Abstract
BACKGROUND
Dilated cardiomyopathy and hypertrophic cardiomyopathy arise from mutations in many genes. TTN, the gene encoding the sarcomere protein titin, has been insufficiently analyzed for cardiomyopathy mutations because of its enormous size.
METHODS
We analyzed TTN in 312 subjects with dilated cardiomyopathy, 231 subjects with hyper-trophic cardiomyopathy, and 249 controls by using next-generation or dideoxy sequencing. We evaluated deleterious variants for cosegregation in families and assessed clinical characteristics.
RESULTS
We identified 72 unique mutations (25 nonsense, 23 frameshift, 23 splicing, and 1 large tandem insertion) that altered full-length titin. Among subjects studied by means of next-generation sequencing, the frequency of TTN mutations was significantly higher among subjects with dilated cardiomyopathy (54 of 203 [27%]) than among subjects with hypertrophic cardiomyopathy (3 of 231 [1%], P = 3×10−16) or controls (7 of 249 [3%], P = 9×10−14). TTN mutations cosegregated with dilated cardiomyopathy in families (combined lod score, 11.1) with high (>95%) observed penetrance after the age of 40 years. Mutations associated with dilated cardiomyopathy were overrepresented in the titin A-band but were absent from the Z-disk and M-band regions of titin (P≤0.01 for all comparisons). Overall, the rates of cardiac outcomes were similar in subjects with and those without TTN mutations, but adverse events occurred earlier in male mutation carriers than in female carriers (P = 4×10−5).
CONCLUSIONS
TTN truncating mutations are a common cause of dilated cardiomyopathy, occurring in approximately 25% of familial cases of idiopathic dilated cardiomyopathy and in 18% of sporadic cases. Incorporation of sequencing approaches that detect TTN truncations into genetic testing for dilated cardiomyopathy should substantially increase test sensitivity, thereby allowing earlier diagnosis and therapeutic intervention for many patients with dilated cardiomyopathy. Defining the functional effects of TTN truncating mutations should improve our understanding of the pathophysiology of dilated cardiomyopathy. (Funded by the Howard Hughes Medical Institute and others.)
Gene mutation is an important cause of cardiomyopathy. Mutations in eight sarcomere-protein genes cause hypertrophic cardiomyopathy, detected in 40 to 70% of patients.1,2 Variations in more than 40 genes, most of which encode components of the sarco-mere, the cytoskeleton, or the nuclear lamina, have been shown or posited to cause dilated cardiomy-opathy.3,4 Clinical evaluation identifies 30 to 50% of patients with dilated cardiomyopathy as having a relative who is affected or likely to be affected,5–7 implicating a genetic cause. However, pathogenic mutations have been found in only 20 to 30% of patients.8
