Summary
Lysine methylation of histone proteins regulates chromatin dynamics and plays important roles in diverse physiological and pathological processes. However, beyond histone proteins, the proteome-wide extent of lysine methylation remains largely unknown. We have engineered the naturally occurring MBT domain repeats of L3MBTL1 to serve as a universal affinity reagent for detecting, enriching, and identifying proteins carrying a mono- or di-methylated lysine. The domain is broadly specific for methylated lysine (“pan-specific”) and can be applied to any biological system. We have used our approach to demonstrate that SIRT1 is a substrate of the methyltransferase G9a both in vitro and in cells, to perform proteome-wide detection and enrichment of novel methylated proteins, and to identify candidate in-cell substrates of G9a and the related methyltransferse GLP. Together, our results demonstrate a powerful new approach for global and quantitative analysis of methylated lysine, and they represent the first systems biology understanding of lysine methylation.
Introduction
Post-translational modification of histone and non-histone proteins by the addition of one, two, or three methyl groups to the ε-nitrogen of lysine residues is proposed to play important roles in key signal transduction pathways (Bannister and Kouzarides, 2011; Greer and Shi, 2012; Huang and Berger, 2008; Margueron and Reinberg, 2010). Besides histone proteins, where lysine methylation has been extensively studied, only a relatively small number of other proteins are known to be modified by lysine methylation (Huang and Berger, 2008; Su and Tarakhovsky, 2006). However, as there are greater than 50 potential lysine methyltransferases (KMTs) and approximately 25 lysine demethylases (KDMTs) in the human genome (Greer and Shi, 2012; Kooistra and Helin, 2012; Petrossian and Clarke, 2011), it is highly likely that regulation of hundreds or thousands of proteins by lysine methylation remains to be discovered.
A common strategy for proteome-wide identification of post-translationally modified proteins has relied on the availability of modification-specific antibodies with no dependence on surrounding residues (“pan-specific”). Such an approach has been successful for modifications including phosphorylation, acetylation, and arginine methylation (Choudhary et al., 2009; Ong et al., 2004; Zhang et al., 2005). In contrast, this type of strategy has not been possible for lysine methylation due to the absence of truly pan-specific antibodies (Ong et al., 2004). In addition, the ability to investigate newly discovered lysine methylation events has been limited by the difficulty and cost of raising modification-specific antibodies.
To address this challenge we hypothesized that naturally occurring methyl-lysine binding domains may have the combination of broad sequence specificity and high methyl selectivity required for proteomic analysis. Here we describe an affinity reagent engineered from the three Malignant Brain Tumor domain repeats (3xMBT) of L3MBTL1 and show that it can serve as a tool for detecting, enriching, and identifying mono- and di-methylated lysine on individual proteins and on a proteomic scale. This reagent is highly specific for mono- and di-methylated lysine, and it binds to these residues with essentially no dependence on the surrounding protein sequence (Li et al., 2007; Min et al., 2007; Nady et al., 2012). The 3xMBT domain can be used as a global affinity reagent to detect methylated lysine on a wide range of protein and peptide targets. We have used this approach to show that the lysine deacetylase SIRT1 is methylated in vivo by G9a (also called EHMT2 and KMT1C), and that 3xMBT can be used for screening and identifying potential G9a methylated substrates on a large-scale protein array platform (Levy et al., 2011b). In proteome-wide pull-down 3xMBT specifically enriches over three hundred proteins and allows direct identification of methylated lysine on a subset of those proteins. Finally, we have developed a cell-based proteomic strategy for substrate discovery of lysine methylation regulatory enzymes. We have used this strategy to identify over twenty known and candidate substrates of the KMTs G9a and GLP (Rathert et al., 2008) in cells by examining changes in global lysine methylation following treatment with a specific inhibitor of these enzymes (Kubicek et al., 2007; Vedadi et al., 2011). This is the first proteome-wide analysis of methylated lysine. Our approach will provide a powerful new tool for studying the systems biology of lysine methylation.
Results
3xMBT recognizes methylated lysine with broad sequence specificity
We first aimed to identify a protein domain with broad specificity for methylated lysine. Based on available structural and biochemical data (Li et al., 2007; Min et al., 2007; Nady et al., 2012) we postulated that the 3xMBT domain of L3MBTL1 would be likely to act as a pan-specific reagent for mono- and di-methylated lysine. In its biological context L3MBTL1 plays a role in chromatin compaction mediated by interactions with histone 4 mono- and di- methylated at lysine 20 (H4K20me1/2) and histone H1B mono- and di-methylated at lysine 26 (H1BK26me1/2) (Trojer et al., 2007). It also binds to methylated non-histone proteins such as p53, pRb, and the DNA replication machinery (Gurvich et al., 2010; Saddic et al., 2010; Trojer et al., 2007; West et al., 2010). Previous work has shown that the domain interacts with methylated lysine through a hydrophobic binding pocket, and that hydrogen bonding between the carboxylate of aspartate 355 and the methylammonium proton of mono- or di-methyl lysine confers specificity over tri-methylation (Li et al., 2007; Min et al., 2007). Mutation of aspartate 355 to asparagine (3xMBTD355N) has been shown to abrogate binding to methyl-lysine without affecting the overall structure of the domain (Li et al., 2007; Min et al., 2007). Importantly, the isolated 3xMBT domain forms no significant contacts with the side-chains of amino acids surrounding the methylated residue, providing a structural basis for sequence-independent binding to methyl-lysine. The specificity of L3MBTL1 for physiologically-relevant methylated proteins such as H4 and Rb is thought to be conferred by the 3xMBT binding to methyl-lysine in combination with non-methyl sensitive interactions mediated by regions outside of the 3xMBT domain (Trojer et al., 2007).
