Abstract
Two commonly used promoters to ubiquitously express transgenes in zebrafish are the Xenopus laevis elongation factor 1 α promoter (XlEef1a1) and the zebrafish histone variant H2A.F/Z (h2afv) promoter. Recently, transgenes utilizing these promoters were shown to be silenced in certain adult tissues, particularly the central nervous system. To overcome this limitation, we cloned the promoters of four zebrafish genes that likely are transcribed ubiquitously throughout development and into the adult. These four genes are the TATA box binding protein gene, the taube nuss-like gene, the eukaryotic elongation factor 1-gamma gene, and the beta-actin-1 gene. We PCR amplified approximately 2.5 kb upstream of the putative translational start site of each gene and cloned each into a Tol2 expression vector that contains the EGFP reporter transgene. We used these four Tol2 vectors to independently generate stable transgenic fish lines for analysis of transgene expression during development and in the adult. We demonstrated that all four promoters drive a very broad pattern of EGFP expression throughout development and the adult. Using the retina as a well-characterized component of the CNS, all four promoters appeared to drive EGFP expression in all neuronal and non-neuronal cells of the adult retina. In contrast, the h2afv promoter failed to express EGFP in the adult retina. When we examined EGFP expression in the various cells of the blood cell lineage, we observed that all four promoters exhibited a more heterogenous expression pattern than either the XlEef1a1 or h2afv promoters. While these four ubiquitous promoters did not express EGFP in all the adult blood cells, they did express EGFP throughout the CNS and in broader expression patterns in the adult than either the XlEef1a1 or h2afv promoters. For these reasons, these four promoters will be valuable tools for expressing transgenes in adult zebrafish.
Keywords: Zebrafish, Central nervous system, Ubiquitous, Promoter, Tol2, Adult expression pattern
Introduction
Zebrafish have become a popular model system for studying early development (Malicki et al. 1996a, b; Lewis and Eisen 2003; Udvadia and Linney 2003; Zhang et al. 2003; Kim et al. 2007; Kondo 2007; Lesaffre et al. 2007). In addition, zebrafish have also become an attractive model for studying neurodegeneration (Grunwald et al. 1988; Furutani-Seiki et al. 1996; Tomasiewicz et al. 2002; Vihtelic et al. 2005), regeneration of various tissues and organs (Vihtelic and Hyde 2000; Poss et al. 2002; Bai et al.2005; Iovine et al. 2005; Lee et al. 2005; Whitehead et al. 2005; Thummel et al. 2006a; Nakatani et al. 2007), adult behaviors (Orger et al. 2004; Muto et al.2005; Ninkovic and Bally-Cuif 2006; Bretaud et al.2007; Yu et al. 2007) and aging (Gerhard 2007). To fully exploit zebrafish in these studies, great emphasis is being placed on generating fish that express desired transgenes in either specific subsets of cells or a wide range of tissues and cells.
Many of the earliest zebrafish transgenes took advantage of heterologous promoters that provided ubiquitous expression throughout the embryo, such as the well-defined Xenopus laevis XlEef1a1 promoter. Recently, we demonstrated that EGFP expression from this Xenopus XlEef1a1 promoter (XlEef1a1) was lost in the fin, retina and central nervous system as the embryo matures, however, expression could be reinitiated in the adult fin and retina if these tissues were induced to regenerate (Thummel et al. 2006b). Because tissue regeneration involves the production of progenitor cells that proliferate and differentiate into the new cells and tissue, re-expression of transgenes from the XlEef1a1 promoter may provide a method to identify progenitor or stem cells.
Over the years, several groups demonstrated that zebrafish sequences could be used to direct EGFP expression in either a cell or tissue-specific pattern during embryonic development (Higashijima et al.1997; Long et al. 1997; Meng et al. 1997). In addition, many cell-specific promoters continued to express EGFP in the correct spatial manner into adulthood (Gibbs and Schmale (2000); Kennedy et al. 2001; Udvadia and Linney 2003; Fimbel et al. 2007; Kassen et al. 2007). Unfortunately, there are not a large number of well-defined promoters that provide a broad expression pattern in both the embryo and adult.
Efforts to identify zebrafish promoters that would provide ubiquitous expression in the developing embryo and into adulthood have also met with limited success. The zebrafish histone variant 2A.F/Z (h2afv) promoter was reported to direct ubiquitous EGFP expression in the developing embryo and into adulthood (Pauls et al. 2001). Like the XlEef1a1 promoter, however, EGFP expression from the zebrafish h2afv promoter is also lost or reduced in the adult fin and retina (Thummel et al. 2006b). The zebrafish beta-actin promoter was shown to drive ubiquitous expression of transgenes in the embryo, but was not analyzed for expression in adult tissues (Higashijima et al. 1997). Regardless of its expression in the adult, the size of this 17 kb promoter makes it difficult to generate transgenes. The 0.8 kb zebrafish acidic ribosomal phosphoprotein (P0) promoter produced ubiquitous transient expression in the embryo, but a stable transgenic line was not generated (Ju et al. 1999). To uncover the mechanisms underlying transgene silencing, such as differential methylation, additional ubiquitous promoters that are either not silenced or silenced in different tissues must be identified. Thus, although promoters such as h2afv and XlEef1a1 are widely referred to as ubiquitous promoters, when used in zebrafish transgenesis, they fail to drive EGFP in a ubiquitous manner.
The limited number of candidate promoters that are easily manageable and can drive transgene transcription throughout development and into adulthood severely limits the ability of researchers to fully exploit emerging technologies. Many questions about tissue generation during embryonic development or the regeneration of damaged adult tissues, such as the retina, heart or fin, are now becoming accessible to a variety of molecular and genetic techniques. For example, the Cre/lox site-specific recombination system has been used to examine the cell lineage processes in endocrine pancreas development (Herrera et al. 1998, 2002; Herrera 2002). This involves the expression of a reporter transgene, such as EGFP, from a cell-specific promoter. Expression of the Cre recombinase from a similar cell-specific promoter drives a recombination event that places the reporter transgene under the transcriptional control of a ubiquitous promoter. When these cells migrate and begin to differentiate, they continue to express the transgene from the ubiquitous promoter, while the surrounding cells lack transgene expression due to their failure to initially express Cre. This type of technology, which would be extremely powerful to study embryonic development in the transparent zebrafish embryo or tissue regeneration in the adult zebrafish is dependent on the generation of ubiquitous promoters.
We identified and characterized four different zebrafish ubiquitous promoters in order to determine whether they could drive EGFP in a broader manner than the two previously described ubiquitous promoters, h2afv and XlEef1a1. We analyzed the TATA box binding protein (tbp), the taube nuss-like (tbnl), the eukaryotic elongation factor 1-gamma (eef1g), and the beta-actin-1 (bactin1). We PCR amplified approximately 2.5 kb of the region upstream of the putative translational start site for each of the four genes and independently cloned them into a modified Tol2 expression vector that contains the EGFP reporter transgene (Kawakami and Shima 1999; Kawakami et al. 2000; Thummel et al. 2005). Each expression construct was injected into zebrafish embryos to generate several stable and independent transgenic lines. We characterized the EGFP expression from several independent single-insert lines and compared this expression to both the heterologous Xenopus XlEef1a1 promoter and the zebrafish h2afv promoter in both the embryo and adult tissues. We found that the tbp, tbnl, eef1g, bactin1 promoters drove EGFP in a similar ubiquitous pattern in the embryo as the previously defined ubiquitous promoters, however, they also expressed EGFP in a broader pattern in adult tissues, such as the central nervous system, than either the XlEef1a1 or the h2afv promoters did. While these promoters apparently expressed EGFP in all the neuronal and non-neuronal cell types of the adult retina, they only expressed EGFP in a subset of cell types in the blood cell lineage. Thus, we identified four new zebrafish promoters that are widely expressed in both the embryo and adult, which should be valuable in expressing transgenes throughout the lifetime of the fish.