TTN, the gene encoding titin, has been implicated in cardiomyopathy but has been incompletely studied, owing to technical challenges posed by the large size of its coding sequence (approximately 100 kb). TTN mutations have been definitively linked to dilated cardiomyopathy in three families,9–11 but not to hypertrophic cardiomyopathy. In addition, TTN mutations have been implicated in congenital myopathies involving cardiac and skeletal muscle, hereditary myopathy with early respiratory failure, tibial muscular dystrophy, and limb-girdle muscular dystrophy.12–15
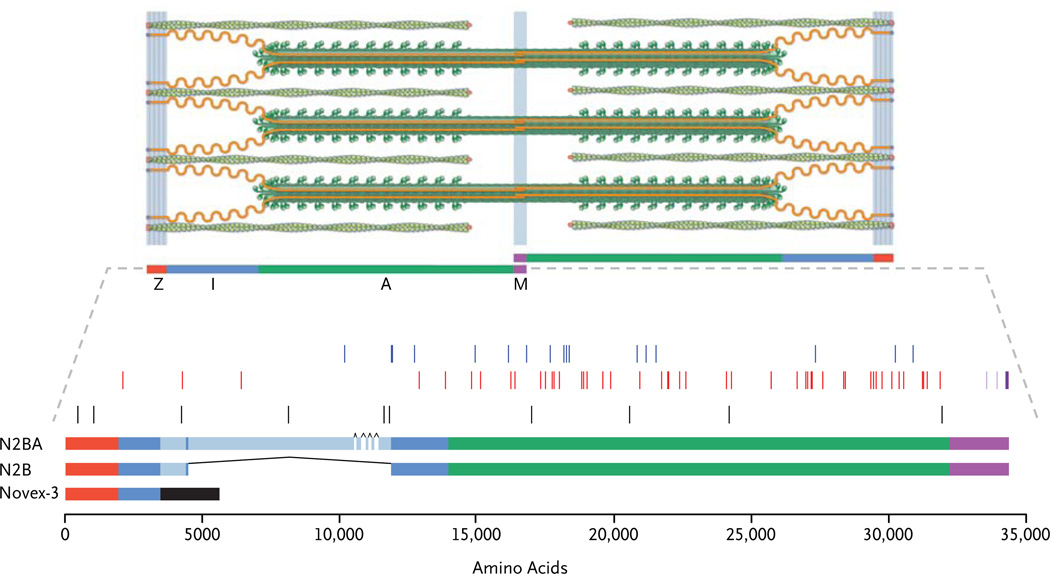
Titin is the largest human protein (composed of approximately 33,000 amino acids) and the third most abundant striated-muscle protein.16 Two titin molecules together span the sarcomere (about 2 µm in width) and are anchored at the Z-line and M-line (Fig. 1).18 Titin is necessary for sarcomere assembly19,20 and in striated muscle provides most of the passive force21,22 and modulates the active contractile force.23,24 There are many different isoforms of titin; those in the heart are predominantly N2B or N2BA (Fig. 1).25 TTN also encodes a separate cardiac isoform, novex-3 titin, which is only 5600 amino acids in length, lacks the A-band and M-band segments of titin,26 and is less abundant in cardiac tissue than full-length titin.
Figure 1. Spatial Distribution of Truncating Mutations in Titin.
The cardiac sarcomere (top) consists of titin (orange), the third major filament, in addition to the thick filaments (green rods with globular heads) and thin filaments (green coiled ovals). Regions of the sarcomere are demarcated as follows: Z-disk (red), I-band (blue), A-band (green), and M-band (purple). The sarcomere depiction is adapted from Granzier and Labeit.17 TTN mutations are shown as thin vertical bars (middle), with overlapping mutations appearing as thicker bars. Splicing and copy-number mutations (blue) and nonsense and frameshift mutations (red) in subjects with dilated cardiomyopathy are shown, including two frameshift mutations previously reported to be linked to dilated cardiomyopathy (Table 14 in the Supplementary Appendix). All types of truncating mutations in controls and subjects with hypertrophic cardiomyopathy (black) are also indicated, as are truncating mutations previously identified in patients with congenital myopathy (light purple) or limb-girdle muscular dystrophy (dark purple). Titin isoforms and sequence variants are shown relative to the titin sequence (UniProt sequence Q8WZ42, www.uniprot.org) (bottom). Analysis of the N2BA class of cardiac titin isoforms excluded exons (black carets) with little evidence for cardiac expression (see Table 15 in the Supplementary Appendix). In cardiac tissue, TTN isoforms N2BA and N2B span the sarcomere, whereas novex-3 titin is shorter and less abundant (the exon specific to novex-3 titin is shown as a black bar).
We undertook filter-based hybridization capture followed by next-generation sequencing27 or traditional dideoxy sequencing to assess the contribution of TTN mutations to cardiomyopathies, analyzing DNA from 312 subjects with idiopathic dilated cardiomyopathy, 231 subjects with hypertrophic cardiomyopathy, and 249 control subjects.