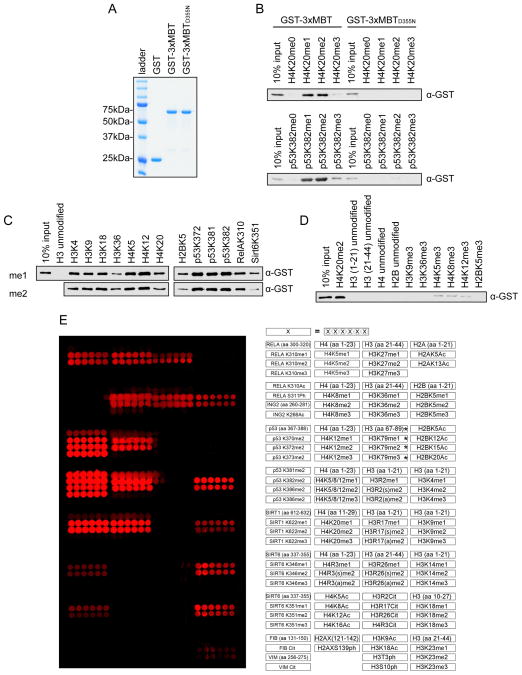
We first expressed the isolated 3xMBT domain of L3MBTL1 and the 3xMBTD355N mutant as GST fusions (Figure 1A) and characterized their binding to methylated peptides. As in previous studies, the wild-type domain, but not the point-mutant, co-precipitates with mono- and di-methylated peptides from p53 and H4, both known binding partners of L3MBTL1 (Figures 1B and S1A) (Min et al., 2007; Nady et al., 2012; West et al., 2010). This interaction is largely independent of GST (e.g. Flag can be used as the epitope tag as well) and can be detected through both N- and C-terminal tagging of the domain (Figure S1B). 3xMBT also co-precipitates with thirteen different mono- and di-methylated peptides drawn from variety of histone and non-histone proteins (Figures 1C and S1C). In contrast, 3xMBT does not bind a series of non-methylated peptides, and binds very weakly to some tri-methylated peptides (Figures 1D and S1D). We next probed 3xMBT on a peptide array displaying over 100 unique peptides with different post-translational modifications (PTMs) (Bua et al., 2009; Kuo et al., 2012). The domain bound specifically to the 41 peptides containing mono- and di-methylated lysine, and did not bind to the dozens of unmodified peptides, peptides containing tri-methylated lysine, or peptides with other PTMs (Figure 1E). In comparison, commercially available pan-methyl antibodies tested using the same array format failed to recognize several methylated peptides and cross-reacted with peptides bearing other post-translational modifications (Levy et al., 2011b). Together, these data demonstrate that highly preferential binding of 3xMBT to mono- and di-methylated lysine is a generalizable characteristic across diverse amino acid sequences.
Figure 1.
3xMBT recognizes methylated lysine with broad sequence specificity. (A) Coomassie stain of glutathione S-transferase (GST) alone, GST-tagged 3xMBT, and GST-tagged 3xMBTD355N. (B) 3xMBT, but not methyllysine-binding mutant 3xMBTD355N, binds to mono- and di-methylated H4K20- and p53K382-containing peptides. Western blot analysis of peptide pull-downs with GST-3xMBT and GST-3xMBTD355N and the indicated biotinylated peptides. H4 peptides: amino acids 1–23. p53 peptides: amino acids 367–388. (C) 3xMBT coprecipitates with a panel of biotinylated mono- and di-methylated peptides with varying sequences. Peptides are labeled by their methylated residues; all peptides are roughly 20 amino acids in length, with only the indicated residues methylated. (D) 3xMBT does not bind to a series of non-methylated peptides. Pull-down experiments with the indicated peptides as in (C). (E) 3xMBT binds preferentially to diverse mono- and di-methylated peptides. Microarrays spotted with over 100 distinct histone and non-histone peptides as indicated in the array key on the right were probed with GST-3xMBT. Red spots indicate positive binding. 3xMBT does not bind unmodified peptides, tri-methylated peptides, or peptides bearing other post-translational modifications. *H3K79 peptides have low solubility and therefore are not transferred efficiently to the array surface. See also Figure S1.
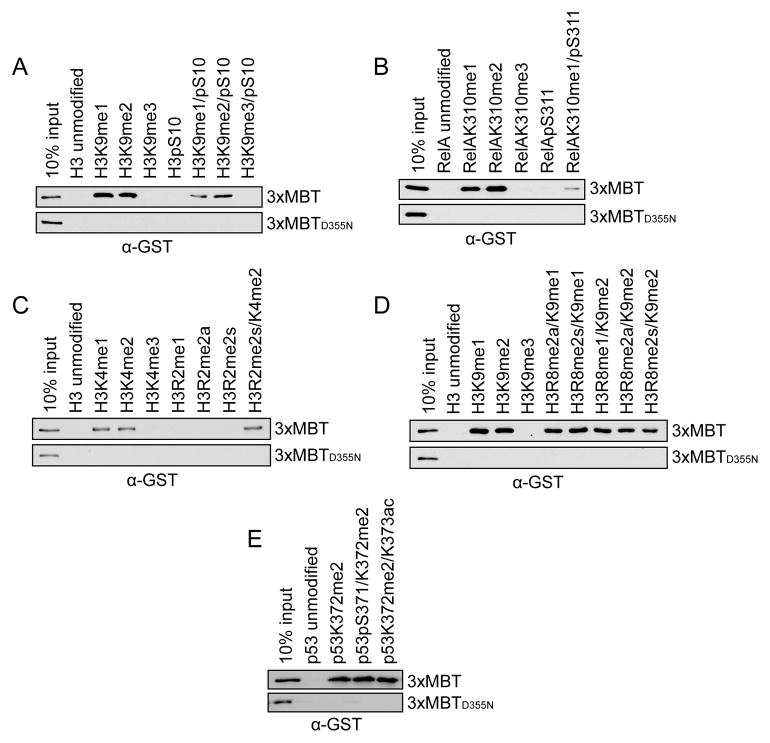
Secondary modification of adjacent residues are well known to affect the binding affinity of many methyl-lysine binding proteins and antibodies that recognize specific methylated lysines (Bock et al., 2011; Dhayalan et al., 2011; Fuchs et al., 2011; Levy et al., 2011a; Ramón-Maiques et al., 2007; Rothbart et al., 2012; Varier et al., 2010). To test whether nearby lysine acetylation, serine phosphorylation, and arginine methylation events influence binding of 3xMBT to mono- and di-methylated lysine, peptide pull-down assays were performed with 3xMBT and a series of singly- or doubly-modified peptides (Figures 2A–E; see Figures S2A–E for peptide loading controls). Consistent with previous reports (Li et al., 2007), phosphorylation on a C-terminally adjacent serine decreased, but did not eliminate, binding of 3xMBT to mono- or di-methylated lysine (Figures 2A and 2B). Arginine methylation one or two residues removed from the lysine methylation site had little effect on binding by 3xMBT (Figures 2C and 2D). Similarly, acetylation of a neighboring lysine and phosphorylation of an N-terminally adjacent serine did not affect 3xMBT binding (Figure 2E). We conclude that 3xMBT preferentially recognizes mono- or di-methylated lysine even in the presence of several different types of common secondary post-translational modifications.
Figure 2.
3xMBT recognizes mono- and di-methyl lysine in the presence of secondary posttranslational modifications. (A–E) Western blot analysis of peptide pull-down assays with GST-3xMBT and GST-3xMBTD355N and the indicated biotinylated peptides. All modified peptides have the same base sequence and length as the unmodified peptide control. (A) Phosphorylation of H3S10 modestly decreases binding of 3xMBT to mono- and di-methyl H3K9. (B) Phosphorylation of RelA at S311 decreases, but does not abolish, binding of 3xMBT to RelAK310me1. (C) Symmetric dimethylation of H3R2 does not block 3xMBT binding to H3K4me2-containing peptides. (D) Different states of methylation of H3R8 have little effect on binding of 3xMBT to H3K9me1- and H3K9me2-containing peptides. (E) 3xMBT binds p53K372me2 in the presence of adjacent serine phosphorylation or lysine acetylation. See also Figure S2.
