional studies, suggests an important role for miR-145 induction in response to lenalidomide. Further data may help broaden the understanding of the pathophysiology of del(5q) and the mechanism of action of targeted agents.
Supplementary Material
Acknowledgments
We would also like to thank Dr. Martin Jadersten for his help in the statistical analysis of the clinical cohort.
Footnotes
Funding
This work was funded by a grant from the Canadian Institutes of Health Research (MOP 89976), and a collaborative research agreement with Celgene Corp.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Raza A, Reeves JA, Feldman EJ, Dewald GW, Bennett JM, Deeg HJ, et al. Phase 2 study of lenalidomide in transfusion-dependent, low-risk, and intermediate-1 risk myelodysplastic syndromes with karyotypes other than deletion 5q. Blood. 2008;111(1):86–93 [DOI] [PubMed] [Google Scholar]
- 2.List A, Dewald G, Bennett J, Giagounidis A, Raza A, Feldman E, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456–65 [DOI] [PubMed] [Google Scholar]
- 3.Fenaux P, Giagounidis A, Selleslag D, Beyne-Rauzy O, Mufti G, Mittelman M, et al. A randomized phase 3 study of lenalidomide versus placebo in RBC transfusion-dependent patients with Low-/Intermediate-1-risk myelodysplastic syndromes with del5q. Blood. 2011;118(14):3765–76 [DOI] [PubMed] [Google Scholar]
- 4.Le Bras F, Sebert M, Kelaidi C, Lamy T, Dreyfus F, Delaunay J, et al. Treatment by Lenalidomide in lower risk myelodysplastic syndrome with 5q deletion-The GFM experience. Leuk Res. 2011;35(11):1444–8 [DOI] [PubMed] [Google Scholar]
- 5.Wei S, Chen X, Rocha K, Epling-Burnette PK, Djeu J Y,, Liu Q, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci USA. 2009;106(31):12974–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58 [DOI] [PubMed] [Google Scholar]
- 7.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar MS, Narla A, Nonami A, Mullally A, Dimitrova N, Ball B, et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood. 2011;118(17):4666–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliva EN, Cuzzola M, Nobile F, Ronco F, D'Errigo MG, Lagana C, et al. Changes in RPS14 expression levels during lenalidomide treatment in Low- and Intermediate-1-risk myelodysplastic syndromes with chromosome 5q deletion. Eur J Haematol. 2010;85(3):231–5 [DOI] [PubMed] [Google Scholar]
- 10.Moutouh-de Parseval LA, Verhelle D, Glezer E, Jensen-Pergakes K, Ferguson GD, Corral LG, et al. Pomalidomide and lenalidomide regulate erythropoiesis and fetal hemoglobin production in human CD34+ cells. J Clin Invest. 2008;118(1):248–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert BL, Galili N, Tamayo P, Bosco J, Mak R, Pretz J, et al. An Erythroid Differentiation Signature Predicts Response to Lenalidomide in Myelodysplastic Syndrome. PLoS Med. 2008;5(2):e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.List AF, Estes M, Williams A, Sekharam M, Ozawa U, Gao G, et al. Lenalidomide (CC-5013; Revlimid(R)) Promotes Erythropoiesis in Myelodysplastic Syndromes (MDS) by CD45 Protein Tyrosine Phosphatase (PTP) Inhibition. ASH Annual Meeting Abstracts. 2006;108(11):1360 [Google Scholar]
- 13.Pellagatti A, Jadersten M, Forsblom AM, Cattan H, Christensson B, Emanuelsson EK, et al. Lenalidomide inhibits the malignant clone and up-regulates the SPARC gene mapping to the commonly deleted region in 5q- syndrome patients. Proc Natl Acad Sci USA. 2007;104(27):11406–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci USA. 2011;108(22):9184–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim A F,, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71(15):5214–24 [DOI] [PubMed] [Google Scholar]
- 17.Fuse M, Nohata N, Kojima S, Sakamoto S, Chiyomaru T, Kawakami K, et al. Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int J Oncol. 2011;38(4):1093–101 [DOI] [PubMed] [Google Scholar]
- 18.Starczynowski DT, Kuchenbauer F, Wegrzyn J, Rouhi A, Petriv O, Hansen CL, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39(2):167–78 e4 [DOI] [PubMed] [Google Scholar]
- 19.Cheson BD, Bennett JM, Kantarjian H, Pinto A, Schiffer CA, Nimer SD, et al. Report of an international working group to standardize response criteria for myelodysplastic syndromes. Blood. 2000;96(12):3671–4 [PubMed] [Google Scholar]
- 20.Cheson BD, Greenberg PL, Bennett JM, Lowenberg B, Wijermans PW, Nimer SD, et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108(2):419–25 [DOI] [PubMed] [Google Scholar]
- 21.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tehranchi R, Woll PS, Anderson K, Buza-Vidas N, Mizukami T, Mead AJ, et al. Persistent malignant stem cells in del(5q) myelodysplasia in remission. N Engl J Med. 2010;363(11):1025–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.