METHODS
SUBJECTS
Studies were performed according to institutional guidelines and the U.K. Human Tissue Act guidelines and with the approval of local ethics committees. The 312 subjects with idiopathic dilated cardiomyopathy were from three cohorts (see Table 3 in the Supplementary Appendix, available with the full text of this article at NEJM.org): 92 subjects recruited at Brigham and Women’s Hospital (group A), 71 subjects recruited during evaluation for cardiac transplantation at the Royal Brompton and Harefield National Health Service Trust (group B), and 149 subjects prospectively recruited in Colorado or Italy into a Familial Dilated Cardiomyopathy Registry (group C). Groups A and C had a high frequency of familial disease. The 231 subjects with hypertrophic cardiomyopathy were recruited at Brigham and Women’s Hospital or the Mayo Clinic. Dilated cardiomyopathy and hyper-trophic cardiomyopathy were diagnosed on the basis of published criteria.28,29 The 249 control subjects without cardiomyopathy were recruited from multiple sites. No subject within a cohort had a known familial relationship with any other subject in the cohort. For more information on the cohorts, see the Supplementary Appendix.
DNA SEQUENCING AND GENOTYPING
Genomic DNA from each subject was used to construct DNA libraries. The DNA libraries were enriched for TTN by means of filter-based hybridization capture27 with minor modifications (Tables 1 and 2 in the Supplementary Appendix) and were studied with the use of single-end or paired-end sequencing (with the Illumina Genome Analyzer II or HiSeq).30 The TTN sequence was assessed in group C by means of traditional Sanger dideoxy sequencing performed at the Department of Genome Sciences, University of Washington.
SEQUENCE ANALYSES
Next-generation sequence data were analyzed by means of a custom pipeline that integrated existing tools, including Novoalign (www.novocraft.com) and the Genome Analysis Toolkit,31 as well as Perl (with the use of Bio-Samtools) and R32 scripts. Primary analyses of TTN variations were performed among subjects studied by the same approach (Table 1) in order to control for differences in variant detection. The amino acid positions of titin variants were identified with the use of the UniProt titin sequence (Q8WZ42), and mutations were reported with the use of Human Genome Variation Society nomenclature (Table 4 in the Supplementary Appendix). Variant confirmation and genotyping were performed by means of one or more of these methods: polymerase-chain-reaction amplification followed by dideoxy sequencing; restriction digestion and gel electrophoresis26; and RNA sequencing of cardiac tissue.33
Table 1.
Subjects with TTN Truncating Variants, According to Cohort.
| Type of Mutation | Subjects with Dilated Cardiomyopathy (N = 312) |
Subjects with Hypertrophic Cardiomyopathy (N = 231) |
Controls (N = 249) |
P Value* | |
|---|---|---|---|---|---|
| All Subjects |
Selected Subjects |
||||
| number of subjects | |||||
| Nonsense† | 28 | 0 | 0 | 1×10 −10 | 5×10−11 |
| Frameshift | 19 | 2 | 2 | 0.0004 | 1×10−5 |
| Splicing† | 19 | 1 | 5 | 0.001 | 4×10−6 |
| Copy number | 1 | 0 | 0 | NA | NA |
| Any mutation | 67 | 3 | 7 | <2×10−16 | <2×10−16 |
P values were calculated on the basis of either all subjects or selected subjects (those studied by means of next-generation sequencing). Forty subjects with dilated cardiomyopathy who were excluded from TTN sequencing were included in these comparisons, for a total of 352 subjects with dilated cardiomyopathy. The one copy-number mutation was excluded from the calculations.
Three nonsense and three splicing mutations each occurred in two subjects with dilated cardiomyopathy. An additional subject with dilated cardiomyopathy had two different splicing mutations.
STATISTICAL ANALYSIS
Association, cross-cohort, and cross-group analyses were performed with the use of Fisher’s exact test, exact conditional tests of independence, or goodness-of-fit tests, unless otherwise specified. The uniformity of the spatial distribution of mutations was assessed by means of a chi-square goodness-of-fit test incorporating the size of each region. The clinical characteristics of each group in the dilated cardiomyopathy cohort were compared with the use of two-tailed, unpaired t-tests. Kaplan–Meier curves were computed by means of the “survfit” function in R and were compared by means of the “coxph” function in R.32
We calculated two-point lod scores for 19 families with dilated cardiomyopathy (Fig. 2 and Table 13 in the Supplementary Appendix) by using FASTLINK software, computed with the settings θ = 0, phenocopy rate = 0.005,34 and indicated disease penetrances.35 An indeterminate status was assigned to family members 40 years of age or younger who did not meet clinical criteria for dilated cardiomyopathy28 and to family members with confounding cardiac diagnoses.