3xMBT recognizes methylated proteins by Far Western analysis and in pull-down assays
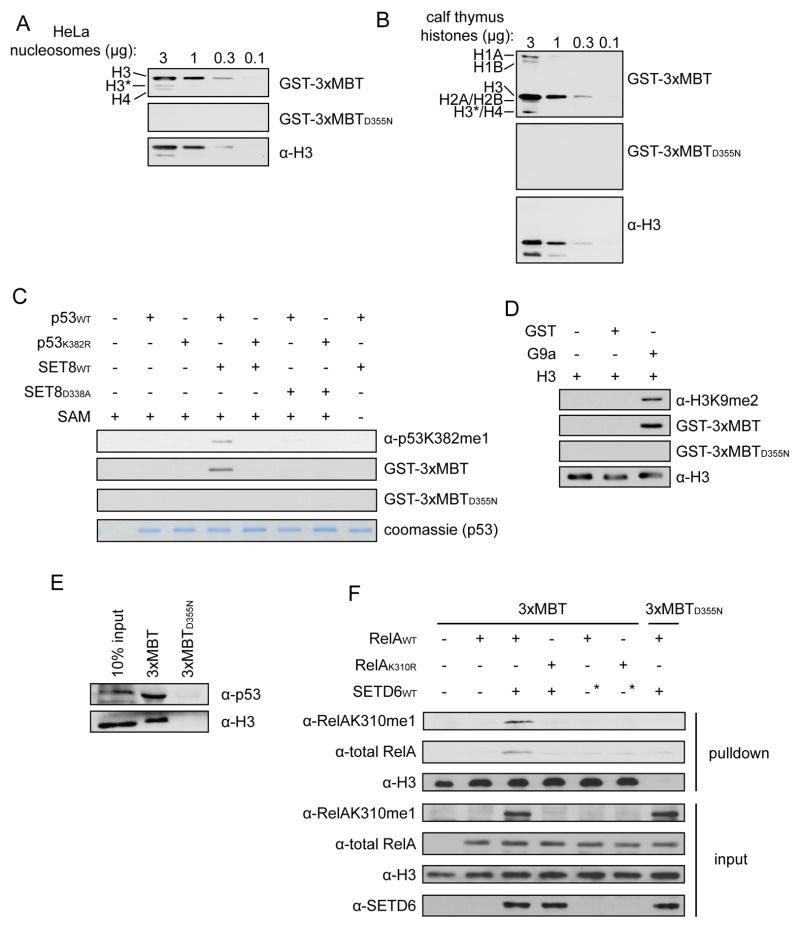
Two common uses of modification-specific antibodies are Western blotting and immunoprecipitation (IP). We therefore asked whether the 3xMBT domain could be adapted in lieu of an antibody for these types of techniques. In Far Western assays 3xMBT, but not 3xMBTD355N, detected histones from purified HeLa nucleosomes (Figure 3A) and calf thymus histones (Figures 3B, S3A and S3B), suggesting that the intact methyl-lysine binding pocket specifically recognizes endogenous methylated histones.
Figure 3.
3xMBT detects multiple methylated proteins in common affinity-based assays. (A and B) 3xMBT, but not 3xMBTD355N, recognizes endogenous histones in Far Western assays. Far Western analysis of (A) purified HeLa nucleosomes and (B) purified calf thymus histones serially diluted to the indicated quantities and probed with GST-3xMBT and GST-3xMBTD355N. Binding of GST fusion proteins was detected with α-GST antibody. *H3 indicates an H3 degradation product that migrates near H4. H3 blots are shown as a loading control. (C) 3xMBT, but not 3xMBTD355N, specifically detects monomethylated p53 by Far Western. In vitro methylation reactions with wild-type SET8 or catalytically inactive SET8D338A on recombinant wild-type p53 or p53 carrying K382R substitution (p53K382R) were probed with α–p53K382me1 antibody as previously described (Shi et al., 2007) or the indicated GST fusion as in (A). Co-factor SAM was included in the reactions as indicated. Coomassie stain of p53 is shown below as a loading control. (D) 3xMBT, but not 3xMBTD355N, specifically detects recombinant H3 dimethylated at lysine 9 (H3K9me2). In vitro methylation reactions of recombinant H3 with recombinant G9a SET domain, detected as described in (C). Total H3 is shown as a loading control. (E) 3xMBT, but not 3xMBTD355N, precipitates known methylated proteins H3 and p53 from HeLa nuclear extract (NE). Western blot of GST-3xMBT and GST-3xMBTD355N NE pull-downs probed with the indicated antibodies. (F) 3xMBT, but not 3xMBTD355N, precipitates methylated RelA from cells. Pull-down assays of NE from 293T cells co-expressing SETD6 (or two different negative control constructs, the second of which is indicated by the asterisks) and wild-type RelA or RelA carrying K310R substitution (RelAK310R) and probed with the indicated antibodies. α-RelAK310me1 antibody was previously described (Levy et al., 2011a). See also Figure S3.
To test if the Far Western signal is indeed specific for methylated lysine, we probed for in vitro methylation of recombinant proteins. 3xMBT, but not 3xMBTD355N, detected recombinant p53 when mono-methylated at lysine 382 by SET8 (also named PR-Set7 and SETD8), similar to a previously described anti-p53K382me1 antibody (Figures 3C and S3C)(Shi et al., 2007). This binding is lost when the methylation event is blocked by mutating p53 lysine 382 to arginine, by using a catalytically-inactive SET8 mutant, or by withholding the methyl-donating cofactor S-adenosyl methionine (SAM) (Figures 3C and S3C). Similarly, 3xMBT recognizes recombinant H3 in a Far Western when H3 is dimethylated by G9a at lysine 9 (Figure 3D) (Shinkai and Tachibana, 2011). Based on these data we conclude that the 3xMBT can be used in Far Western assays to detect various mono- and di-methylation events.
We next asked whether 3xMBT could specifically capture methylated proteins from cellular extract. Immobilized 3xMBT precipitates both H3 (an abundant methylated protein) and p53 (a low abundance methylated protein) from nuclear extracts (Figure 3E). To test if the pull-down by 3xMBT is methylation-dependent, pull-downs were conducted using nuclear extract from 293T cells co-transfected with the KMT SETD6 and its substrate RelA; SETD6 mono-methylates RelA at lysine 310 (RelAK310me1) (Levy et al., 2011a). RelA was precipitated by 3xMBT in a manner dependent upon intact K310 and expression of wild-type SETD6 (Figure 3F). Similar results were observed in 3xMBT pull-downs of methylated p53 from cells overexpressing p53 and SET8 (Figure S3D). Taken together, we conclude that 3xMBT can be used to affinity purify methylated proteins from cellular extracts.
