Abstract
MicroRNA have been demonstrated to be deregulated in multiple myeloma. We have previously reported that miR-214 is down-regulated in multiple myeloma compared to in normal plasma cells. The functional role of miR-214 in myeloma pathogenesis was explored by transfecting myeloma cell lines with synthetic microRNA followed by gene expression profiling. Putative miR-214 targets were validated by luciferase reporter assay. Ectopic expression of miR-214 reduced cell growth and induced apoptosis of myeloma cells. In order to identify the potential direct target genes of miR-214 which could be involved in the biological pathways regulated by this microRNA, gene expression profiling of the H929 myeloma cell line transfected with precursor miR-214 was carried out. Functional analysis revealed significant enrichment for DNA replication, cell cycle phase and DNA binding. miR-214 directly down-regulated the expression of PSMD10, which encodes the oncoprotein gankyrin, and ASF1B, a histone chaperone required for DNA replication, by binding to their 3'-untranslated regions. In addition, gankyrin inhibition induced an increase of P53 mRNA levels and subsequent up-regulation of CDKN1A (p21Waf1/Cip1) and BAX transcripts, which are direct transcriptional targets of p53. In conclusion, MiR-214 functions as a tumor suppressor in myeloma by positive regulation of p53 and inhibition of DNA replication.
Introduction
MicroRNA (miRNA, miR) are small non-coding RNA that regulate gene expression at the post-transcriptional level and are involved in critical biological processes, including cellular growth and differentiation. Previous studies have shown that miRNA expression is deregulated in myeloma cells as compared to in normal plasma cells. Moreover, several miRNA have been involved in the pathogenesis of multiple myeloma (MM).1-3 In this sense, it has recently been shown that there is a mechanism of p53 regulation through miRNA acting on MDM2 expression; thus, miR-192, 194 and 215 re-expression in myeloma cell lines induced degradation of MDM2 with subsequent p53 up-regulation and cell growth inhibition.4 In addition, miR-15a and 16 have also been shown to regulate myeloma proliferation by inhibiting AKT and MAP-kinases, and by reducing bone marrow angiogenesis.5
We have previously reported the down-regulation of 11 miRNA (miR-375, miR-650, miR-214, miR-135b, miR-196a, miR-155, miR-203, miR-95, miR-486, miR-10a and miR-196b) in MM samples compared to in normal plasma cells.6 Interestingly, only miR-214 and miR-375 were significantly under-expressed in all the different cytogenetic subgroups (including IGH translocations, RB deletions and normal fluorescence in situ hybridization). miR-214 deregulation has been observed in different types of cancer. Overexpression of miR-214 has been reported in several tumors, such as melanoma, ovarian cancer and gastric cancer.7-9 In contrast, miR-214 was found to be down-regulated in breast cancer, resulting in increased cell proliferation and invasion.10 Likewise, low miR-214 expression levels were associated with metastasis and invasion of cervical tumors11 and it has also been recently described that miR-214 is down-regulated in primary central nervous system lymphomas.12
In order to investigate the potential involvement of miR-214 in myeloma pathogenesis, we explored the functional role of miR-214 in myeloma cells. We found that ectopic expression of this miR-214 reduced cell growth and induced apoptosis of myeloma cells. This effect was mediated by interfering with the p53 signaling pathway through downregulation of p28/gankyrin protein and by inhibition of replication via decreasing the level of histone chaperone Asf1b.
Design and Methods
Cells and culture conditions
The human myeloma cell lines, NCI-H929 and MM1S, were acquired from the American Type Culture Collection (ATCC) and JJN3 and RPMI-8226 from the Deuthche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). It was previously described that H929 and MM1S cells have wild-type (wt) P53 and JJN3 and RPMI-8226 have mutated/null P53.4 All cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and antibiotics (Gibco). Cells were routinely checked for the presence of mycoplasma with MycoAlert kit (Lonza GmBH) and only mycoplasma-free cells were used in the experiments. The phenotypic and cytogenetic identities of the cell lines were verified by flow cytometry and fluorescence in situ hybridization before the experiments. Bone marrow samples were obtained from eight patients with myeloma and four healthy subjects undergoing bone marrow harvest for allogeneic transplantation. CD138+ plasma cells were isolated (purity >95%) from the bone marrow samples using the AutoMACS automated separation system (Miltenyi-Biotec). All patients as well as healthy donors provided written informed consent in accordance with the Helsinki Declaration, and the research ethics committee of the University Hospital of Salamanca approved the study.
Transfection with synthetic microRNA
Human myeloma cell lines were transfected with Pre-miR™ miRNA precursors pre-miR-214 or Pre-miR™ miRNA negative, non-targeting control#1 (Ambion) at a 50 nM concentration, using the nucleofector II system (Amaxa) with the C-16 program for H929 and JJN3, and the G-16 program for MM1S and RPMI-8226. Transfection efficiency was assessed with Block-iT™ Fluorescent Oligo (Invitrogen) by flow cytometry (Online Supplementary Figure S1). Target gene down-regulation was monitored using the Pre-miR™ miRNA Precursor Starter Kit (Ambion).
RNA extraction and quantitative real-time polymerase chain reaction analysis
RNA was extracted from the cell lines using Trizol reagent (Invitrogen) according to the standard protocol. The RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). To detect mature miR-214 expression levels, a TaqMan quantitative real-time polymerase chain reaction (qRT-PCR miRNA) assay (Applied Biosystem) was performed. The relative expression of mature miR-214 levels normalized to the RNU43 endogenous control was determined using the 2-ΔCt method. Each measurement was performed in triplicate.