RESULTS
STUDY SUBJECTS
Idiopathic dilated cardiomyopathy was diagnosed in the 312 subjects from the three independent groups: group A, group B, and group C (Tables 9, 10, and 11, respectively, in the Supplementary Appendix). Hypertrophic cardiomyopathy was diagnosed in 231 subjects; 249 subjects without known cardiomyopathy served as controls. Summary characteristics of each group and cohort are presented in Table 3 in the Supplementary Appendix.
TTN GENETIC VARIATION
DNA Sequencing
Using genomic DNA isolated from subjects with dilated cardiomyopathy in groups A and B and from the subjects with hypertrophic cardiomyopathy and controls, we captured and performed next-generation sequencing of 145 kb of the TTN gene, including all annotated exons and splice sites. For each subject, more than 97% of the targeted bases were sequenced at least 20 times (data not shown). From genomic DNA isolated from subjects with dilated cardiomyopathy, we determined TTN sequences by means of traditional dideoxy sequencing.
After excluding TTN variants with frequencies of 0.01 or greater in the 1000 Genomes Project (www.1000genomes.org/data) or present in all 792 study subjects, we identified 951 rare missense (minor allele frequency <0.01), nonsense, frame-shift, or splicing or copy-number TTN variants that are predicted to change the titin amino acid sequence (Tables 4 and 5 in the Supplementary Appendix). Every study subject had approximately 1 rare missense variant (range of averages per group or cohort, 0.91 to 1.45 per subject).
Since the TTN transcript novex-3 (Fig. 1) is thought not to interact with the sarcomere M-band, and its expression in the heart is approximately one twentieth that of full-length titin isoforms (unpublished data), we did not study variants that exclusively altered novex-3 transcripts. We prioritized for analysis four types of variants — i.e., nonsense, frameshift, splicing, and copynumber — that (unlike most missense variants) are predicted to have a profound effect on the structure of full-length titin polypeptides (Fig. 1). We classify these as TTN truncating variants.
TTN Variants in Subjects with Hypertrophic Cardiomyopathy and in Controls
In the subjects with hypertrophic cardiomyopathy, we found three TTN truncating variants: two frame-shift variants and one splicing variant (Table 1, and Tables 6 and 7 in the Supplementary Appendix). In each of the three affected subjects, concurrent analyses revealed a pathogenic mutation in the well-established hypertrophic cardiomyopathy genes encoding β-myosin heavy chain (MYH7) or myosin binding protein C (MYBPC3) (data not shown). No family members were available for segregation analyses. In control subjects, we identified two frameshift and five splicing variants (Table 1, and Tables 6 and 7 in the Supplementary Appendix). The frequency of TTN truncating variants did not differ significantly between subjects with hypertrophic cardiomyopathy and controls (1% and 3%, respectively; P = 0.34).