Materials and methods
Animal rearing
Zebrafish (Danio rerio) were raised at 28°C in a 14 h light: 10 h dark regime using standard husbandry techniques (Westerfield 1995). Prior to sacrificing the fish for tissue dissection for EGFP analysis or DNA isolation, fish were deeply anesthetized with 2-phenoxyethanol (1:500 dilution). All experiments were conducted in accordance with the protocols approved by the animal use committee at the University of Notre Dame and the ARVO statement on the use of animals in vision research.
RNA extraction and RT-PCR amplification
Total RNA was extracted from adult zebrafish or isolated retinal samples using Trizoltm (Invitrogen, Carlsbad, CA). The RT-PCR was performed on each sample using a Superscript one step for long templates kit (Invitrogen). PCR amplification of the cDNA was done with the following cycling conditions: 45°C for 30 min, 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, 68°C for 1 min with a final elongation at 68°C for 10 min.
Primer design for amplification of promoter sequences
The four promoters selected for analysis corresponded to the TATA box binding protein gene (tbp, ZBD-GENE-030616-563), the taube nuss-like gene (tbnl, formerly called the TATA box binding protein associated factor gene or tbnl, ZBD-GENE-040426-1013), the eukaryotic elongation factor 1-gamma gene (eef1g, ZBD-GENE-020423-3), and the beta-actin-1 gene (bactin1, ZBD-GENE-000329-1). For each promoter, one primer was generated immediately upstream of the AUG translation initiation codon. The second primer was designed to amplify a 2.5 kb upstream product. Primers for amplification of the tbp promoter contain the XhoI and SalI restriction sites, while the tbnl, bactin1 and eef1g primers were engineered to contain XhoI and BamHI restriction sites. The following primer sequences were used with the restriction site underlined: tbp- forward primer 5′CTCGAGAGTTATGTACCATGAGATTAGGC 3′, reverse primer 5′ GTCGACAACAACAGAGCAGT GGTTGAGGG 3′; tbnl- forward primer 5′ CTCGAGTGCATAACTGTGATCACAAGC 3′, reverse primer 5′ GGATCCTTCCTCTGATGTTTAGCATT CGC 3′; eef1g- forward primer 5′ CTCGAGTCATGTTATTGACACCACTACCG 3′, reverse primer 5′ GGATCCGACGAGAGAAAGGAAGAACGAGC3′; bactin1- forward primer 5′ GGATCCTTGTGACGTCTGGCCGTGGCG 3′, reverse primer 5′ CTCGAGATTGCCCCAAGCTGAAGGCAGGAC 3′. Of the four new promoter constructs, only the tbp expression vector contained the β-globin intron from the original Tol2 expression construct (Kawakami et al. 1998,.2000) between the promoter and the EGFP open reading frame.
DNA isolation, promoter amplification and gel extraction
High molecular weight genomic DNA was isolated from AB and transgenic fish as previously described (Westerfield 1995), with minor modifications. Following incubation of the tissue in the Genomic DNA Extraction Buffer (10 mM Tris pH8, 100 mM EDTA pH 8, 0.5% SDS, 200 μg/ml Proteinase K), the samples were extracted once with 1 ml phenol and centrifuged (10 min at 13,500 rpm). The aqueous phase was removed, extracted with 1:1 phenol/chloroform, and separated as before. This step was repeated and followed by an extraction with 1 ml of chloroform. After centrifugation, the aqueous phase was removed and the DNA was precipitated by addition of one volume of 95% ethanol. High molecular weight DNA was removed by spooling on a glass rod (World Precision Instruments; Sarasota, FL), washed in 70% ethanol and air dried for 1 min.
The 2.5 kb promoter elements were amplified from AB genomic DNA using the primers described in the previous section and Platinum Taq Hi Fidelity enzyme (Invitrogen, Carlsbad, CA) with the following cycles: 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 62.5°C for 1 min, 68°C for 8 min with a final elongation at 68°C for 10 min. Amplified products were cloned into the TOPO-TA pCR4 vector (Invitrogen).
Cloning of amplified promoter sequences
Promoter fragments for each gene were sub-cloned from the pCR4 vector into a modified Tol2 expression vector (Thummel et al. 2005) with T4 DNA ligase (New England Biolabs; Beverly, MA). Expression vectors were transformed into DH5α competent cells and screened for positive clones by restriction enzyme digestion. A large-scale isolation of each positive clone was performed using a Qiagen maxiprep kit (Qiagen; Valencia, CA) followed by an extraction with phenol/chloroform (1:1). Samples were re-extracted with chloroform and precipitated with 95% ethanol. The precipitated expression vectors were washed with 70% ethanol, air dried and re-suspended in nuclease free water (Invitrogen), after which an OD260/280 was taken.
Generation of transgenic animals
Tol2 transposase mRNA was in vitro transcribed from the pCSTZ2.8 plasmid (gift of K. Kawakami) using an SP6 mMessage mMachine kit (Ambion; Foster City, CA). Expression constructs were injected into 1–4 cell stage embryos as previously described (Thummel et al. 2005). Positive embryos (mosaic for EGFP expression) were raised to adulthood and outcrossed to the AB strain. Founder fish were isolated by the presence of EGFP expression in the F1 offspring. EGFP-positive F1 fish were outcrossed to AB to generate independent F2 families. Each line continued to express EGFP through either the F3 or F4 generation.
Southern analysis of transgenic fish
High molecular weight genomic DNA, extracted as described above, was subjected to Southern analysis. A restriction enzyme that cuts a single site within the transgene was used, BglII for the Tg(tbp:EGFP)nt17 transgenic fish and StuI for the Tg(tbnl:EGFP)nt16 transgenic fish. Genomic DNA (10 μg) was digested with either of the two enzymes for 5 h and precipitated with the addition of 5 M sodium acetate and 100% ethanol. Digested DNA was pelleted, washed with 70% ethanol, air dried and re-suspended in 20 μl of TE (pH8.0). Digested DNA was separated overnight on a 1% agarose gel and then transferred to a nitrocellulose membrane. The DNA was fixed to the membrane using an oven at 80°C for 30 min. The membrane was hybridized with a 900 bp digoxygenin-labled EGFP PCR product using the PCR DIG probe synthesis kit (Roche; Indianapolis, IN). Detection of the labeled probe was performed using the Dig wash and block buffer set (Roche) and Blue X-ray film (RPS Imaging; Michigan City, IN).