To detect the target genes (mRNA expression), total RNA (1 μg) was reverse transcribed to cDNA using SuperScript™ III First-Strand Synthesis SuperMIx (Invitrogen). For PSMD10, ASF1B and P53 expression, iQ™ SYBR® Green Supermix kit (BioRad) was used. SYBR green qRT-PCR was performed using the Bio-Rad iQ5 PCR detection system with the following gene-specific primers: GAPDH, forward 5' GCTTCGCTCTCTGCTCCTCCTGT-3' and reverse 5'- ACTGGACTGACCTGGTCCAC-3'; PSMD10, forward 5'-GGTGTCCCAAGGAGCAAGTA-3' and reverse 5' ACACTGGGGACAACAACACA-3'; ASF1B, forward 5'-GAG-GCTTTCTTGGTCAGTGC-3' and reverse 5'-TCCTGCTGAT-CACACCTCTG-3'; P53, forward 5' GGCCCACTTCACCG-TACTAA-3' and reverse 5'-GTGGTTTCAAGGCCAGATGT-3; MAPK3, forward 5'- ACTATGACCCGACGGATGAG-3′ and reverse 5'- CTAACAGTCTGGCGGGAGAG-3′; SCAMP3, forward 5′-TTTGAGACCCGGGAGCCACCA-3′ and reverse 5′-AAAGCCCAAAGCCTGCGCCA-3′; PEMT, forward 5′-TCGTCGGTGACCTGTGGGACT-3′and reverse 5′-CCTGCGT-GAAGCAGTGCGAG-3′; CDC7AL, forward 5′-TGGC-GACTCGCTACCAGATCCC-3′ and reverse 5′-GCGCACATC- CTGCTGTTTCCCT-3′; SEC24C, forward 5′-GGAGCCGGGA-GATAATCTGAA-3 and reverse 5′-GGCAGCCTTTGCATCT-GAAC-3′; ORC1L, forward 5′- TCTCAAGCCTAGAACGCCAC-3′ and reverse 5′- TGGCCTGTTAGCTTCTGCAA-3′. Expression of BAX, MCM2, MCM4, MCM6, MCM7 was assessed using Dharmacon Solaris qPCR and CDKN1A(p21/Waf1) using TaqMan expression assays, each normalized to the respective Dharmacon Solaris or TaqMan GAPDH expression assay. Relative gene expression was calculated using the 2-ΔCt method.
5-aza-2′-deoxycytidine treatment
JJN3 and H929 cells were subcultured at a density of 7x105 cells/mL (viability > 95 %) the day before the experiment. These cells were then exposed to 5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich) 1 μM for 72 h. The corresponding amount of dimethlysulfoxide (DMSO; Sigma-Aldrich) was added to the negative controls.
Cell proliferation assays
Cell viability was evaluated by a trypan blue exclusion test (Invitrogen) and by a CellTiter-Glo® luminescent cell viability assay based on the amount of ATP present (Promega), in accordance with the manufacturer's protocol. Cell growth was examined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (Sigma-Aldrich) as previously described.13 BrdU incorporation was measured using a BrdU Cell Proliferation Kit according to the manufacturer's protocol (Millipore). BrdU was incorporated over 24 h.
Apoptosis assays
Apoptosis was measured using an annexin V-fluorescein isoth-iocyanate/propidium iodide (PI) double staining (Immunostep) according to the manufacturer's procedure. Mitochondrial membrane depolarization was evaluated using DiOC6(3)/PI dual staining (Sigma-Aldrich). Cells were acquired on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with the CellQuest program. Caspases 3/7, 8 and 9 activity was evaluated by Caspase-Glo® 3/7, Caspase-Glo® 8 and Caspase-Glo® 9 Assays (Promega), respectively, according to the manufacturer's protocol. In the assays 20,000 cells/well were incubated with the reagents at room temperature for 1 h in the case of caspase 3/7 and 2 h for caspases 8 and 9. Light intensity was measured in each well using a Tekan Infinite® F500 microplate reader.
Cell cycle analysis
Cells were washed in phosphate-buffered saline, fixed in 70% ethanol overnight, washed with phosphate-buffered saline again and resuspended in 500 μL of PI/RNASE staining solution (Immunostep) and incubated for 20 min at room temperature in the dark. Samples were analyzed by FACSCalibur flow cytometry. For each sample a signal from 30,000 cells was acquired and the cell cycle was analyzed with ModFit™ software.
Immunoblotting
Whole cell lysates were collected using RIPA buffer (Sigma-Aldrich) containing protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, Roche) and phosphatase inhibitors (Phosphatase Inhibitor Cocktail A and B, Santa Cruz Biotechnology). Protein concentration was measured using the Bradford assay (BioRad). Protein samples (40 μg/lane) were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.45 mm polyvinyli-dene fluoride (PVDF) membranes (BioRad). The primary antibodies used for immunoblotting were anti-Asf1b and anti-p-Mdm2 (Cell Signaling), anti-gankyrin, anti-p53, anti-Mdm2 and anti-p21 (Santa Cruz Biotech), and anti-β-actin (Sigma-Aldrich) as an internal control for protein loading. The membranes were then washed and subsequently incubated with the secondary horseradish per-oxidase-linked anti-mouse IgG and anti-rabbit IgG antibodies (PierceNet) (1:10000). Chemiluminescence was detected using the Amersham ECL Plus™ Western Blotting Detection Reagent (GE Healthcare).