Subjects with Dilated Cardiomyopathy
Nonsense and Frameshift Variants
We identified 44 unique nonsense or frameshift variants that altered full-length titin in 47 subjects with dilated cardiomyopathy (Table 1, and Table 6 in the Supplementary Appendix). In the cohorts and groups studied by means of next-generation sequencing, the proportion of subjects with these variants was higher among the subjects with dilated cardiomyopathy in groups A and B (21 of 92 subjects [23%] and 14 of 71 [20%], respectively) than among the subjects with hypertrophic cardiomyopathy (2 of 231 [1%], P = 2×10−12) or the control subjects (2 of 249 [1%], P = 3×10−13). Twelve of the 149 subjects (8%) with dilated cardiomyopathy in group C, studied by means of traditional dideoxy sequencing, had such variants. We observed strong cosegregation (lod score, 9.3) of nonsense and frameshift variants with clinical status among 60 members of 16 families affected by dilated cardiomyopathy (Fig. 2 and Tables 12 and 13 in the Supplementary Appendix), indicating an odds of approximately 1 in 109 that the segregation of these TTN variants occurred by chance. One subject who presented with dilated cardiomyopathy at 17 years of age had one rare variant on each TTN allele: a previously described pathogenic missense mutation9 and a nonsense variant inherited from her mother (Fig. 2 in the Supplementary Appendix). In all other families, there was coinheritance of dilated cardiomyopathy and a single TTN frame-shift or nonsense variant. Among the 32 family members who were more than 40 years of age and for whom data were available, the penetrance of TTN truncating mutations was more than 95%.
Splicing Variants
We identified 17 TTN variants in 19 subjects with dilated cardiomyopathy that are predicted to alter RNA splicing, including 11 that altered absolutely conserved splice-site nucleotides (Table 1, and Table 7 in the Supplementary Appendix). RNA sequencing of cardiac tissue specimens from two subjects with splicing variants (one an adult child from a consanguineous marriage with a homozygous splicing variant [data not shown], the other described in Fig. 3 in the Supplementary Appendix) confirmed aberrant TTN splicing. Among subjects studied with the use of next-generation sequencing, we observed significant enrichment in splicing variants in the subjects with dilated cardiomyopathy in group A (with 15 of 92 subjects [16%] affected) and group B (with 3 of 71 subjects [4%] affected) as compared with the subjects with hypertrophic cardiomyopathy (with only 1 of 231 subjects [<1%] affected, P = 7×10−8) and the control subjects (with 5 of 249 subjects [2%] affected, P = 9×10−6). We found complete cosegregation of splicing variants and dilated cardiomyopathy (lod score, 1.8, indicating an association that was 60 times as likely as that due to chance) among 11 members of three families (Fig. 2 and Table 13 in the Supplementary Appendix).
Copy-Number Variants
We assessed copy number across TTN in the subjects with dilated cardiomyopathy in groups A and B by comparing the distribution of sequence reads among subjects (Fig. 4 in the Supplementary Appendix). The traditional dideoxy sequencing used for samples from the subjects with dilated cardiomyopathy in group C did not permit these analyses. One copy-number variant was identified in a subject with dilated cardiomyopathy. This tandem insertion of 28 kb (spanning introns 71 to 124) was predicted to incorporate a 13% internal duplication of a portion of titin; its presence was confirmed in the proband, and it was absent in two healthy relatives (Fig. 5 in the Supplementary Appendix). No TTN copy-number variations were observed among the subjects with hypertrophic cardiomy-opathy or the control subjects (480 in all).
CONSEQUENCES OF TTN TRUNCATING VARIATIONS
Because TTN nonsense, frameshift, splicing, and copy-number variants that are predicted to substantially alter titin structure were significantly more frequent among subjects with dilated cardiomyopathy than among subjects with hypertrophic cardiomyopathy (P = 3×10−16) or controls (P = 9×10−14) and were coinherited with dilated cardiomyopathy in families (combined lod score, 11.1), we concluded that these variants cause dilated cardiomyopathy. Six TTN mutations were each present in two subjects; results of analyses of one subject pair were consistent with a shared haplo-type (data not shown).
To conservatively estimate the frequency of truncating TTN mutations in subjects with dilated cardiomyopathy, we increased our population of subjects with dilated cardiomyopathy in group A by adding 40 subjects who were recruited concurrently with the other subjects in the group and whose TTN sequences were not analyzed because studies revealed a pathogenic mutation in another dilated cardiomyopathy gene. After this addition, the frequencies of TTN truncating mutations among the subjects with dilated cardiomyopathy in groups A and B were 28% and 24%, respectively (P = 0.74). Subjects in group C, whose samples were studied with the use of a different DNA-sequencing platform, had significantly fewer TTN mutations (9%, P<0.001). The frequency of TTN mutations did not differ significantly between subjects with and those without a family history of dilated cardiomyopathy (P = 0.36) (Table 3 in the Supplementary Appendix).