Utility of 3xMBT to identify previously uncharacterized methylation events
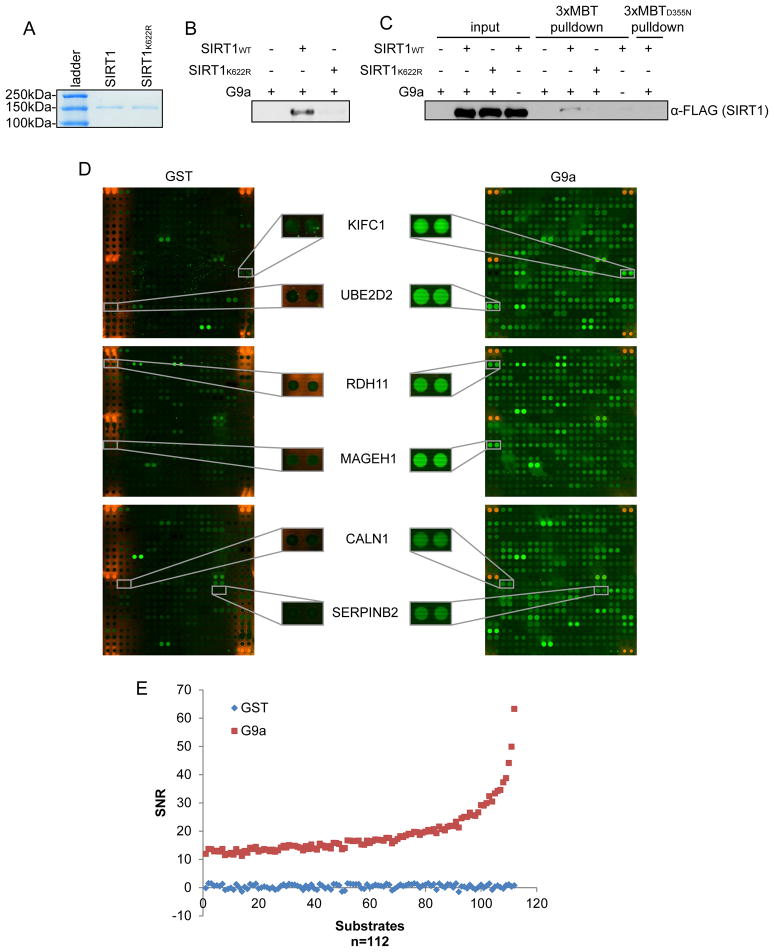
As the generation of site- and state-specific antibodies for the analysis of candidate novel lysine methylation events is expensive and often challenging (Egelhofer et al., 2011; Fuchs et al., 2011), we reasoned that the 3xMBT domain could serve as a readily available and inexpensive reagent to initiate investigation of newly identified methylated proteins. Through in vitro screening of a panel of enzymes as described in (Levy et al., 2011a), we found that the KMT G9a methylates the NAD-dependent lysine deactylase SIRT1 at lysine 622, a previously uncharacterized modification event (Figures 4A and 4B; data not shown). We next asked whether G9a catalyzes this reaction in vivo. We prepared extracts from 293T cells expressing G9a and 3xFlag-SIRT1 or 3xFlag-SIRT1K622R for pull-down assays using 3xMBT and 3xMBTD355N affinity resins. We found that 3xFlag-SIRT1 is specifically precipitated by 3xMBT, but not 3xMBTD355N, only when G9a is co-expressed and the SIRT1 target lysine K622 is intact (Figure 4C). These data indicate that SIRT1 is methylated by G9a in cells and demonstrate how 3xMBT can be used to uncover and characterize new biological methylation events without requiring the time- and cost-consuming generation of a methyl-specific antibody.
Figure 4.
3xMBT detects a novel methylation event on the lysine deacetylase SIRT1 and on candidate G9a substrates. (A) Coomassie stain of full-length recombinant GST-tagged SIRT1 and SIRT1K622R purified from Sf9 cells. (B) G9a methylates SIRT1 as lysine 622. Autoradiograph of in vitro methylation assays on SIRT1 and SIRT1K622R using recombinant G9a SET domain. (C) 3xMBT, but not 3xMBTD355N, precipitates methylated SIRT1 from cells. GST-3xMBT or GST-3xMBTD355N pull-down assays as in (Figure 2E) of NE from 293T cells expressing 3xFlag-SIRT1 or 3xFlag-SIRT1K622R and G9a or control vector. Pull-downs were probed with anti-Flag antibody to detect SIRT1. (D) 3xMBT detects G9a-methylated substrates on protein microarrays. Representative blocks of human Invitrogen ProtoArrays® methylated in vitro using GST alone (left) or GST-tagged recombinant G9a SET domain (right). Methylation was detected by probing the arrays with 3xFlag-3xMBT, followed by α-Flag antibody and species-matched fluorescent secondary antibody. Magnified regions show examples of G9a-methylated proteins detected by Flag-3xMBT. A list of all candidate G9a-methylated proteins is shown in Table S1. (E) Scatter plot comparing GST array signal-to-noise ratio (SNR) and G9a SNR for ranked ProtoArray® hits shown in Table S1.
We next asked whether 3xMBT could be used to identify other potential G9a substrates in the context of an on-chip protein array methylation system that we previously described (Levy et al., 2011b). On-chip in vitro methylation assays using recombinant GST-G9a SET domain, or as a negative control, GST alone, were performed on Invitrogen ProtoArrays® bearing ~9,500 unique recombinant human proteins (Figure 4D). Positively methylated proteins were detected by probing the arrays with Flag-tagged 3xMBT, followed by α-Flag antibody and fluorescent secondary antibody. Using a very strict threshold (see Supplemental Experimental Procedures) followed by manual inspection, we identified 112 proteins that were detected by Flag-3xMBT on the G9a-methyated array but not on the control array, indicating that these hits are potential G9a substrates (Figures 4D and 4E; Table S1). Previously, G9a targets could not be identified in the protein array format using antibodies because of restricted affinity of antibodies recognizing di-methylated lysine, the state catalyzed by G9a. Thus, our results demonstrate that 3xMBT can be used in combination with a protein array platform to elucidate candidate substrates of mono- and di-methyltransferase KMTs.
We note that several previously reported G9a targets were not detected in the ProtoArray® experiment, likely due to a combination of limitations associated with protein array approaches, including (1) absence of the target, (2) presentation of substrates in vitro on arrays versus in cells, and (3) other inherent issues with protein arrays such as candidate substrates being truncated proteins or improperly folded (Levy et al., 2011b). Thus, in vitro protein array approaches are likely to be most effective in helping focus on substrates when combined with other strategies, such as chemical biological approaches (Islam et al., 2012; Islam et al., 2011), and cell-based assays for KMT substrate identification such as the proteomic strategy described below.