Gene expression profiling
Total RNA was amplified, labeled and hybridized to Human Gene 1.0 ST Array (Affymetrix) as previously reported.6 The integrity of the RNA was verified with an Agilent 2100 Bioanalyzer. Full microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xjcdlmem-keeemps&acc=GSE35948). Bioinformatic analysis was carried out using DNA-Chip Analyzer software (DChip).14 The comparison criteria used in dChip analysis were fold change E/B>2 or B/E>2, mean difference E-B>100 or B-E>100 and the lower 90% confidence bound of fold-change was used. Gene-annotation enrichment analysis was performed with version 6.7 of the Database for Annotation, Visualization and Integrated Discovery (DAVID)(http://david.abcc.ncifcrf.gov/).15,16
Computational microRNA target prediction
Three different databases (Targetscan http://www.targetscan.org/,17,18 miRDB http://mirdb.org/,19,20 and miRANDA http://www.microrna.org/)21 were used to identify potential miR-214 target genes.
Luciferase reporter assay
The double-stranded oligonucleotides corresponding to the wild-type (WT-3'UTR) or mutant (MUT-3'UTR) miR-214 binding site in the UTR of PSMD10 (NM_002814) or ASF1B (NM_018154) were synthesized (Sigma-Aldrich) and ligated between the PmeI and Xbal restriction sites of the pmirGLO vector (Promega). The oligonucleotides sequences are presented in Online Supplementary Table S1. For luciferase assays, HEK293 cells were transfected with 500 ng of the above constructs and co-transfected with 25 nM miRNA precursor molecule by nucleofection using the HEK293 cell line program in the Amaxa II nucleofector system. Twenty-four hours after transfection cells were collected and Firefly and Renilla luciferase activities were measured using the Dual-Glo® Luciferase Assay System (Promega) according to the manufacturer's protocol. Measurements were performed on a Tekan Infinite® F500 microplate reader. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
The two-sided Student's t test was used to analyze differences in experiments. Data are reported as mean values ± SD of at least triplicate determinations. The Mann-Whitney U test was used to identify statistically significant differences between normal plasma cells and those from patients with MM. P values < 0.05 were considered statistically significant. All statistical analyses were conducted using the SPSS 15.0 statistical package (SPSS).
Results
Involvement of DNA methylation in down-regulation of microRNA-214
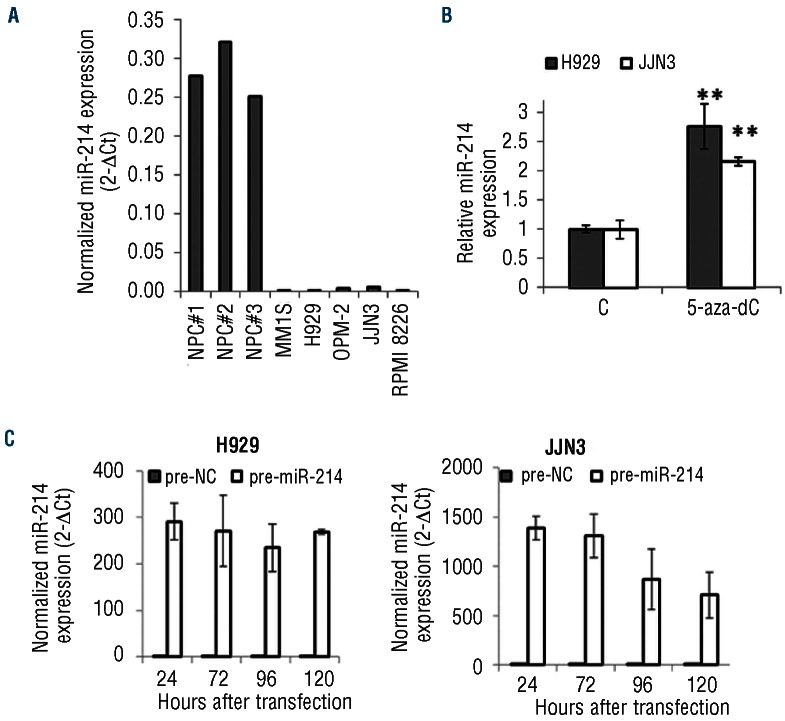
In order to corroborate the down-regulation of miR-214 previously reported in primary MM cells, we measured the level of miR-214 in a set of MM cell lines (MM1S,H929, OPM-2, JJN3 and RPMI 2886) and compared it to the level in normal plasma cell samples (n=3) using miRNA TaqMan qRT-PCR. miR-214 was down-regulated in all MM cell lines tested (Figure 1A). Since the 1q region, in which miR-214 is located, is commonly gained in MM, we hypothesized that epigenetic mechanisms could be involved in the under-expression of miR-214 in MM. To address this question we treated H929 and JJN3 cells with the DNA demethylating agent 5-aza-dC and quantified the expression of miR-214. After 5-aza-dC treatment, miR-214 expression was increased more than 2-fold in both myeloma cell lines (Figure 1B).
Figure 1.
Expression level of miR-214 in myeloma cells. (A) Comparison of miR-214 expression by qRT-PCR between normal plasma cells (NPC) and MM cell lines. (B) Expression of miR-214 after 72 h of 5-aza-dC treatment. Data represent the average of three independent experiments ±SD. (C) Level of miR-214 after transfection with negative, non-targeting control and pre-miR-214 in H929 (left panel) and JJN3 (right panel) cells. Expression of miR-214 was normalized to RNU43 and presented as 2(-ΔCt).