Analysis of EGFP expression
Images of transgene expression from wholemounts of embryos, fry, and adults were obtained using a Leica MZIII dissecting scope with an EGFP III filter. EGFP expression was documented using a Spot RTke camera (Diagnostic Instruments, Inc.; Sterling Heights, MI) at the same settings for each time point except where noted.
Cryosectioning of zebrafish tissues was done to determine what tissues expressed EGFP in both fry and adult fish. Tissues were fixed in either 4% PFA or ethanolic formaldehyde (9 parts 95% EtOH:1 part 1× PBS) and confocal images of frozen sections were taken using a Leica TCS Sp5 Broadband Confocal System (Leica Microsystems; Wetzlar, Germany) as previously described (Vihtelic et al. 2001).
Western analysis of EGFP expression
Total protein for immunoblot analysis was harvested from AB and ubiquitous transgenic embryos by homogenizing at least 25 embryos in extraction buffer (1× PBS [pH 7.4]/1% Triton X-100) and incubating on ice for 1 hr. The total protein equivalent of 0.5 embryos was combined with 4× sample loading buffer and 10× reducing buffer (Invitrogen). The sample was incubated at 70°C for 10 min, electrophoresed through a 4–12% SDS-PAGE (Invitrogen), and transferred to PVDF. The membrane was blocked for 2 h in PBS, 5% nonfat dry milk, and 0.1% Tween-20. The membrane was then incubated with either a mouse anti-GFP monoclonal antibody (Abcam; Cambridge, MA; 1:5000) or a mouse monoclonal anti-actin (clone JLA20; 1:10,000; Cal-Biochem, San Diego, CA). The ECL-Plus detection system (GE Healthcare; Piscataway, NJ) was used to visualize primary antibody binding.
Fluorescence-activated cell sorting
Fluorescence-activated cell sorting (FACS) analysis was completed essentially as previously described (Traver 2004). Briefly, kidney marrow was dissected from transgenic and wild-type animals and resuspended in 0.9× PBS containing 5% fetal bovine serum. Samples were filtered (40 micron filter) and propidium iodide was added to exclude dead cells during fluorescence activated cell sorting (FACS). Blood cell populations were separated based on side (forward scatter) and granularity (side scatter) defining red blood cells, myelomonocytes, lymphocytes, and progenitors (Traver 2004). Fluorescence of the EGFP protein was also measured at the time of FACS.
Results
Generation of Tg(bactin1:EGFP)nt14, Tg(eef1g:EGFP)nt15, Tg(tbnl:EGFP)nt16 and Tg(tbp:EGFP)nt17 transgenic fish lines
To identify promoters that are ubiquitously expressed, we selected 10 different genes that encode basal transcription or translation factors and cytoskeletal proteins. To determine if these genes were present in the adult retina and body, we performed RT-PCR on total retinal RNA and total body RNA (data not shown). This analysis revealed that four genes, the TATA box binding protein (tbp) gene, the taube nuss-like (tbnl) gene, the eukaryotic elongation factor 1 gamma (eef1g) gene, and the β-actin 1 (bactin1) gene were expressed at relatively high levels in both RNA populations. We PCR amplified an approximately 2.5 kb product from wild-type AB zebrafish genomic DNA that is upstream from the start of the open reading frame. These PCR fragments were cloned into a modified Tol2 expression vector containing the EGFP reporter transgene (Fig. 1) and co-injected with Tol2 transposase mRNA into 1–4 cells stage embryos. Embryos that exhibited mosaic EGFP expression were raised to adulthood and out-crossed to AB fish. EGFP-positive F1 embryos were identified, raised to adulthood, and used to generate several families for each of the constructs. Two lines of data revealed that nearly all the families contained single copies of the transgene. First, approximately 50% of the F2 progeny from each family expressed EGFP. Second, single transgene inserts were identified on genomic Southern blots for several F3 individuals in the Tg(tbp:EGFP)nt17 and Tg(tbnl:EGFP)nt16 transgenic fish lines (data not shown). Surprisingly, every independent transgenic line exhibited maternal EGFP expression when a female carrier was crossed to the AB line. Therefore, the subsequent analysis of EGFP expression during early development (bud stage to 7 dpf) was performed on progeny of male carrier fish outcrossed to the AB line. We also compared the EGFP expression in these transgenic lines with the previously described Tg(XlEef1a1:EGFP)nt12 and Tg(h2afv:EGFP)nt13 lines (Thummel et al. 2006b).
Fig. 1.
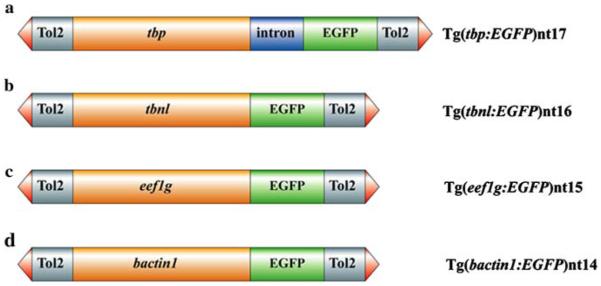
Structure of the tbp:EGFP, tbnl:EGFP, eef1g:EGFP and bactin1:EGFP transgenes. Each promoter is 2.5 kb (orange boxes). The Tol2 sequences that flank each transgene include the terminal inverted repeats (red arrowheads) and approximately 500 bp (grey boxes). The EGFP transgene is 900 bp in length (green boxes). The tbp:EGFP transgene contains the β-globin intron (a), which is 644 bp and represented as the blue box
Expression of the tbp:EGFP, tbnl:EGFP, eef1g:EGFP and bactin1:EGFP transgenes is ubiquitous in early development
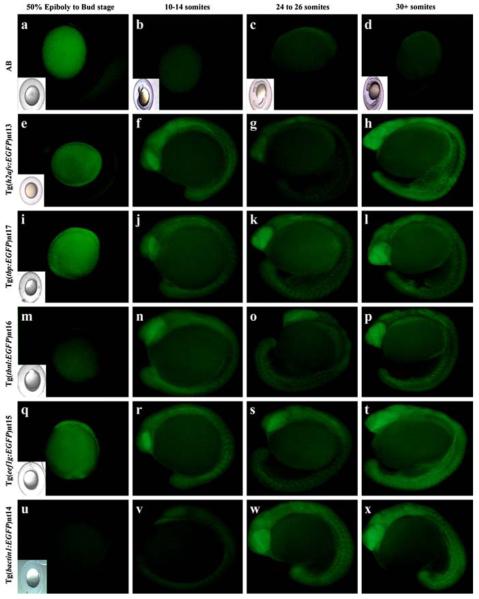
EGFP expression was not detected in embryos produced from carrier males prior to the shield stage of development in any of the transgenic lines analyzed. However, EGFP expression was detected in the Tg(h2afv:EGFP)nt13, Tg(tbp:EGFP)nt17, and Tg(eef1g:EGFP)nt15 transgenic embryos by the bud stage at 10 h post fertilization (hpf) (Fig. 2, Panels e, i, and q, respectively). By the 10–14 somite stage, EGFP was expressed ubiquitously throughout the embryo in all five transgenic lines (Fig. 2, Panels f, j, n, r, and v), although expression was relatively weak in the Tg(bactin1:EGFP)nt14 line (Fig. 2v). From the 24–26 somite stage through the 30+ somite stage, the EGFP continued to be broadly expressed in all five transgenic lines (Fig. 2, Panels g, h, k, l, o, p, s, t, w and x). At the 24–36 h time point, expression of EGFP persisted throughout the embryo, except for the fin fold of all five transgenic lines (Fig. 3, Panels b–f, arrowheads). EGFP expression was not detected in wild-type AB fish at any of these developmental stages (Fig. 2, Panels a-d; Fig. 3a). We independently confirmed the EGFP expression at 24 hpf and 7 dpf by immunoblots (Fig. 4). At 24 hpf, the h2afv:EGFP, tbp:EGFP, and bactin1:EGFP transgenes expressed EGFP at the highest levels, while the tbp:EGFP, eef1g:EGFP, bactin1:EGFP, and tbnl:EGFP transgenes exhibited the greatest amount of EGFP by 7 dpf. As expected, EGFP was not detected in the wild-type AB protein fraction at either time point on immunoblots (Fig. 4). The immunoblots also revealed that a larger EGFP protein was expressed from the tbnl:EGFP transgene relative to the other four transgenes. Upon re-examining the DNA sequence of this transgene, we identified a translation initiation codon upstream and in frame with the EGFP open reading frame. This upstream AUG codon is consistent with the increased molecular weight of the EGFP translational fusion in the Tg(tbnl:EGFP)nt16 transgenic line.