3xMBT enriches the methyl-lysine proteome
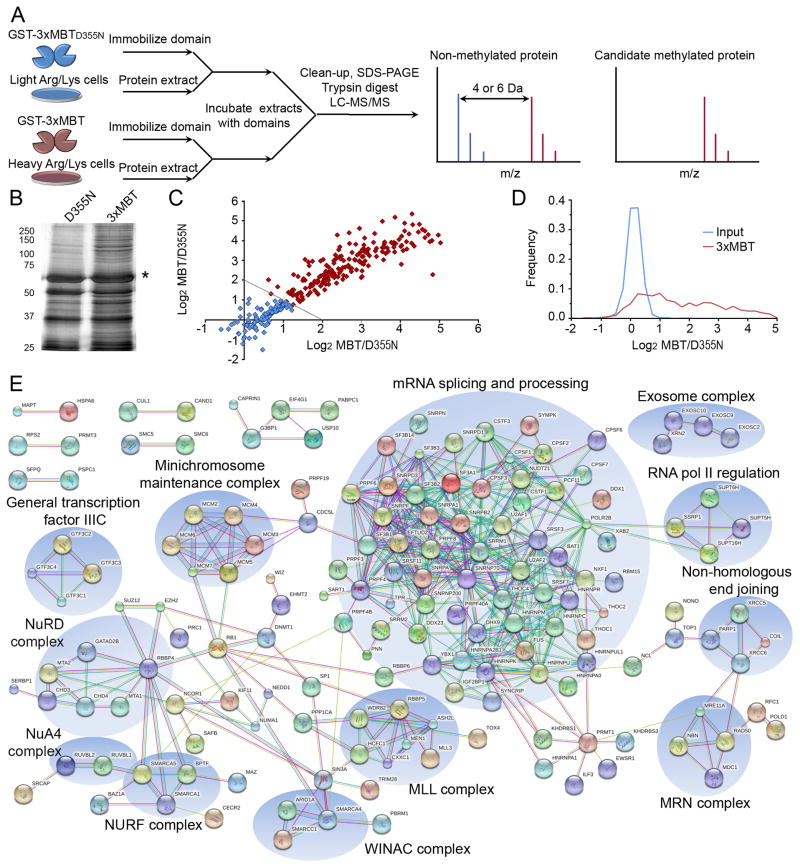
We postulated that we could apply quantitative mass spectrometry in combination with the differential methyl-lysine recognition properties of 3xMBT versus the 3xMBTD355N mutant to enrich an entire lysine methylation proteome (see schematic, Figure 5A). To this end, we first performed protein pull-downs from nucleoplasmic extract of 293T cells using the 3xMBT domain or the 3xMBTD355N negative control. In multiple independent experiments, 3xMBT consistently captured more proteins than 3xMBTD355N (Figure 5B), indicating that the intact methyl-lysine binding pocket is responsible for a large fraction of bound proteins.
Figure 5.
Protein pull-down using 3xMBT reproducibly enriches for the methyl-lysine proteome. (A) Protocol schematic for proteome-wide capture and identification of candidate methylated proteins using 3xMBT. Peptides derived from specifically enriched proteins show a distinctive signal in the SILAC label associated with 3xMBT, while peptides from non-specifically bound proteins (equal binding to 3xMBT and 3xMBTD355N resins) have the same intensity in both SILAC conditions. (B) Abrogating the methyl-lysine binding site of 3xMBT greatly reduces the amount of protein captured from nuclear extract. Silver stain of total proteins present in GST-3xMBT and GST-3xMBTD355N pull-downs from 293T nuclear extracts. Asterisk indicates GST fusion band. (C) Specific proteins are reproducibly and quantitatively enriched by 3xMBT relative to 3xMBTD355N. Axes represent the quantitative ratio (log base 2) for 3xMBT over 3xMBTD355N in independent experiments. Each point represents a protein identified in both experiments, and enrichment in these experiments is correlated with R2=0.86. (D) Most proteins identified following enrichment by 3xMBT (approximately 60%) are specifically enriched by the methyl-lysine binding domain. Probability density plots show 3xMBT enrichment for all 544 identified proteins identified in pull-down experiments compared to the SILAC ratios of proteins identified from input material. (E) Proteins enriched by 3xMBT tend to be functionally related based on protein interaction networks. The diagram shows all high-confidence protein-protein interactions from the STRING interaction database between proteins enriched to at least 2-fold by 3xMBT relative to 3xMBTD355N. See also Tables S2 and S3.
We next used stable isotopic labeling by amino acids in cell culture (SILAC) (Ong et al., 2002) and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to quantitatively measure proteins enriched by binding to 3xMBT relative to the 3xMBTD355N negative control (see schematic, Figure 5A). This experiment consisted of protein pull-downs in which immobilized 3xMBT or 3xMBTD355N was incubated with proteins containing either L-lysine/L-arginine or D4-L-lysine/13C6-L-arginine (“light” and “heavy” respectively), and proteins bound in each pull-down were combined for analysis by LC-MS/MS. We conducted “forward” and “reverse” labeling experiments where 3xMBT was matched with heavy or light lysate respectively, and only proteins observed in both labeling directions were included in the dataset for downstream analysis. The entire label swap experiment was conducted twice to determine inter-experiment reproducibility (total of four mass spectrometry analyses). A total of 544 proteins were identified in at least one pair of label-swap experiments. The SILAC ratio for 3xMBT pull-down relative to 3xMBTD355N was highly correlated between the two experiments (R2=0.86, Figure 5C), showing that enrichment is quantitative and reproducible.
We found that a large fraction of proteins were quantitatively enriched by 3xMBT relative to 3xMBTD355N. For example, 313 of the 544 proteins (57.5%) were enriched at 2-fold by 3xMBT versus the mutant while only 3 of 544 proteins (0.6%) exceeded that threshold in the other direction (Figure 5D; Table S2). For comparison, 0.4% of proteins identified in the input material fell outside a SILAC ratio of 2. These data strongly argue that the proteins identified in the 3xMBT pull-down are enriched in a methyl-dependent manner.
For functional analysis of enriched proteins we used the set of 313 proteins enriched 2-fold by 3xMBT. Gene Ontology (GO) and the String protein interaction database were used to identify biological functions and protein complexes over-represented among enriched proteins relative to their frequency among proteins identified in the input material (see Methods). Over-represented GO terms include mRNA processing (enriched at 2.9-fold, p = 7e-20), transcription (2.1 fold, p=1.5e-4), and RNA and DNA helicase activity (enriched at 4.3-fold and 4.0-fold, p = 0.014 and 0.011) (all p-values Bonferroni-corrected, Table S3) (Huang et al., 2009a, b). Analysis of protein interactions using the String Database shows that proteins bound by the 3xMBT domain group into related clusters (Figure 5E) (Szklarczyk et al., 2011), the largest of which is composed of proteins involved in RNA processing. Smaller clusters include the DNA replication-associated minichromosome maintenance helicase complex, proteins involved in the DNA damage response, and a number of chromatin modifying complexes.