Over-expression of microRNA-214 reduces growth of myeloma cells
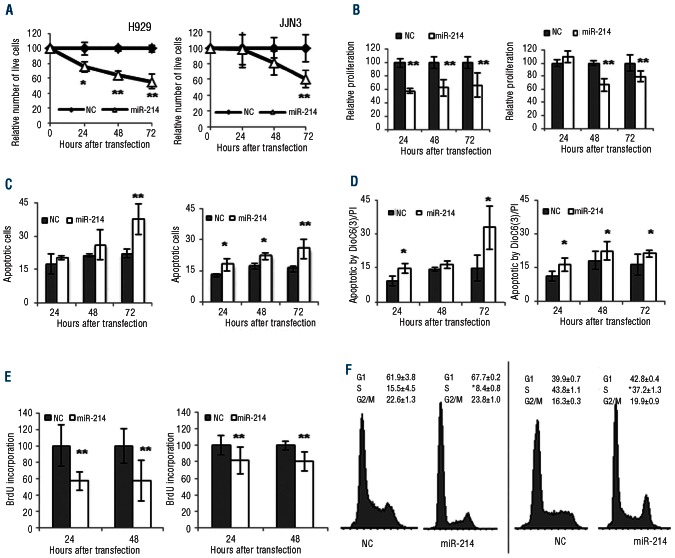
To investigate the functional role of miR-214 expression in MM, we carried out gain-of-function experiments in H929 and MM1S (wt P53) and JJN3 and RPMI-8226 (P53 mutated/null). The miR-214 over-expression persisted for up to 5 days (Figure 1C). Our results showed that over-expression of miR 214 inhibited cell growth, as assessed by trypan blue counting and quantification of ATP levels, in the four cell lines with respect to in the non-targeting control although the growth inhibitory effect started 24 h later in P53 mutated/null cells (JJN3 and RPMI-8226) than in P53 wt cells (H929 and MM1S) (Figure 2A and B, and Online Supplementary Figure S2A, B).
Figure 2.
Overexpression of miR-214 reduces cell growth and induces apoptosis in myeloma cells. H929 cell line (left panel) and JJN3 cell line (right panel). (A) Relative number of live cells in a time course evaluated by trypan blue counting after miR-214 or non-targeting control (NC) transfection. Data are the average ± SD of three independent experiments. (B) Proliferation evaluated by ATP quantitation (CellTiter-Glo®) of myeloma cells transfected with miR-214. The results are presented as the mean ± SD of three different experiments and considering the result after non-targeting control (NC) transfection as 100%. (C) Apoptosis examined by annexin V/PI assay and (D) mitochondrial membrane depolarization after miR-214 or NC transfection. Data are the mean ± SD of three separate experiments. (E) BrdU incorporation 24 and 48 h after miR-214 transfection. The results were quantified as described for panel B. (F) Cell cycle changes after 24 h of transfection with miR-214 or NC. Results are the mean of three independent experiments. *P<0.05; **P<0.01.
To assess whether the decrease of cell growth was the result of apoptosis induction, annexin V-positive cells were quantified as well as DiOC6(3) negative-cells in order to determine the involvement of mitochondria in cell death. Ectopic transfection of miR-214 induced apoptosis at 72 h at significant levels in H929 and JJN3 cell lines (P<0.01) (Figure 2C). In addition, using DiOC6(3), a decrease in mitochondrial membrane potential was observed after miR-214 transfection (Figure 2D). To evaluate the role of caspases in the induction of apoptosis, the levels of activity of caspase 3/7, 8 and 9 were analyzed in H929 and JJN3 cell lines after transfection with miR-214 or non-targeting control. Transfection with miR-214 induced executioner caspases 3 and 7 in both cell lines, although this effect was less strong for P53 mutated/null JJN3 cells (Online Supplementary Figure 3A). Caspase 8 (Online Supplementary Figure 3B) and caspase 9 (Online Supplementary Figure 3C) were also activated by miR-214 in both cell lines (JJN3 and H929) suggesting the activation of both extrinsic and intrinsic pathways of apoptosis.
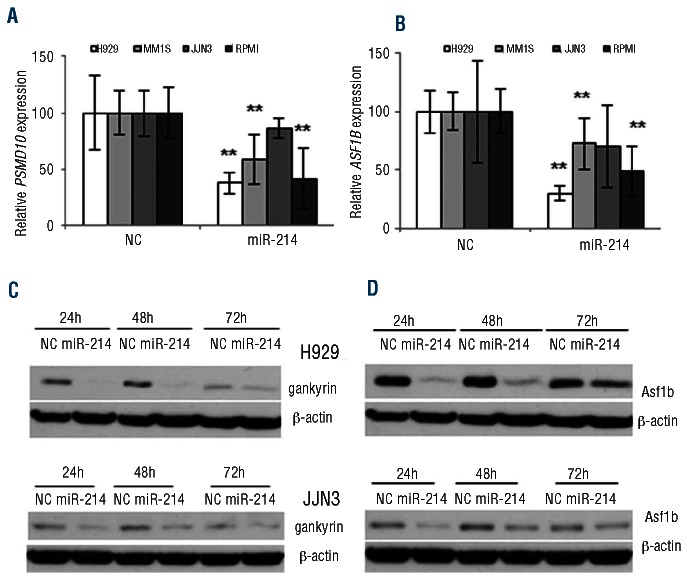
Figure 3.
Validation of miR-214 targets in myeloma cells. (A) mRNA level measured by qRT-PCR of PSMD10 and (B)ASF1B in H929 cells 24 h after transfection with miR-214 precursor or NC normalized to GAPDH. **P<0.01. The results are presented as the mean ± SD of three different experiments. (C) Level of Psmd10 (gankyrin) and (D) Asf1b proteins by western blotting in H929 and JJN3 cells after transfection with miR-214 or NC at 24, 48 and 72 h. The panels show the results of an experiment that was repeated twice.