Fig. 2.
Developmental time course of EGFP expression in transgenic embryos. AB embryos were collected at the same time points from either 50% epiboly through the 30+ somite stage as the transgenic lines. EGFP fluorescence was not detected in any of the AB embryos (a–d). EGFP expression was present from the bud stage through the 30+ somite stage in the Tg(h2afv:EGFP)nt13 (e–h), Tg(tbp:EGFP)nt17 (i–l), and Tg(eef1g:EGFP)nt15 lines (q–t). EGFP expression was detected in the Tg(tbnl:EGFP)nt16 and Tg(bactin1:EGFP)nt14 lines from the 10–14 somite through the 30+ somite stages (Panels m–p and u–x, respectively) Embryos that are difficult to see in the fluorescent field have an inset of a bright field image
Fig. 3.
EGFP expression between 24 and 36 hpf in the transgenic embryos. The AB embryos possessed only a very low level of autofluorescence in the yolk sac and anal yolk extension at 24–36 hpf (Panel a, arrows). Furthermore, no fluorescence was detected in the fin fold (Panel a, arrowhead). A bright field image showing the AB embryo is inset. The EGFP expression was ubiquitous in the Tg(h2afv:EGFP)nt13, Tg(tbp:EGFP)nt17, Tg(tbnl:EGFP)nt16, Tg(eef1g:EGFP) nt15, and Tg(bactin1:EGFP)nt14 lines (Panels b–f), except for the absence of EGFP expression in the fin folds (arrowheads)
Fig. 4.
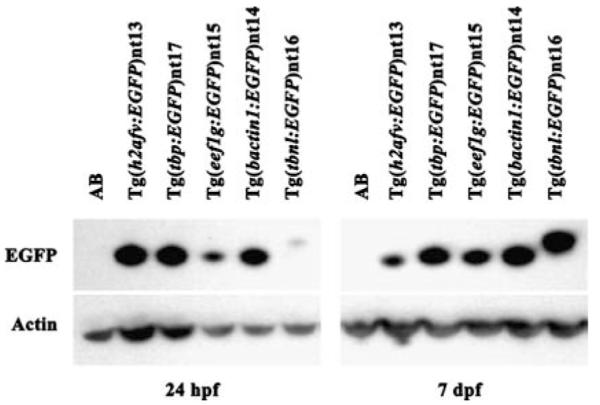
Immunoblot of EGFP expression at 24 hpf and 7 dpf. Total protein extracts from AB and five transgenic lines were isolated at 24 hpf and 7 dpf, electrophoresed through 4–12% SDS-PAGE, and transferred to PVDF membrane. Duplicate membranes were incubated with either anti-GFP or anti-actin antibodies, which served as a loading control. The immunoblots revealed a decrease in EGFP expression from 24hfp to 7 dpf in the Tg(h2afv:EGFP)nt13 line and increased expression in the Tg(eef1g:EGFP)nt15 and the Tg(tbnl:EGFP)nt16 lines. The Tg(tbnl:EGFP)nt16 line also revealed an EGFP protein that was slightly larger than the EGFP in the other transgenic lines. This increased molecular weight is due to an AUG codon that is in frame and upstream of the EGFP open reading frame, which yields a fusion protein
EGFP remained broadly expressed in all five transgenic lines, at 7 dpf but continued to be silenced in the fin fold
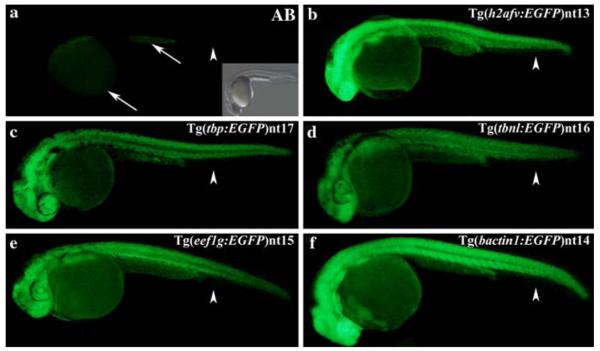
The fin fold, which will develop into the fins, remained EGFP-negative in all five transgenic lines at 7 dpf (Fig. 5, Panels b–f). The EGFP expression in the Tg(h2afv:EGFP)nt13 transgenic line was less intense in the body (Fig. 5b, arrows), but remained strong in the brain and along the spinal cord (Fig. 5b, arrowheads). In contrast, EGFP expression in the other four transgenic lines continued to be strong and remained uniform in the body, brain and spinal cord (Fig. 5, Panels c–f).
Fig. 5.
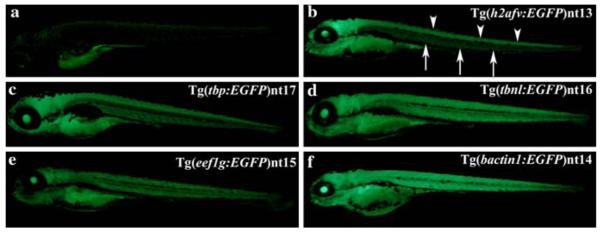
Ubiquitous transgene expression at 7 dpf. Similar to 24 hpf, a low level of autofluorescence was detected at 7 dpf in the AB fry (Panel a). The Tg(h2afv:EGFP)nt13 transgenic fish expressed EGFP in the brain and spinal cord (Panel b, arrowheads), while the body had a much lower level of EGFP expression (arrows). In comparison, the other four transgenic lines exhibited broader expression throughout the body, brain and spinal cord (Panels c–f). EGFP continued to be absent from the fin fold in all transgenic lines
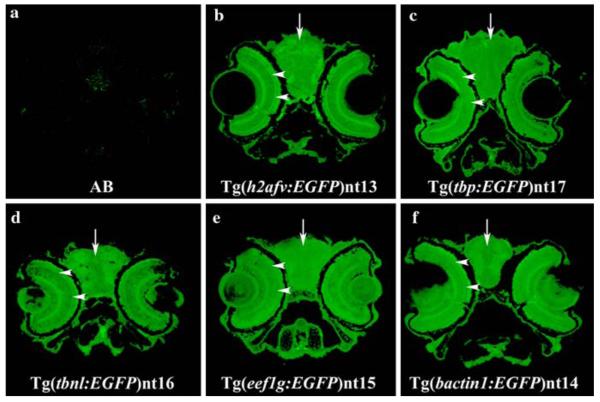
We immunolocalized EGFP expression in transverse head sections of 7 dpf fish in the different transgenic lines (Fig. 6). EGFP expression was detected in the retina (arrowheads), brain (arrows) and most other tissues of the head in all five transgenic lines (Fig. 6, Panels b–f), but was not detected in the wild-type AB head (Fig. 6a). By 7 dpf, the retinal laminar pattern has formed and the retina is functional (Schmitt and Dowling 1994,1999; Malicki et al. 1996a). At this age, EGFP-positive cells were present throughout all the retinal cell layers in every transgenic line.