Quantitative analysis of the dynamic methyl-lysine proteome
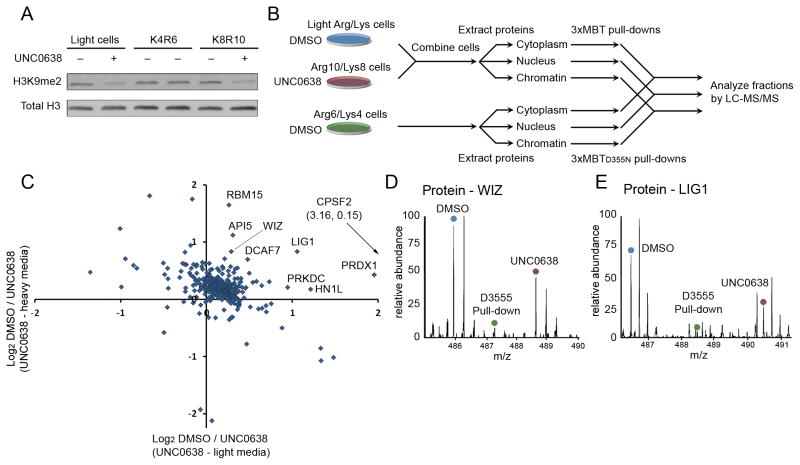
A powerful application of proteome-wide analysis is identification of dynamic lysine methylation during biological processes or in response to inhibition of a KMT or lysine demethylase. To determine whether this approach can be used to identify candidate methyltransferase substrates in their cellular context we treated 293T cells with UNC0638, a potent and selective inhibitor of the KMTs G9a and GLP (Vedadi et al., 2011). We confirmed that in cells treated with the drug, levels of the G9a target H3K9me2 were reduced (Figure 6A). Triple SILAC labeling coupled with quantitative mass spectrometry was then used to identify changes induced upon drug treatment among the proteins specifically captured by 3xMBT from cytoplasmic, nuclear and chromatin fractions (see schematic, Figure 6B).
Figure 6.
Candidate physiologic substrates of G9a and GLP are identified by targeted inhibition and quantitative proteomic analysis. (A) Treatment with UNC0638 decreases global H3K9me2 levels. Western blot of 293T whole cell extracts ± UNC0638 treatment and probed with the indicated antibodies. (B) A schematic diagram of the proteomic experiment. Cells grown in “light” or “heavy” media were treated with DMSO or the G9a/GLP inhibitor UNC0638 and combined prior to extracting proteins from cytoplasmic, nuclear and chromatin fractions. Cell prepared in “medium” media were treated with DMSO, processed and fractionated in parallel. Light/heavy extracts were subjected to pull-down by 3xMBT, while “medium” extracts were subjected to pull-down by 3xMBTD355N. Bound proteins from each fraction were combined and analyzed by LC-MS/MS. (C) Identification of dozens of candidate in-cell substrates of G9a/GLP. Many proteins show decreased association with 3xMBT following 24 hour treatment of 293T cells with UNC0638 relative to DMSO treatment. Only proteins that were also enriched by 3xMBT relative to 3xMBTD355N are shown. Axes show log2 ratios for levels from vehicle control over treated cells, and each axis represents an independent biological replicate. (D) MS1 spectrum showing that the amount of WIZ, a known G9a substrate, captured by 3xMBT from cell lysate is reduced following treatment with UNC0638. The source of the relevant peak is indicated. (E) MS1 spectrum showing reduction in LIG1 captured by 3xMBT following treatment with UNC0638 as in (D). See also Figure S4 and Table S4.
First, the analysis identified an additional 370 proteins enriched by 3xMBT compared to 3xMBTD355N in two biological replicates (Table S4; Figure S4A). These proteins are strongly enriched for functions in mRNA splicing, translation and cell cycle (Figure S4B), and they span the cytoplasmic and nuclear compartments (Figure S4C). Among enriched proteins were 23 candidate G9a/GLP substrates showing reduced association with 3xMBT in response to UNC0638 treatment (Table S4). A known G9a substrate, the protein WIZ, has one of the highest scores in this analysis (Rathert et al., 2008)(Figure 6C–D; Table S4). Among novel candidate substrate proteins, DNA ligase 1 (LIG1) scores especially strongly and contains the amino acid sequence ARKT, an optimal target site for G9a (Rathert et al., 2008)(Figure 6E; Table S4). Of the 23 candidate substrates, the majority are not present or intact on ProtoArrays, though two hits, MTA1 and RBM15 showed G9a-dependent signal on the protein arrays (Figure S4E).
Multiple methylation sites present on H3 besides K9 (e.g. H3K4) are not perturbed in response to UNC0638 treatment (Vedadi et al., 2011). Thus, the amount of H3 captured by 3xMBT does not decrease appreciably in our analysis and the specific peptide containing K9 after trypsin digestion is not amenable to LC-MS/MS analysis. In addition, we did not reliably detect peptides from other potential G9a substrates, such as SIRT1 (see above) or G9a itself (Rathert et al., 2008) due to limitations in the sensitivity and dynamic range of the mass spectrometer – a limitation of available technology and not inherent to 3xMBT as a proteomic tool. However, direct investigation of low abundance peptides identified that 3xMBT capture of a peptide from ACIN1, a known G9a substrate (Rathert et al., 2008), was decreased strongly following inhibitor treatment (Figure S4D). Taken together, the combination of the 3xMBT strategy, SILAC-based quantitative mass spectrometry, and chemical inhibition of KMTs, has allowed us to identify many known and candidate physiologic substrates of G9a and GLP, and argues that such an approach can generally be used to study lysine methylation dynamics at a proteomic level (see Discussion).
Direct identification of methylated proteins
Proteins identified in our 3xMBT pull-down experiments are being enriched in a methyl-dependent fashion, indicating that they are either directly methylated or in a complex containing a methylated protein(s). To determine specific methylation sites, we analyzed the LC-MS/MS data for peptides directly showing methylation of enriched proteins. 102 modified peptides were detected within the 3xMBT pull-down experiments using MaxQuant software. However, automated identification of methylated lysine by MS/MS is particularly difficult because the mass shift for methylation is identical to the difference between certain pairs of amino acids (e.g. glycine and alanine). Therefore to rule out any potential MaxQuant-indicated false positives we manually verified the MS/MS spectra from the 102 potentially modified peptides (see Supplemental Experimental Procedures for false-discovery rate calculations) (Nichols and White, 2009). We also conducted an independent 3xMBT pull-down from cells metabolically labeled with D313C-L-methionine. The isotopic label adds 4 Dalton to post-translational methylation, giving it a distinctive mass shift. We identified a total of 26 lysine residues that are unambiguously methylated using strict criteria set to rule out any potential false positives: 18 as mono-methyl, 3 as di-methyl and 5 as both mono and di-methyl (Table 1 and Table S5; Figure S5). In every case the highest scoring match for each methylated peptide was observed with the isotopic label corresponding to capture by 3xMBT as opposed to 3xMBTD355N. Notably, twenty-one of the twenty-six methylated residues have not been previously reported. Many additional real methylation events on proteins identified following pull-down with 3xMBT are almost certainly present in our data and could be identified by additional approaches (see Discussion). Taken together, our results highlight the power of using native methyl-lysine-binding domains as affinity reagents in combination with quantitative mass spectrometry to reveal novel methylation events.
Table 1.