To gain additional insights into the nature of the growth inhibitory effects after miR-214 transfection, we assessed cell proliferation. MiR-214 over-expression reduced the proportion of BrdU-positive cells by 58% in H929 cells and by 80% in JJN3 cells (P<0.01) in comparison with non-targeting controls (Figure 2E). This result was consistent with the decrease in S phase in cells transfected with miR-214 compared to the non-targeting control (P<0.05) (Figure 2F).
Identification of microRNA-214 targets by microarray analysis
To elucidate the molecular basis of the miR-214 down-regulation, we compared the gene expression profile of H929 cells transfected with synthetic miR-214 and that of the non-targeting control. Overall, 64 genes were differentially expressed by >2-fold after ectopic (69 probesets) transfection of miR-214 (Online Supplementary Table S2). Among them, 44 (69%) were under-expressed. Functional analysis using DAVID software revealed significant enrichment (P<0.05 after Benjamini's multiple test correction) for DNA replication, cell cycle phase and DNA binding. In order to identify miR-214 predicted targets among the genes down-regulated by miR-214 transfection, we crossed these genes with databases of putative direct targets of this miR-214. We only considered those mRNA predicted by two of the three different algorithms used (TargetScan, miRDB and miRANDA). Genes that were down-regulated most significantly and were predicted as miR-214 targets are presented in Online Supplementary Table S3. The expression of these genes after miR-214 transfection was further validated by qRT-PCR in four MM cell lines (Online Supplementary Figure S4).
Validation of microRNA-214 targets in myeloma cells
Among the genes predicted as miR-214 targets which were down-regulated after transfection of H929 cells with miR-214, we chose to validate in more detail PSMD10 and ASF1B. PSMD10, also named 26S proteasome non-ATPase regulatory subunit 10, acts as an oncoprotein (p28/gankyrin) involved in the negative regulation of tumor suppressors RB1 and P53.22ASF1B, which is required for proper DNA replication and nucleosome formation, was the most down-regulated mRNA after miR-214 transfection.23
To validate gene expression profiling results, PSMD10 and ASF1B expression was assessed by qRT-PCR and western blotting after transfecting MM cell lines with miR-214 precursor. PSMD10 and ASF1B mRNA levels were found to be dramatically decreased in H929, MM1S and RPMI-8226 cells transfected with miR-214 in comparison with the levels in the non-targeting control (Figure 3A,B). When we assessed the effect of miR-214 on protein levels, western blotting showed a decrease of gankyrin (encoded by PSMD10) and Asf1b proteins after transfection (Figure 3C and D for H929 and JJN3, Online Supplementary Figure S5 for MM1S and RPMI-8226). To further validate these results on primary MM samples, we evaluated PSMD10 and ASF1B expression in myeloma cell samples obtained from eight MM patients who, according to previous miRNA profiling analysis, had low miR-214 levels.6 The expression values of PSMD10 and ASF1B were significantly higher in MM cases than in normal plasma cells: median value of 0.042 (range, 0.029-0.074) versus median, 0.009 (range, 0.006-0.025) for PSMD10 (P=0.004), and median value of 0.019 (range, 0.009-0.033) versus median, 0.009 (range, 0.002-0.013) for ASF1B (P=0.048) (Online Supplementary Figure S6).
PSMD10 and ASF1B are direct targets of microRNA-214
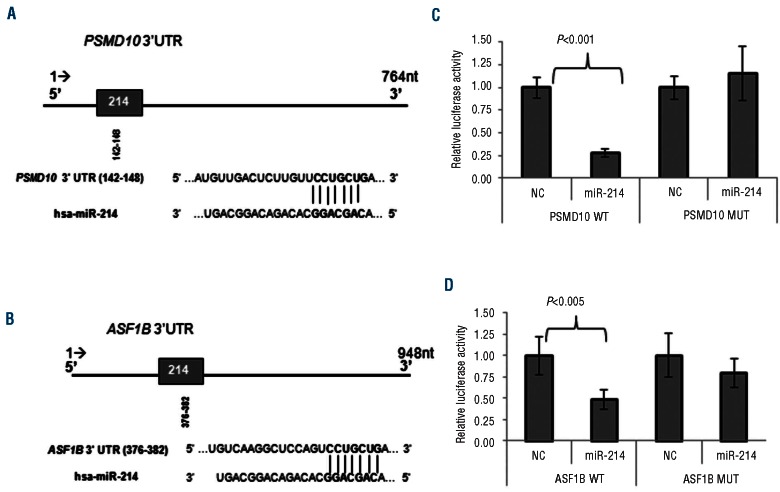
According to the prediction algorithms, miR-214 has one putative binding site (8-mer) in the PSMD10 3'UTR (Figure 4A) and another (8-mer) in the ASF1B 3'UTR (Figure 4B). It should be noted that based on the TargetScan 5.2 algorithm, it was predicted that mir-214 had the strongest effect on PSMD10 expression among all miRNA (total context score: -0.39). To determine whether PSMD10 and ASF1B transcripts were direct targets of miR-214, we carried out luciferase reporter assays in HEK293 cells. We cloned the 3'UTR of PSMD10 and ASF1B harboring the complementary sequence to the miR-214 seed sequence in a reporter plasmid vector. In parallel, the same PSMD10 and ASF1B sequences with four point mutations were cloned in the same reporter plasmid. Data from these experiments revealed that luciferase activity of the wt PSMD10 and ASF1B 3'UTR luciferase reporter in cells co-transfected with pre-miR-214 was significantly lower (P<0.01) than that of those transfected with non-targeting control miRNA, whereas luciferase activity of constructs in which the miR-214 binding site was mutated was not affected by miR-214 over-expression (Figure 4C,D). Taken together these results demonstrate that miR-214 modulates PSMD10 and ASF1B expression through the consensus miR-214 binding site in the PSMD10 and ASF1B 3'UTR.