Fig. 6.
Ubiquitous transgene expression throughout the head 7 dpf. At 7 dpf, EGFP expression was immunolocalized throughout the brain, retina, and other tissues of the head in all five transgenic lines (Panels b–f, arrows and arrowheads, respectively). The lens in each transgenic line appears to have a distinct EGFP expression pattern that was not further analyzed in this paper. However, EGFP appears to be expressed in the lens of Tg(tbp:EGFP)nt17, Tg(tbnl:EGFP)nt16, Tg(eef1g:EGFP)nt15, and Tg(bactin1:EGFP)nt14 lines (Panels c–f), but not in the Tg(h2afv:EGFP)nt13 transgenic fish (Panel b). As expected, AB fish stained with the same antibody did not show any substantial EGFP staining in the head (Panel a)
EGFP expression persisted in the adult in all five transgenic lines
EGFP expression was still detected in adults of all five transgenic lines (Fig. 7, Panels b–f), but not in the AB line (Fig. 7a). The Tg(h2afv:EGFP)nt13 transgenic line expressed a lower level of EGFP that was unevenly distributed relative to the other transgenic lines (Fig. 7b). The bactin1:EGFP, eef1g:EGFP, tbnl:EGFP, and tbp:EGFP transgenes continued to drive EGFP expression throughout the adult body, with some differences in EGFP intensity. In most cases, the variation involved a more intense expression in the reproductive organs, especially in female carrier fish (Fig. 7c, arrow). Adult fins continued to lack EGFP expression in all the transgenic lines, except for a subset of the Tg(bactin1:EGFP)nt14 and Tg(tbnl:EGFP)nt16 lines, that contained EGFP expression in the bony fin rays (Table 1).
Fig. 7.
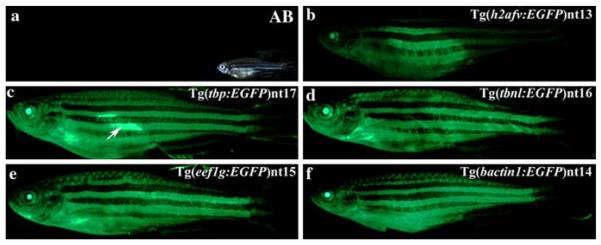
EGFP expression persists in a ubiquitous pattern in the adult transgenic fish, with the exception of the adult fins. EGFP expression is readily visible from the jaw to the girdle of the tail fin in the Tg(tbp:EGFP)nt17, Tg(tbnl:EGFP)nt16, Tg(eef1g:EGFP)nt15, and Tg(bactin1:EGFP)nt14 lines (Panels c–f). In contrast, the Tg(h2afv:EGFP)nt13 transgenic fish (Panel b), exhibited dramatically reduced EGFP expression in the anterior and posterior regions. In several fish, the female reproductive organs expressed elevated levels of EGFP (Panel c, arrow). There was no fluorescence detected in the AB fish (Panel a). A brightfield image of the adult AB fish is shown (Panel a, inset)
Table 1.
Summary of EGFP expression in the Tg(tbp:EGFP)nt17, Tg(tbnl:EGFP)nt16, Tg(eef1g:EGFP)nt15, and Tg(bactin1:EGFP)nt14 lines
| Transgene | Lens | Retina | Brain | Body | Fins | Heart | Gills | Blood |
|---|---|---|---|---|---|---|---|---|
| tbp:EGFP | + | + | + | + | − | + | + | − |
| tbnl:EGFP | + | + | + | + | + | + | + | − |
| eef1g:EGFP | + | + | + | + | − | + | + | − |
| bactin1:EGFP | + | + | + | + | + | + | + | − |
Expression of EGFP in the adult fish was determined by examination of each tissue for fluorescence. The + represents tissues that were scored as positive for EGFP expression and − represents no detectable EGFP expression. EGFP expression in the fins of the Tg(tbnl:EGFP)n16 and Tg(bactin1:EGFP)nt14 transgenic lines was restricted to the bony fin rays
To further analyze the expression of EGFP in the transgenic lines, several different tissues were dissected from adult fish and examined for EGFP expression (Table 1). To visualize the distribution of EGFP in the body, we also cut 1–2 cm thick transverse sections through the body and examined them under the dissecting scope (Fig. 8a). The tbp:EGFP, tbnl:EGFP, eef1g:EGFP and bactin1:EGFP transgenes drove EGFP expression throughout the muscle tissue (Fig. 8, Panels d–g). These sections confirmed the differences in EGFP intensity between the different lines that were seen previously with the live adult fish (Fig. 7). Furthermore, all four of these transgenes consistently produced higher EGFP expression levels than the Tg(h2afv:EGFP)nt13 transgenic line (Fig. 8c). As expected, we failed to detect fluorescence in sections of AB fish taken at the same settings (Fig. 8b).
Fig. 8.