Methylated residues directly identified in 3xMBT pull-down experiments. 29 methylation events at 24 lysine residues are identified with high confidence from 3xMBT pull-down using by automated analysis followed by manual validation. Reported methylation sites appear in the NCBI protein database (www.pubmed.org) or PhosphoSitePlus (www.phosphosite.org) as of November 12, 2012. Multiple proteins are listed if the same peptide occurs in more than one context. H3 and NRK do not have ratios because they were only identified in the methionine-labeling experiment, MS1 signal for ERC1/2 was too low to be reliably quantified.
| Protein | Residue (methylation state) | Protein Ratio 3xMBT/D355N | Reported |
|---|---|---|---|
| CMAS | 399 (mono) | 4.2 | No |
| DDX1 | 234 (mono) | 11.2 | No |
| eEF1A1/eEF1A2 | 55 (mono, di) | 5.2/1.5 | Yes |
| eEF1A1 | 165 (mono, di) | 5.2 | Yes |
| 384 (di) | Yes | ||
| 408 (mono) | No | ||
| eEF-2 | 594 (mono) | 4.4 | No |
| ERC1/2 | 319/315 (di) | - | No |
| FAM50A | 5 (mono) | 3.7* | No |
| GTF2I | 219 (mono) | 1.5 | No |
| H3 | 79 (mono, di) | - | Yes |
| hnRNP D/hn RNP A/B | 129 (mono) | 22.5/13.4 | No |
| hnRNP K | 139 (mono) | 8.0 | No |
| LIG1 | 795 (mono) | 10.7 | No |
| MCM4 | 216 (mono) | 6.6 | No |
| MCM7 | 449 (mono, di) | 6.7 | No |
| MSH6 | 1126 (mono) | 1.2 | No |
| NHP2L1 | 33 (mono) | 4.4* | No |
| NRK | 818 (di) | - | No |
| RBM25 | 97 (mono) | 7.7 | No |
| TD-60 | 293 (mono) | 1.7 | No |
| SAFB1/SAFB2 | 226/225 (mono) | 9.9 | No |
| SCML2 | 123 (mono) | 1.7 | No |
| SSRP1 | 143 (mono) | 14.6 | No |
| TUBB2C/TUBB4† | 19 (mono) | 2.1/1.9 | No |
| WIZ | 967 (mono, di) | 5.3 | Yes |
Discussion
Lysine methylation is a modification that has been extensively characterized on histone proteins, but about which relatively little is known in the context of non-histone proteins (Su and Tarakhovsky). Notably, there are more than 50 candidate KMTs and 25 KDMTs in the human genome, and the majority of these enzymes are implicated in the etiology of diverse human diseases (Greer and Shi, 2012; Kooistra and Helin, 2012; Petrossian and Clarke, 2011; Varier and Timmers, 2011). However, the physiologic substrate specificities for many of these KMTs have yet to be discovered, highlighting the need for proteomic strategies to identify novel methylated proteins (Luo, 2012). Prior to our work, a robust method to characterize lysine methylation on a proteome-wide scale had not been reported. Traditional proteomic approaches have been unable to address these questions because of the difficulty in developing pan-specific antibodies for methylated lysine that have the high selectivity and broad specificity required for proteome-wide analysis. Bioinformatics analysis of high-throughput proteomics dataset has identified methylation occurring on a number of abundant proteins in yeast (Pang et al., 2010), further supporting the notion that methylation is a common regulatory modification and complementing the strategy described here to experimentally enrich methylated lysine.
In the present study we have shown that the naturally occurring 3xMBT domain from L3MBTL1 can be engineered as a general affinity reagent recognizing mono- and di-methylated lysine. 3xMBT is able to detect several known methylation events in vitro and in cells, and its methyl selectivity and lack of sequence specificity, along with the point mutant negative control, make it a powerful tool for proteome-wide enrichment of protein complexes containing methylated lysine. Global proteomic analysis using 3xMBT shows that this PTM occurs widely across the proteome and is likely involved in a much broader range of biological processes than previously realized. Further, we applied the characterization of cellular lysine methylomes to demonstrate how this strategy can be combined with a chemical biological approach to identify new candidate substrates of the KMTs G9a and GLP. Although some known G9a substrates were identified, a change in overall mono- and di-methylation of histone H3 was not observed. This highlights the constraint that total methylation may not change appreciably when a protein is methylated at high levels on multiple lysine residues or by several KMTs. Regardless, the proof-of-concept shows that this strategy can be applied to elucidate physiologic substrates of the numerous known and orphan lysine methyltransferases and demethylases present throughout eukaryotic genomes.
3xMBT complements existing tools and strategies for studying lysine methylation, such as radioactive labeling of the methyl moiety and use of methylation-specific antibodies. The 3xMBT domain can be reproducibly expressed in E.coli, and production of the domain from a recombinant plasmid allows epitope tags and detection strategies to be quickly exchanged, increasing the functionality of the domain when a particular tag is incompatible with the rest of the experimental design.
Though the domain does not appear to prefer specific amino acids at flanking positions, it does appear to require the presence of flanking residues for efficient binding; it prefers methylated lysine centered within a peptide rather than at the terminus (K. Moore, S. Carlson, and O. Gozani, unpublished observations). This prevents our approach from being applied to trypsin-digested peptides, rather than proteins, in the manner often used for proteomic analysis of modifications such as acetylated lysine (Choudhary et al., 2009) and phosphorylated tyrosine (Zhang et al., 2005). We postulate that alternative strategies for protein digestion may generate longer peptides that are more suitable for direct capture by 3xMBT, allowing direct identification of more methylated residues. We envision that the use of native broad-specificity methyl-lysine binding domains can be expanded beyond the 3xMBT of L3MBTL1 to provide new tools to distinguish between mono- and di-methylated lysine, and to study tri-methylated lysine. For example, the 3xMBT domain can be engineered to be selective for binding to only mono-methylated lysine or only di-methylated lysine (Nady et al., 2012). Specifically, Nady et al. found that mutating threonine 411 to glutamine results in a preference for monomethyl lysine, while mutating leucine 361 to phenylalanine results in a preference for dimethyl lysine. In addition, multiple domains could be combined to generate “poly” domain reagents with defined or expanded specificity; this approach might be most helpful for detection of tri-methyated protein as we have yet to identify a highly sequence promiscuous trimethyl-lysine binding domain (K. Moore, S. Carlson, and O. Gozani, unpublished observations).
The 3xMBT domain represents a new tool for rapid analysis of methylated lysine. It can be applied to any organism or biological system, and will be applicable to measurement of lysine methylation dynamics during biological processes or resulting from perturbations such as inhibition, knock-down or over-expression of a KMT. Global proteomic analysis will be a powerful tool to investigate the biological function of lysine methyltransferases and demethylases, especially in the many cases where physiological targets of these enzymes have not yet been determined. Quantitative studies will help to integrate lysine methylation into broader signaling networks and contribute to a fuller understanding of how protein lysine methylation regulates diverse cellular processes.