Figure 4.
miRNA-214 targets the 3'UTR of ASF1B and PSMD10. (A) Schematic diagram of the miR-214 predicted site on the PSMD10 and (B) ASF1B 3'UTR. 3'UTR of both genes have one binding site for miR-214 predicted by TargetScan which is the exact match to positions 2-8 of the mature miRNA (the seed + position 8) followed by an ‘A’. (C) Luciferase activity in HEK293 cells co-transfected with pre-miR-NC or pre-miR-214 and plasmid pmiR-Glo with the putative miR-214 binding site of PSMD10 and (D) ASF1B cloned downstream of the luciferase reporter gene. Luciferase activity was normalized using Renilla. The results are presented as the mean ± SD of three different experiments.
MicroRNA-214 increases the level of p53 by down-regulation of PSMD10
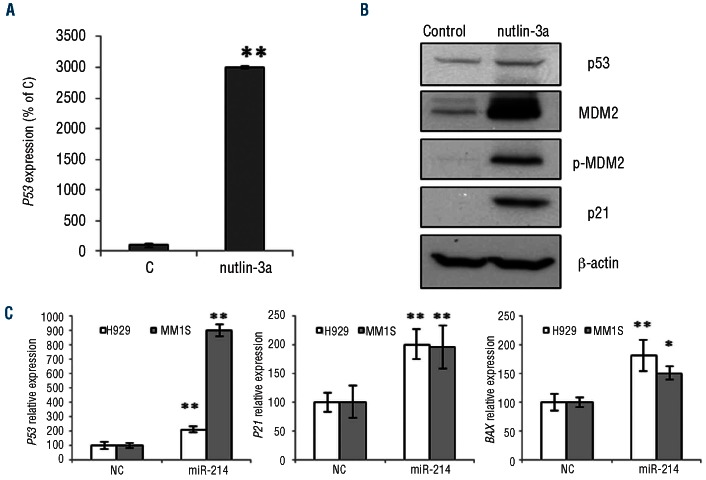
PSMD10 encodes the gankyrin protein which functions as a negative regulator of p53 by modulating MDM2 activity.22 This fact prompted us to investigate the effect of the interaction between miR-214 and gankyrin on p53 status. First, as an internal control, we used a small antagonist of the MDM2/p53 interaction, Nutlin-3a, which leads to stabilization of p53.24 Nutlin-3a inhibits the MDM2/p53 interaction by binding to the part of the MDM2 protein that interacts with p53.25 After incubating H929 cells with Nutlin-3a, we observed an increase in P53 mRNA and in the level of the protein (Figure 5A,B). The activation of p53 induced MDM2 and pMDM2 accumulation, and p21 expression, as expected (Figure 5B). Further, we investigated the effect of miR-214 over-expression and consequent gankyrin down-regulation on the level of p53.26 Ectopic expression of miR-214 in H929 and MM1S cells, which express wt P53, resulted in an increase of P53 mRNA levels (Figure 5C). Subsequently, downstream transcriptional targets such as, CDKN1A (p21Waf1/Cip1) and BAX were activated (Figure 5C).
Figure 5.
Effect of miR-214 restoration on the P53 pathway. (A)P53 mRNA expression normalized to GAPDH using qRT-PCR in H929 cells incubated with Nutlin-3a compared to untreated cells (control, C). Experiments were performed in triplicate. (B) Western blotting of p53, MDM2, p-MDM2 and p21 protein expression in H929 cells treated with Nutlin-3a. Results are representative of one experiment that was repeated twice. (C) mRNA level of P53, BAX, and CDKN1A (P21Cip1/Waf1) assessed by qRT-PCR after H929 and MM1S transfection with miR-214 precursor or NC. The results are shown as an average of three experiments after normalization to GAPDH. *P<0.05, **P<0.01.
MicroRNA-214 can interfere with the formation of the replication initiation complex
It has been documented that Asf1b, together with MCM 2, 4, 6 and 7 proteins and histones H3-H4, forms a complex that is required for the formation of the replication fork.27 The eukaryotic MCM2-7 complex is recruited onto origins of replication during the G1 phase of the cell cycle and acts as the main helicase at the replication fork during the S phase.28 Since gene expression profiling of cells transfected with miR-214 revealed down-regulation of many genes involved in the replication process, such as ASF1B, MCM2 and ORC1L (Online Supplementary Table S3), we evaluated the level of MCM2-7 transcripts by qRT-PCR. We detected not only down-regulation of the expression of MCM2, but also of MCM 4, 6 and 7 genes, after miR-214 transfection (Online Supplementary Figure S7A). Moreover, we confirmed the down-regulation of MCM2 and MCM4 in JJN3, MM1S and RPMI-8226 cells (Online Supplementary Figures S4E and S7B).