TTN truncating mutations found in subjects with dilated cardiomyopathy were nonrandomly distributed within titin (Fig. 1): they were over-represented in the A-band region as compared with either the remainder of N2BA (P=0.0004) or N2B (P = 0.01) and were absent from the Z-disk and M-band regions (P = 0.006 and P = 0.001 for the comparison with N2BA and with N2B, respectively). The spatial distribution of the 10 variants found in the subjects with hypertrophic cardiomyopathy and the control subjects (Fig. 1) was distinct from that of the variants found in the subjects with dilated cardiomyopathy (P = 0.001). The variants in the subjects with hypertrophic cardiomyopathy and the control subjects were less enriched for the A-band region of titin (40% vs. 84% for mutations in the subjects with dilated cardiomyopathy, P = 0.006) and included Z-band variants (20% vs. 0%, P = 0.01).
CLINICAL CHARACTERISTICS
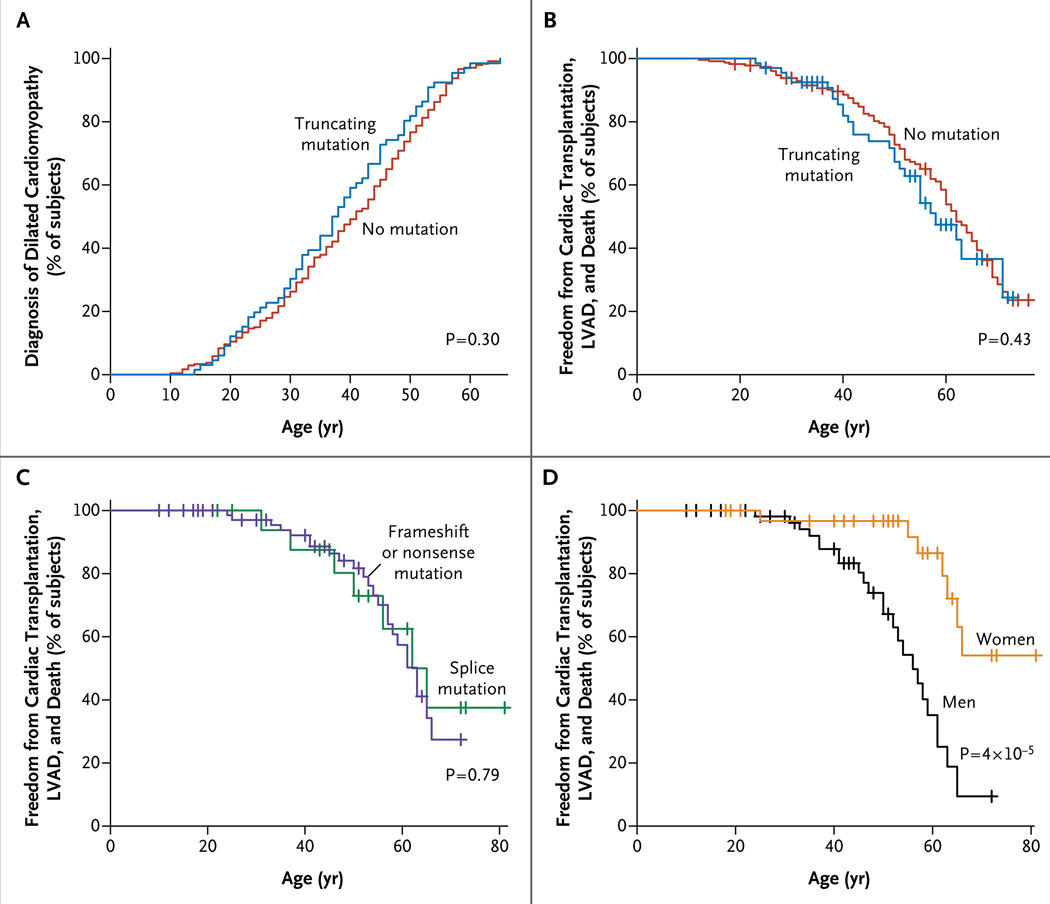
There were no significant differences between subjects with and those without TTN truncating mutations with respect to the age at diagnosis, left ventricular end-diastolic dimensions, ejection fraction, or rates of cardiac transplantation, implantation of a left ventricular assist device, and death from cardiac causes (Table 2 and Fig. 2A and 2B).