Experimental Procedures
Materials
The three MBT repeats of L3MBTL1 (amino acids 190–530 of accession NP_056293.4) were expressed as a GST fusion from the pGEX6P1 vector (West et al., 2010). Recombinant proteins were expressed and purified as previously described (West et al., 2010).
MBT Far Western and protein pull-down
For Far Western assays, proteins were separated by SDS-PAGE and transferred to a PVDF membrane. Following blocking, membranes were incubated overnight with the indicated domain. Domain binding was determined using an epitope tag-specific antibody and appropriate HRP-conjugated secondary antibody.
GSH-Sepharose coupled domain beads were generated for pull-down by incubating GSH-Sepharose resin with E. coli lysate containing sufficient 3xMBT for bead saturation. Bead-coupled 3xMBT was incubated with nuclear extract overnight and proteins that precipitated with the resin were analyzed by SDS-PAGE.
SIRT1 screen and characterization of methylation
Recombinant GST-SIRT1 was expressed by baculoviral transduction of the Sf9 insect cell line as previously described (Levy et al., 2011a). A panel of enzymes was tested for ability to methylate recombinant SIRT1 using 3H-SAM in in vitro methylation reactions (Kuo et al., 2011). SIRT1K622R was generated by site-directed mutagenesis (Stratagene). For the pull-down experiment, 293T cells were transfected with a 10:1 enzyme:substrate DNA ratio. 150 μg of nuclear proteins were used for pull-downs with 3xMBT or 3xMBTD355N coupled GSH-Sepharose as described above.
Protein array
Assay was conducted as previously described (Levy et al., 2011b). Briefly, a ProtoArray® (Invitrogen, version 5.0) was incubated overnight with 50 μg of GST or recombinant GST-tagged G9a SET domain and the cofactor SAM. Methylation was visualized by probing with 3xFlag-3xMBT, followed by α-Flag M2 antibody and an α-mouse fluorescent secondary antibody.
Protein pull-down and mass spectrometry
Protein pull-downs were performed as described above using 600 μg of nuclear extract prepared with isotopic labels as indicated. After eluting with glutathione, 3xMBT domain was removed by dialysis into high salt and rebinding to GSH-sepharose. Bound proteins were separated by SDS-PAGE and analyzed by in-gel tryptic digest followed by LC-MS/MS. Peptides were identified using MaxQuant version 1.2.2.5 (Cox and Mann, 2008) and candidate methylated peptides were verified by manual inspection.
Supplementary Material
Highlights.
3xMBT domain isa “pan -specific” tool for detecting mono- and di-methylated lysine.
3xMBT allows for rapid analysis of novel methylated proteins in vitro and in vivo.
Proteomic analysis shows that lysine methylation occurs widely across the proteome.
A cell-based proteomic assay allows global analysis of dynamic lysine methylation.
Acknowledgments
We thank A. Kuo for assistance with the peptide array and A. Wilkinson for assistance with the protein array. Peptides in Figure 2 were a gift from EpiCypher. Some mass spectrometry was conducted by C. Adams and A. Chien at The Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University supported by NCRR S10RR027425. S.M.C was supported in part by an NRSA Cancer Biology Training Grant PHS NRSA 5T32 CA09302-35 and a postdoctoral fellowship 123711-PF-13-093-01-TBE from the American Cancer Society Illinois Division. This work was supported in part by grants from the NIH to O.G. (R01 GM079641), K.F.C. (R01 AG028867) and R.G.J. (4R00HL103768). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No DGE-1147470 to K.E.M. K.E.M. is also supported by a Hubert Shaw and Sandra Lui Stanford Graduate Fellowship. O.G. is a recipient of an Ellison Senior Scholar in Aging Award. O.G. is a co-founder of EpiCypher, Inc.
Footnotes
Author Contributions
K.E.M. characterized the 3xMBT domain and performed in vivo SIRT1 experiments and the G9a protein array. S.M.C. developed and performed proteomic experiments and analyzed data. P.C. screened for methylation of SIRT1. Mass spectrometry was conducted in the lab of A.W-Y. S.M.C., N.C. and A. W-Y. optimized and conducted 2D HPLC and tandem mass spectrometry. R.G.J. conducted mass spectrometry for the G9a chemical biology experiment. K.F.C. supervised work. O.G. supervised work and designed experiments. K.E.M., S.M.C. and O.G. wrote the paper.
Data Availability
Mass spectrometry raw data are available on ProteomeCommons (www.proteomecommons.org), Tranche hash Du4W2Xy89mv0/q/IfXukqgavWlMNPHfIz9+KFMVaedngkV0ekgiNBqII2LMFPKZ3ffZ7qgK+ILOYUuc8I5uzysuFH00AAAAAAAABuA==.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock I, Kudithipudi S, Tamas R, Kungulovski G, Dhayalan A, Jeltsch A. Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC Biochem. 2011;12:48. doi: 10.1186/1471-2091-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen M, Rehman M, Walther T, Olsen J, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvich N, Perna F, Farina A, Voza F, Menendez S, Hurwitz J, Nimer SD. L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proc Natl Acad Sci U S A. 2010;107:22552–22557. doi: 10.1073/pnas.1017092108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Islam K, Bothwell I, Chen Y, Sengelaub CA, Wang R, Deng H, Luo M. Bioorthogonal Profiling of Protein Methylation (BPPM) Using Azido Derivative of S-adenosyl-L-methionine. J Am Chem Soc. 2012 doi: 10.1021/ja2118333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam K, Zheng W, Yu H, Deng H, Luo M. Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling. ACS Chem Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol. 2011a;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Liu CL, Yang Z, Newman AM, Alizadeh AA, Utz PJ, Gozani O. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin. 2011b;4:19. doi: 10.1186/1756-8935-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- Nady N, Krichevsky L, Zhong N, Duan S, Tempel W, Amaya MF, Ravichandran M, Arrowsmith CH. Histone recognition by human malignant brain tumor domains. J Mol Biol. 2012;423:702–718. doi: 10.1016/j.jmb.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Nichols AM, White FM. Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. Methods Mol Biol. 2009;492:143–160. doi: 10.1007/978-1-59745-493-3_8. [DOI] [PubMed] [Google Scholar]
- Ong S, Blagoev B, Kratchmarova I, Kristensen D, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong S, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- Pang CN, Gasteiger E, Wilkins MR. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics. 2010;11:92. doi: 10.1186/1471-2164-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrossian TC, Clarke SG. Uncovering the human methyltransferasome. Mol Cell Proteomics. 2011;10:M110.000976. doi: 10.1074/mcp.M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR, Rubin SM, Gozani O, Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Tarakhovsky A. Lysine methylation and ‘signaling memory’. Curr Opin Immunol. 2006;18:152–157. doi: 10.1016/j.coi.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J. 2010;29:3967–3978. doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem. 2010;285:37725–37732. doi: 10.1074/jbc.M110.139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross P, Pappin D, Rush J, Lauffenburger D, White F. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.