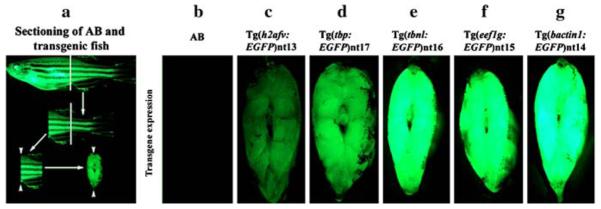
Visualizing distribution of EGFP expression in the transgenic fish body. EGFP expression was analyzed in tissue posterior to the anus/urogenital pore and anterior to the dorsal fin. The white lines represent where transverse cuts were made in each fish (Panel a). Arrowheads indicate the tissues that were visualized for EGFP expression in the transgenic lines. The tbp, tbnl, eef1g and bactin1 promoters directed EGFP expression ubiquitously in the body muscle (Panels d–g). While EGFP expression was seen in the body muscle from Tg(h2afv:EGFP)nt13 transgenic zebrafish, the reduced expression in some regions is obvious (Panel c). There was no EGFP expression detected in comparably treated AB fish taken at the same settings (Panel b)
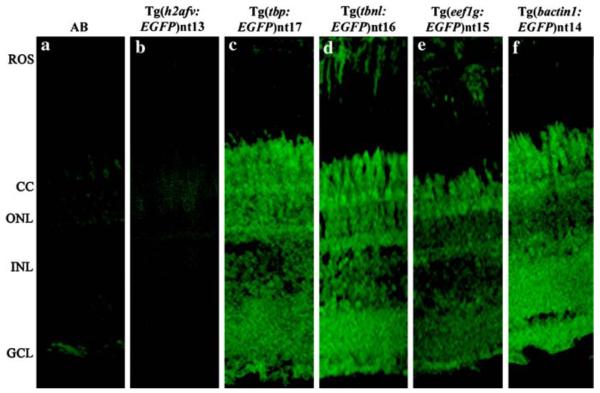
We confirmed the previous report (Thummel et al., 2006b) that the h2afv:EGFP transgene was silenced in the adult retina (Fig. 9b). In contrast, immunolocalization revealed that the bactin1:EGFP, eef1g:EGFP, tbnl:EGFP and tbp:EGFP transgenes drove EGFP expression in all the layers of the adult retina through the F3 or F4 generation (Fig. 9, Panels c–f). As expected, there was no detectable EGFP expression in either the adult AB or Tg(h2afv:EGFP)nt13 retinal sections (Fig. 9a–b).
Fig. 9.
EGFP is expressed in the retina from the new transgenes. EGFP was immunolocalized in the retinal sections of wild-type fish (Panel a) and the five transgenic lines (Panels b-f). Only a low level of autofluorescence was observed in the AB and Tg(h2afv:EGFP)nt13 lines (Panels a and b, respectively), which confirmed the silencing of the h2afv:EGFP transgene (Thummel et al. 2006b). In contrast, EGFP was immunolocalized throughout the retina in the Tg(tbp:EGFP) nt17, Tg(tbnl:EGFP)nt16, Tg(eef1g:EGFP)nt15, and Tg(bactin1:EGFP)nt14 lines (Panels c-f). ROS, rod outer segments; CC, cone cell layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer
EGFP expression in different blood cell types in the various transgenic lines
To examine the expression of the transgenes in the hematopoetic cell lineage, FACS analysis was preformed on three adult fish of each of the six different transgenic lines (Table 2). The h2afv:EGFP transgene was expressed in the greatest number of different blood cells, with 92% of the myelomonocytic cells, 93% of the progenitor cells and 82% of the white blood cells expressing EGFP (Table 2). The XlEef1a1:EGFP transgene expresses EGFP in 50% of the progenitor cells, 42% of the lymphocytes, and 40% of the white blood cells. Of the four new transgenes, the tbnl:EGFP transgene exhibits the highest level of EGFP expression in the blood cells, including 30% of the myelomonocytic cells, 27% of the progenitor cells, and 25% of the white blood cells. However, all four of the new transgenes labeled only very low percentages of the red blood cells (0.4–2.2%). Thus, the ubiquitous nature of the h2afv promoter in the hematopoetic cell lineage makes it a good candidate for characterizing the leukocyte cell populations or in studying hematopoetic cell transplantation into fish.
Table 2.
Summary of EGFP expression in blood cell types
| Line | Total EGFP | EGFP+ lymp | EGFP+ myel | EGFP+ RBC | EGFP+ Prog | EGFP+ WBC |
|---|---|---|---|---|---|---|
| Tg(XlEef1a1:EGFP)nt12 | 32.20% | 41.80% | 32.20% | 13.60% | 49.50% | 39.50% |
| Tg(h2afv:EGFP)nt13 | 58.0 ± 4.5% | 65.5 ± 9.3% | 92.0 ± 6.5% | 18.5 ± 5.2% | 93.0 ± 2.0 | 81.7 ± 7.0% |
| Tg(tbp:EGFP)nt17 | 9.5 ± 5.4% | 8.6 ± 5.3% | 13.1 ± 7.3% | 2.0 ± 0.7% | 14.5 ± 7.5% | 12.1 ± 6.9% |
| Tg(tbnl:EGFP)nt16 | 16.2 ± 12.6% | 17.8 ± 11.2% | 30.2 ± 17.4% | 2.2 ± 2.0% | 27.3 ± 16.5% | 24.5 ± 14.6% |
| Tg(eef1g:EGFP)nt15 | 1.1 ± 1.0% | 0.8 ± 1.0% | 1.6 ± 1.1% | 0.4 ± 0.1% | 2.7 ± 4.0% | 2.7 ± 3.9% |
| Tg(bactin1:EGFP)nt14 | 6.8 ± 0.1% | 8.4 ± 5.1% | 11.9 ± 7.3% | 1.0 ± 0.3% | 5.9 ± 2.8% | 8.9 ± 1.2% |
For each transgenic line, except for Tg(XlEef1a1:EGFP)nt12, 3 fish were subjected to FACS analysis to determine which blood cell types were labeled. The percentage of each cell type that expressed EGFP is shown along with the standard error of the mean. Because only one Tg(XlEef1a1:EGFP)nt12 fish was analyzed, a standard error of the mean could not be calculated. Total GFP was the percentage of cells that were EGFP-positive from whole kidney marrow. Distinct cell types were also analyzed for EGFP expression: lymp, lymphocytes; myel, myelomonocytic cells; RBC, red blood cell; Prog, progenitors; WBC, white blood cells. Only the Tg(h2afv:EGFP)nt13 transgenic line contained blood cells of every type that were labeled with EGFP
Discussion
This work describes the identification of four zebrafish promoters and the characterization of their expression through early development and into adulthood. Using sequence from the zebrafish genome sequencing project, we PCR amplified and cloned approximately 2.5 kb of genomic DNA from the tbp, tbnl, eef1g and bactin1 genes. These promoter sequences were cloned into Tol2 expression vectors containing the EGFP reporter transgene. Each of these promoters ubiquitously expressed EGFP from the early stages of development into the adult. While these new promoters expressed EGFP in adult cells that previously failed to express transgenes containing either the heterologous Xenopus laevis XlEef1a1 promoter or the zebrafish h2afv promoter, such as the adult retina and brain, they also failed to express EGFP in the fin and all the blood cell types. While these new promoters still did not generate ubiquitous expression in the adult, they produced a broader adult expression pattern than other promoters that were similarly analyzed.
The Xenopus XlEef1a1 promoter is commonly used to drive ubiquitous transgene expression in early zebrafish development (Chalfie et al. 1994; Johnson and Krieg 1994, 1995; Amsterdam et al. 1995, 1996; Hirsch et al. 2002). An advantage of the XlEef1a1 promoter is its small size and the inclusion of an intron and enhancer sequences, which provide stability and maximal expression to the transgene (Johnson and Krieg 1994; Amsterdam et al. 1995, 1996; Brinster et al. 1988; Le Hir et al. 2003). A considerable disadvantage of the XlEef1a1 promoter is the loss of transgene expression in certain adult zebrafish tissues (Thummel et al. 2006b).