Table 2.
Characteristics of Subjects with Dilated Cardiomyopathy, According to the Presence or Absence of TTN Mutations.
| Characteristic | Group A | Group B | Group C | |||
|---|---|---|---|---|---|---|
| Mutation (N = 37) |
No Mutation (N = 55) |
Mutation (N = 17) |
No Mutation (N = 54) |
Mutation (N = 13) |
No Mutation (N = 136) |
|
| Female sex — no. (%) | 11 (30) | 17 (31) | 2 (12) | 14 (26) | 4 (31) | 56 (41) |
| Familial dilated cardiomyopathy — % | 92 | 79 | 21 | 24 | 85 | 66 |
| Age — yr | 45.9±13.6 | 46.7±14.3 | 48.4±11.0 | 51.7±12.8 | 49.2±12.8 | 49.7±14.0 |
| Age at diagnosis — yr | 37.1±13.2 | 37.3±14.6 | 38.5±9.8 | 41.3±13.4 | 37.6±14.7 | 39.6±12.3 |
| LVEF — % | 26.4±10.8 | 30.7±11.6 | 24.6±11.0 | 24.8±11.7 | 29.3±8.5 | 31.4±11.8 |
| Fractional shortening — % | 19.6±9.9 | 19.6±10.2 | NA | NA | 13.5±3.4 | 16.6±6.9 |
| LVEDD | ||||||
| Absolute diameter — mm | 63.7±10.9 | 61.4±9.7 | 70.8±8.1 | 74.2±10.2 | 65.5±9.2 | 65.6±10.4 |
| Relative diameter — mm/cm2 of body-surface area | 33.0±5.1 | NA | 37.6±6.0 | NA | 34.0±7.1 | NA |
| NYHA III or IV — % | 62 | 41 | 94 | 91 | 46 | 43 |
| ICD — no. (%) | 22 (59) | 30 (55) | 1 (6) | 2 (4) | NA | NA |
| Cardiac transplantation, implantation of VAD, or death from cardiac causes — no. (%) |
14 (38) | 17 (31) | 17 (100) | 47 (87) | 1 (8) | 31 (23) |
Plus–minus values are means ±SD. The values for left ventricular ejection fraction (LVEF), fractional shortening, and left ventricular end-diastolic diameter (LVEDD) are based on echocardiographic measurements. ICD denotes implantable cardioverter–defibrillator, NA not available, NYHA New York Heart Association functional class, and VAD ventricular assist device.
Figure 2. Kaplan–Meier Estimates of Age at Diagnosis and Clinical Progression in Subjects with Dilated Cardiomyopathy Caused by TTN Mutations.
Panel A shows data for the time of diagnosis, and Panels B, C, and D show freedom from cardiac transplantation, implantation of left ventricular assist device (LVAD), and death from cardiac causes. Panels A and B show data for the 67 subjects who had mutations and the 228 who did not. Panels C and D show data for the 94 subjects (from 19 families) who had truncating mutations (Fig. 2 in the Supplementary Appendix; also see the Supplementary Appendix for a description of the effect of sex). Hatch marks indicate subjects with censored data.