Discussion
In the present study, we show that miR-214 could function as a tumor suppressor in myeloma by targeting PMSD10 and ASFB1, which are involved in essential biological pathways. The mechanism responsible for down-regulation of miR-214 is unlikely to be explained by loss of the chromosome region in which the gene encoding this miRNA is located (1q24), since 1q is gained in more than 60% of MM patients. According to our results with 5-aza-dC treatment, it seems possible that epigenetic silencing by DNA methylation could be involved in the down-regulation of miR-214 level in myeloma cells. Methylation of DNA has also been shown to be important in the regulation of several miRNA in MM, such as miR-203, 194-2, 192 and 34a.4,29,30
The restoration of miR-214 in myeloma cell lines decreased proliferation and induced apoptosis. Other miRNA have been reported to provoke similar effects in MM. Thus, miR-15a and 16 regulate cell proliferation of MM cells by inhibiting AKT serine/threonine-protein-kinase (AKT3), ribosomal-protein-S6, MAP-kinases and NF-κB-activator MAP3KIP3.5 MiR-203, which is epigenet-ically silenced in MM, has been reported to act as a tumor suppressor.29 Moreover, ectopic re-expression of miR-29b, down-regulated in MM, resulted in induction of caspase-dependent apoptosis in myeloma cells.31
In order to discover miR-214 target genes that could be involved in the biological pathways regulated by this miRNA, we used a gene expression profiling strategy. PSMD10 was one of the predicted targets down-regulated after miR-214 transfection. Using gain-of-function experiments and luciferase reporter assays we observed that ectopic transfection of miR-214 decreased the level of gankyrin protein by directly targeting PSMD10. Moreover, as a consequence of gankyrin down-regulation in cells with wt P53, we observed an increase of P53 mRNA levels and subsequent up-regulation in CDKN1A (p21Waf1/Cip1) and BAX transcripts, which are direct transcriptional targets of p53.26 One of the oncogenic mechanisms attributed to gankyrin is to recruit the MDM2/p53 complex to the proteasome and foster the turnover of p53 in an MDM2-dependent manner. In this process, gankyrin protein binds to MDM2 protein and enhances the ability of MDM2 to ubiquitinate p53.32 Thus, it has been shown that over-expression of gankyrin blocks p53-dependent apoptosis in human bone osteosarcoma U-2 OS cells that have wt P53.22 When gankyrin was silenced in hepatocellular carcinoma cell lines with wt and mutated P53, a different phenotype was induced in the wt P53 cells, with up-regulation of p53 levels and subsequent activation of caspase 9 and induction of apoptosis.33 Our results showed stronger inhibition of cell growth and a greater pro-apoptotic effect after miR-214 transfection in cell lines with wt and functional P53 than in mutated P53 cell lines. However, apoptosis was also induced in JJN3 cells, which although they do not carry the P53 mutation, do have impaired activity of the p53 pathway.4,34 These data indicate that the role of gankyrin might not be restricted to the interaction with p53. This hypothesis has been suggested in hepatoma cells.35
Mutations of P53 are relatively rare in newly diagnosed MM, occurring in approximately 5% of patients, although the frequency of mutations appears to increase with advancing disease.36,37 On the other hand, deletion (mainly mono-allelic) of P53 occurs in about 10% of MM cases.36,38 In spite of this rather low incidence, p53 may play a broader role in myeloma pathogenesis than previously thought, since an imbalance in p53 with biological consequences in myeloma cells can also be induced through post-transcriptional mechanisms. Thus, Pichiorri et al. observed that the reinforced expression of miRNA-192, -194 and -215 induced dramatic down-regulation of MDM2 together with p53 over-expression and p21 activation. These miRNA can, in turn, be transcriptionally activated by p53 as demonstrated by Nutlin-3a treatment.4 In this regard, Nutlin-3a, a small inhibitor of MDM2, was previously described to stabilize p53 level in myeloma cells by abolishing its degradation forced by MDM2.24 The fact that bortezomib leads to an increase in the level of p53 protein, by inhibition of the proteasome that normally degrades p53, and subsequently to activation of the p53 pathway, underscores the relevance of p53 regulation in the pathogenesis of MM.39 Nutlin-3a and bortezomib have been reported to have a synergistic anti-myeloma effect.40 Although the activity of a single miRNA compared to the use of synthetically generated compounds, such as Nutlin-3a or bortezomib, is gentler, our results support the importance of the p53/MDM2 regulatory mechanism in MM.
A significant enrichment for DNA replication category was found among the genes deregulated in H929 cells after ectopic transfection with miR-214. Interestingly, three of the genes, ASF1B, MCM2 and ORC1L, involved in DNA replication and down-regulated after miR-214 transfection were miR-214 predicted targets. It has been described that Asf1b, together with MCM 2, 4, 6 and 7 proteins and histones H3-H4, forms a complex that is required for the formation of the replication fork.27 The MCM2-7 complex serves as the eukaryotic replicative helicase, the molecular motor that both unwinds duplex DNA and powers fork progression during DNA replication.41 In the present study, qRT-PCR analysis revealed down-regulation of MCM 2, 4, 6 and 7 genes after over-expression of miR-214 in MM cells. The restoration of miR-214 into myeloma cells also decreased the mRNA and protein levels of Asf1b. Moreover, we were able to demonstrate in a direct 3'UTR luciferase assay that this decrease was a result of direct targeting of 3'UTR of ASF1B by miR-214. The function of Asf1b protein, a histone H3-H4 chaperone, is to deliver histones to the replication sites. A decrease of Asf1b has been described in various types of quiescent human cells.42 Asf1b is a specific marker for discriminating between cycling and non-cycling cells and is a marker of proliferation status in breast cancer. Tumor cells transfected with siRNA against Asf1b decreased the ability to form colonies. The genes that were found to be affected after silencing of Asf1b were mostly involved in proliferation, and as a consequence also in cell survival. Moreover, breast cancer patients with high expression of Asf1b had a poorer prognosis and higher level of metastasis.42 Interestingly, Asf1b is one of the proteins involved in the activity of bortezomib, although its precise function requires further validation.43 Taken together, these results indicate that miR-214 interferes in the replication process of myeloma cells.
In conclusion, we demonstrated that the low expression of miR-214 in MM contributes to the loss of cell growth control. The restoration of miR-214 expression in myeloma cells induced apoptosis and inhibited proliferation by targeting the p53/MDM2 interaction and DNA replication pathway.
Supplementary Material
Acknowledgments
The authors would like to thank Encarna Fermiñán and Xabier Agirre for technical assistance.