The four new ubiquitous zebrafish promoters provide a substantial advantage over using the XlEef1a1 and h2afv promoters. First, even without enhancer sequences, the new promoters directed EGFP expression in an indistinguishable pattern compared to XlEef1a1. Second, unlike the XlEef1a1:EGFP and h2afv:EGFP lines, the bactin1:EGFP and tbnl:EGFP transgenes directed EGFP expression in the bony fin rays. Finally, in contrast to the XlEef1a1:EGFP, all four new transgenic lines showed persistent EGFP expression in the adult retina. Thus, the new promoters drove EGFP expression in a similar pattern to the XlEef1a1 promoter during development and, unlike the XlEef1a1 promoter, continued to express the reporter throughout the adult. Furthermore, the Tol2 constructs that contain these four new ubiquitous promoters contain multiple cloning sites to readily accommodate different reporter genes downstream of the promoters. Thus, these four new expression constructs will be very useful for driving the expression of desired transgenes throughout the developing and adult zebrafish.
Transgenes are a potent means to visualize and track cells in vivo. Recently, Cre-mediated recombination between lox sites was demonstrated in zebrafish (Dong and Stuart 2004; Langenau et al.2005; Pan et al. 2005; Thummel et al. 2005; Feng et al. 2007). This opens the possibility to use reporter transgenes to label specific cells, such as stem or progenitor cells, and follow their migration and differentiation in either development or regeneration (Herrera et al. 1998, 2002; Herrera 2002). The ubiquitously transcribed promoters are essential for the persistent expression of the reporter transgene, such as EGFP, as the stem or progenitor cells migrate and differentiate into their final cell fate. Using cell-specific promoters would result in the transient expression of the reporter through this process, which would yield an incomplete picture of what is happening to the initial cells. This requirement to continually express the reporter transgene during either embryonic development or adult tissue regeneration, underscores the need to identify broadly expressed promoters that remain transcriptionally active throughout the adult for these cell lineage tracing studies.
We plan to use these promoters to study the process of transgene silencing in zebrafish. Several studies describe the silencing of zebrafish transgenes either as the fish ages or as the transgene passes through several generations. Using luciferase as a marker gene, Gibbs et al. (1994) showed that DNA methylation resulted in an undetectable level of transgene expression in zebrafish. Gibbs further showed that addition of 5′-azacytidine, a potent methylation blocker, could re-establish luciferase expression in some cases. Furthermore, transient expression of a transgene could be severely influenced by the methylation state of the injected plasmid (Collas 1998). Using regenerating zebrafish caudal fins, we demonstrated that expression of a h2afv:EGFP transgene was associated with low levels of methylation, while the silenced transgene was heavily methylated (Thummel et al. 2006b). Taken together, these data suggest that DNA methylation affects silencing of transgene expression. However, there are no reports of cell-specific promoters exhibiting gene silencing even after 8–10 generations (Kennedy et al. 2001). This suggests that the ubiquitous expression of the transgene may influence the silencing. These new ubiquitous promoters could be used to further investigate the role of methylation on gene silencing and what features lead to some constitutive promoters being silenced and others not. The silencing of the h2afv:EGFP transgene in the retina, but not most of the blood cell types, compared with the silencing of the eef1g:EGFP transgene in most of the blood cell types, but not the retina, provides an excellent opportunity to examine not only what changes in the DNA methylation pattern are associated with transgene silencing, but also what causes various transgenes to be silenced in different tissues. Thus, these new constitutive promoters enlarge the repertoire of zebrafish promoters that will improve our ability to express transgenes in zebrafish and provide critical controls in studies involving the mechanism underlying transgene silencing.
Acknowledgements
The authors thank the Freimann Life Science Center Staff for the care and maintenance of the zebrafish facility and Suzyanne Guzicki for the injection of the expression constructs into zebrafish embryos. We also thank Koichi Kawakami for the generous gift of the pT2KXIG construct containing the Tol2 transposable element and Tol2 transposase gene (pCSTZ2.8) and the Campos-Ortega lab for the gift of the H2A.F/Z promoter. This work was supported by the National Institute of Health (R21-EY017134 to D.R.H).
Abbreviations
- EGFP
Enhanced green fluorescent protein
- RT-PCR
Reverse transcriptase polymerase chain reaction
- PCR
Polymerase chain reaction
- hpf
Hours post-fertilization
- dpf
Days post-fertilization
Contributor Information
Christopher T. Burket, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA
Jacob E. Montgomery, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA
Ryan Thummel, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA.
Sean C. Kassen, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA
Matthew C. LaFave, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA
David M. Langenau, HHMI/Children’s Hospital of Boston, Harvard Medical School, One Blackfan Circle, Karp Building, #7005C, Boston, MA 02115-5713, USA
Leonard I. Zon, HHMI/Children’s Hospital of Boston, Harvard Medical School, One Blackfan Circle, Karp Building, #7005C, Boston, MA 02115-5713, USA Stem Cell Program and Division Hematology/Oncology Children’s Hospital and Dana Farber Cancer Institute, Howard Hughes Medical Institute, Harvard Stem Cell Institute, Harvard Medical School, Boston, MA 02115, USA.
David R. Hyde, Department of Biological Sciences and the Center for Zebrafish Research, University of Notre Dame, Notre Dame, IN 46556, USA
References
- Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev Biol. 1995;171(1):123–129. doi: 10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Lin S, Moss LG, Hopkins N. Requirements for green fluorescent protein detection in transgenic zebrafish embryos. Gene. 1996;173:99–103. doi: 10.1016/0378-1119(95)00719-9. [DOI] [PubMed] [Google Scholar]
- Bai S, Thummel R, Godwin AR, Nagase H, Itoh Y, Li L, Evans R, McDermott J, Seiki M, Sarras MP., Jr Matrix metalloproteinase expression and function during fin regeneration in zebrafish: analysis of MT1-MMP, MMP2 and TIMP2. Matrix Biol. 2005;24(4):247–260. doi: 10.1016/j.matbio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bretaud S, Li Q, Lockwood BL, Kobayashi K, Lin E, Guo S. A choice behavior for morphine reveals experience-dependent drug preference and underlying neural substrates in developing larval zebrafish. J Neurosci. 2007;146(3):1109–1116. doi: 10.1016/j.neuroscience.2006.12.073. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Collas P. Modulation of plasmid DNA methylation and expression in zebrafish embryos. Nuc Acids Res. 1998;26(19):4454–4461. doi: 10.1093/nar/26.19.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Stuart GW. Transgene manipulation in zebrafish by using recombinases. Methods Cell Biol. 2004;77:363–379. doi: 10.1016/s0091-679x(04)77020-x. [DOI] [PubMed] [Google Scholar]
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, Kanki JP, Look TA. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br J Haematol. 2007;138(2):169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27(7):1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani-Seiki M, Jiang YJ, Brand M, Heisenberg CP, Houart C, Beuchle D, van Eeden FJ, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Neural degeneration mutants in the zebrafish, Danio rerio. Development. 1996;123:229–239. doi: 10.1242/dev.123.1.229. [DOI] [PubMed] [Google Scholar]
- Gerhard GS. Small laboratory fish as models for aging research. Ageing Res Rev. 2007;6(1):64–72. doi: 10.1016/j.arr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Gibbs PD, Schmale MC. GFP as a genetic marker scorable throughout the life cycle of transgenic zebra fish. Mar Biotechnol (NY) 2000;2(2):107–125. doi: 10.1007/s101269900014. [DOI] [PubMed] [Google Scholar]
- Gibbs PD, Peek A, Thorgaard G. An in vivo screen for the luciferase transgene in zebrafish. Mol Mar Biol Biotechnol. 1994;3(6):307–316. [PubMed] [Google Scholar]
- Grunwald DJ, Kimmel CB, Westerfield M, Walker C, Strei-singer G. A neural degeneration mutation that spares primary neurons in the zebrafish. Dev Biol. 1988;126(1):115–128. doi: 10.1016/0012-1606(88)90245-x. [DOI] [PubMed] [Google Scholar]
- Herrera PL. Defining the cell lineages of the islets of Langerhans using transgenic mice. Int J Dev Biol. 2002;46(1):97–103. [PubMed] [Google Scholar]
- Herrera PL, Nepote V, Delacour A. Pancreatic cell lineage analyses in mice. Endocrine. 2002;19(3):267–278. doi: 10.1385/ENDO:19:3:267. [DOI] [PubMed] [Google Scholar]
- Herrera PL, Orci L, Vassalli JD. Two transgenic approaches to define the cell lineages in endocrine pancreas development. Mol Cell Endocrinol. 1998;140(1–2):45–50. doi: 10.1016/s0303-7207(98)00028-8. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192(2):289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225(4):522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278(1):208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Krieg PA. pXeX, a vector for efficient expression of cloned sequences in Xenopus embryos. Gene. 1994;147(2):223–226. doi: 10.1016/0378-1119(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Krieg PA. A Xenopus laevis gene encoding EF-1 alpha S, the somatic form of elongation factor 1 alpha: sequence, structure, and identification of regulatory elements required for embryonic transcription. Dev Genet. 1995;17(3):280–290. doi: 10.1002/dvg.1020170313. [DOI] [PubMed] [Google Scholar]
- Ju B, Xu Y, He J, Liao J, Yan T, Hew CL, Lam TJ, Gong Z. Faithful expression of green fluorescent protein (GFP) in transgenic zebrafish embryos under control of zebrafish gene promoters. Dev Genet. 1999;25(2):158–167. doi: 10.1002/(SICI)1520-6408(1999)25:2<158::AID-DVG10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, Burket CT, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225(1–2):17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240(1):239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97(21):11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BN, Vihtelic TS, Checkley L, Vaughan KT, Hyde DR. Isolation of a zebrafish rod opsin promoter to generate a transgenic zebrafish line expressing enhanced green fluorescent protein in rod photoreceptors. J Biol Chem. 2001;276(17):14037–14043. doi: 10.1074/jbc.M010490200. [DOI] [PubMed] [Google Scholar]
- Kim KH, Antkiewicz DS, Yan L, Eliceir KW, Heideman W, Peterson RE, Lee Y. Lrrc10 is required for early heart development and function in zebrafish. Dev Biol. 2007;308(2):494–506. doi: 10.1016/j.ydbio.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M. Bone morphogenetic proteins in the early development of zebrafish. Febs J. 2007;274(12):2960–2967. doi: 10.1111/j.1742-4658.2007.05838.x. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2005;102(17):6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28(4):215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132(23):5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Lesaffre B, Joliot A, Prochiantz A, Volovitch M. Direct non-cell autonomous Pax6 activity regulates eye development in the zebrafish. Neural Develop. 2007;2:2. doi: 10.1186/1749-8104-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69(6):419–449. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124(20):4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996a;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Malicki J, Schier AF, Solnica-Krezel L, Stemple DL, Neuhauss SC, Stainier DY, Abdelilah S, Rangini Z, Zwartkruis F, Driever W. Mutations affecting development of the zebrafish ear. Development. 1996b;123:275–283. doi: 10.1242/dev.123.1.275. [DOI] [PubMed] [Google Scholar]
- Meng A, Tang H, Ong BA, Farrell MJ, Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci USA. 1997;94(12):6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, Smear MC, Kay JN, Page-McCaw PS, Gahtan E, Xiao T, Nevin LM, Gosse NJ, Staub W, Finger-Baier K, Baier H. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1(5):e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Kawakami A, Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev Growth Differ. 2007;49(2):145–154. doi: 10.1111/j.1440-169X.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- Ninkovic J, Bally-Cuif L. The zebrafish as a model system for assessing the reinforcing properties of drugs of abuse. Methods. 2006;39(3):262–274. doi: 10.1016/j.ymeth.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Orger MB, Gahtan E, Muto A, Page-McCaw P, Smear MC, Baier H. Behavioral screening assays in zebrafish. Methods Cell Biol. 2004;77:53–68. doi: 10.1016/s0091-679x(04)77003-x. [DOI] [PubMed] [Google Scholar]
- Pan X, Wan H, Chia W, Tong Y, Gong Z. Demonstration of site-directed recombination in transgenic zebrafish using the Cre/loxP system. Transgen Res. 2005;14(2):217–223. doi: 10.1007/s11248-004-5790-z. [DOI] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev Genes Evol. 2001;211(12):603–610. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J Comp Neurol. 1994;344(4):532–542. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J Comp Neurol. 1999;404(4):515–536. [PubMed] [Google Scholar]
- Thummel R, Bai S, Sarras MP, Jr, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006a;235(2):336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- Thummel R, Burket CT, Brewer JL, Sarras MP, Jr., Li L, Perry M, McDermott JP, Sauer B, Hyde DR, Godwin AR. Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn. 2005;233(4):1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- Thummel R, Burket CT, Hyde DR. Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. TSW Develop Embryol. 2006b;6:65–81. doi: 10.1100/tsw.2006.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasiewicz HG, Flaherty DB, Soria JP, Wood JG. Transgenic zebrafish model of neurodegeneration. J Neurosci Res. 2002;70(6):734–745. doi: 10.1002/jnr.10451. [DOI] [PubMed] [Google Scholar]
- Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2004;76:127–149. doi: 10.1016/s0091-679x(04)76008-2. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Linney E. Windows into development: historic, current, and future perspectives on transgenic zebrafish. Dev Biol. 2003;256(1):1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44(3):289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Yamamoto Y, Springer SS, Jeffery WR, Hyde DR. Lens opacity and photoreceptor degeneration in the zebrafish lens opaque mutant. Dev Dyn. 2005;233(1):52–65. doi: 10.1002/dvdy.20294. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Yamamoto Y, Sweeney MT, Jeffery WR, Hyde DR. Arrested differentiation and epithelial cell degeneration in zebrafish lens mutants. Dev Dyn. 2001;222(4):625–636. doi: 10.1002/dvdy.1217. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio) University of Oregon Press; Eugene: 1995. [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310(5756):1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Yu CJ, Gao Y, Li P, Li L. Synchronizing multiphasic circadian rhythms of rhodopsin promoter expression in rod photoreceptor cells. J Exp Biol. 2007;210:676–684. doi: 10.1242/jeb.02694. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bai S, Zhang X, Nagase H, Sarras MP., Jr The expression of novel membrane-type matrix metalloproteinase isoforms is required for normal development of zebrafish embryos. Matrix Biol. 2003;22(3):279–293. doi: 10.1016/s0945-053x(03)00020-9. [DOI] [PubMed] [Google Scholar]