Funding
This work was partially supported by the Spanish FIS (PI080568 and PS0901897), the "Gerencia Regional de Salud, Junta de Castilla y León" (GRS202/A08 and GRS 702/A/11), and the Spanish Myeloma Network Program (RD06/0020/0006). MES is supported by the Ministerio de Sanidad y Consumo (CA08/00212).
Footnotes
This is an open-access paper. The online version of this article has a Supplementary Appendix
Authorship and Disclosures: Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105(35):12885-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci USA. 2010;107(17):7904-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009;114(25):e20-6 [DOI] [PubMed] [Google Scholar]
- 4.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010; 18(4):367-81 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, Ticona FV, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010; 24(3):629-37 [DOI] [PubMed] [Google Scholar]
- 7.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30(10):1990-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29(24): 3545-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11 (2):136-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derfoul A, Juan AH, Difilippantonio MJ, Palanisamy N, Ried T, Sartorelli V. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis. 2011;32(11):1607-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiang R, Wang F, Shi LY, Liu M, Chen S, Wan HY, et al. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int J Biochem Cell Biol. 2011;43(4):632-41 [DOI] [PubMed] [Google Scholar]
- 12.Fischer L, Hummel M, Korfel A, Lenze D, Joehrens K, Thiel E. Differential microRNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol. 2011;13(10):1090-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiso P, Carvajal-Vergara X, Ocio EM, Lopez-Perez R, Mateo G, Gutierrez N, et al. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer Res. 2006;66(11): 5781-9 [DOI] [PubMed] [Google Scholar]
- 14.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98(1):31-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44-57 [DOI] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15-20 [DOI] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim L P,, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell.2007;27(1): 91-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics.2008;24(3):325-32 [DOI] [PubMed] [Google Scholar]
- 20.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA.2008;14(6): 1012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res.2008;36(Database issue):D149-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8(1):75-87 [DOI] [PubMed] [Google Scholar]
- 23.Peng H, Nogueira ML, Vogel JL, Kristie TM. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci USA. 2010;107(6):2461-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuhmer T, Chatterjee M, Hildebrandt M, Herrmann P, Gollasch H, Gerecke C, et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood. 2005;106(10):3609-17 [DOI] [PubMed] [Google Scholar]
- 25.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303 (5659):844-8 [DOI] [PubMed] [Google Scholar]
- 26.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regu-lated genes. Nat Rev Mol Cell Biol. 2008;9(5):402-12 [DOI] [PubMed] [Google Scholar]
- 27.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318(5858): 1928-31 [DOI] [PubMed] [Google Scholar]
- 28.Costa A, Onesti S. Structural biology of MCM helicases. Crit Rev Biochem Mol Biol. 2009;44(5):326-42 [DOI] [PubMed] [Google Scholar]
- 29.Wong KY, Liang R, So CC, Jin DY, Costello J F,, Chim CS. Epigenetic silencing of MIR203 in multiple myeloma. Br J Haematol. 2011;154(5):569-78 [DOI] [PubMed] [Google Scholar]
- 30.Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31(4):745-50 [DOI] [PubMed] [Google Scholar]
- 31.Zhang YK, Wang H, Leng Y, Li ZL, Yang Y F,, Xiao FJ, et al. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun. 2011; 414(1):233-9 [DOI] [PubMed] [Google Scholar]
- 32.Lozano G, Zambetti GP. Gankyrin: an intriguing name for a novel regulator of p53 and RB. Cancer Cell. 2005;8(1):3-4 [DOI] [PubMed] [Google Scholar]
- 33.Umemura A, Itoh Y, Itoh K, Yamaguchi K, Nakajima T, Higashitsuji H, et al. Association of gankyrin protein expression with early clinical stages and insulin-like growth factor-binding protein 5 expression in human hepatocellular carcinoma. Hepatology. 2008;47(2):493-502 [DOI] [PubMed] [Google Scholar]
- 34.Drexler HG, Fombonne S, Matsuo Y, Hu ZB, Hamaguchi H, Uphoff CC. p53 alterations in human leukemia-lymphoma cell lines: in vitroartifact or prerequisite for cell immortalization? Leukemia. 2000;14(1):198-206 [DOI] [PubMed] [Google Scholar]
- 35.Sun W, Ding J, Wu K, Ning BF, Wen W, Sun HY, et al. Gankyrin-mediated dedifferentia-tion facilitates the tumorigenicity of rat hepatocytes and hepatoma cells. Hepatology. 2011;54(4):1259-72 [DOI] [PubMed] [Google Scholar]
- 36.Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of multiple myeloma. Best Pract Res Clin Haematol. 2007;20(4):571-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lode L, Eveillard M, Trichet V, Soussi T, Wuilleme S, Richebourg S, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010;95(11):1973-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia. 2007;21(1):143-50 [DOI] [PubMed] [Google Scholar]
- 39.Ooi MG, Hayden PJ, Kotoula V, McMillin DW, Charalambous E, Daskalaki E, et al. Interactions of the Hdm2/p53 and proteasome pathways may enhance the antitumor activity of bortezomib. Clin Cancer Res. 2009;15(23):7153-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha MN, Jiang H, Jayakar J, Reece D, Branch DR, Chang H. MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol Ther. 2010;9(11):936-44 [DOI] [PubMed] [Google Scholar]
- 41.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73(4):652-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaitre C, et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30(3):480-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, Blank JL, Peters T, Liu XJ, Rappoli DM, Pickard MD, et al. Genome-wide siRNA screen for modulators of cell death induced by proteasome inhibitor bortezomib. Cancer Res. 2010;70(11):4318-26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.