The diverse clinical manifestations of dilated cardiomyopathy were reflected in subjects in groups A, B, and C. Subjects in group A were more likely to have familial disease than subjects in group B (P = 6×10−13) or group C (P = 0.005). Subjects in group B (whose clinical features were ascertained during evaluation for cardiac transplantation) were more likely to have severe dilated cardiomyopathy than subjects in groups A and C, with significantly increased left ventricular end-diastolic diameter (P = 1×10−9 and P = 3×10−6, respectively) and a lower left ventricular ejection fraction (P = 0.03 and P = 0.0001, respectively) (Table 2, and Fig. 1 in the Supplementary Appendix).
Rates of cardiac transplantation, implantation of a left ventricular assist device, and death from cardiac causes among subjects and family members with TTN mutations did not appear to be influenced by mutation type but were influenced by the sex of the subjects (Fig. 2C and 2D). The mean (±SD) age at the time of these adverse events in 94 mutation carriers within 19 families was 68±5 years for the 33 women and 56±3 years for the 61 men (P = 4×10−5) (Fig. 2 in the Supplementary Appendix).
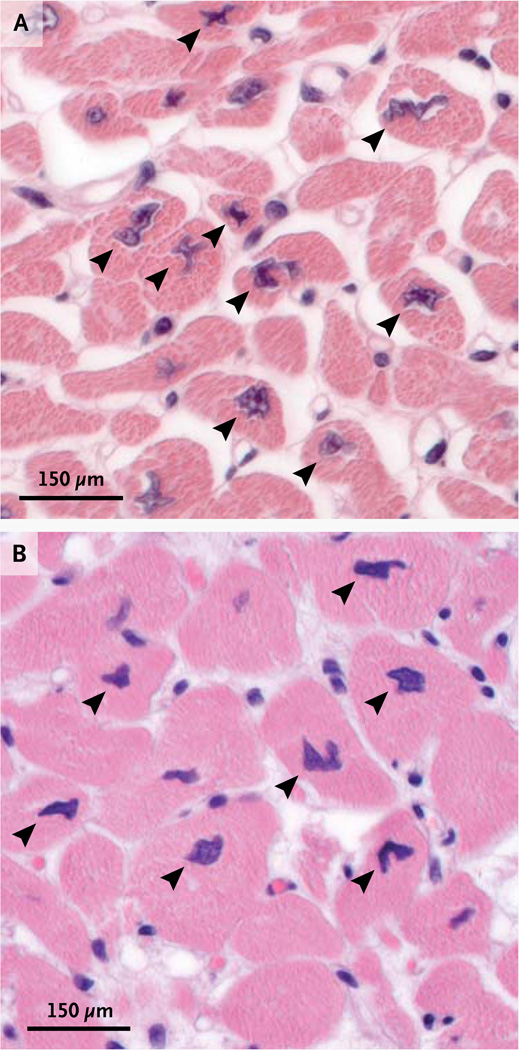
Subjects with TTN truncating mutations had dilated cardiomyopathy that was usually unaccompanied by conduction system or skeletal-muscle disease (Tables 9, 10, and 11 in the Supplementary Appendix), although overt skeletal myopathy occurred in one subject with a homozygous TTN splicing mutation. Histopathological characteristics of cardiac specimens from subjects with TTN truncating mutations were typical of idiopathic dilated cardiomyopathy. Some sections revealed foci of myocytes with bizarre, stellate nuclear morphology, best appreciated in cross section (Fig. 3). Electron-microscopical examination of one specimen obtained at autopsy showed intact sarcomeric structures (Fig. 6 in the Supplementary Appendix).
Figure 3. Histopathological Abnormalities in Cardiac Specimens from Two Subjects with TTN Truncating Mutations.
Specimens from the cardiac interventricular septum in two subjects (Panel A and Panel B, hematoxylin and eosin) show myocyte nuclei with abnormal morphologic characteristics (arrowheads).
DISCUSSION
We identified TTN truncating variants in 67 subjects with dilated cardiomyopathy, 3 subjects with hypertrophic cardiomyopathy, and 7 controls (Tables 1 and 2, and Tables 3 through 7 in the Supplementary Appendix). TTN variants included nonsense and frameshift mutations predicted to cause protein truncation, variants of splice donor or acceptor sites predicted to cause exon skipping or to include intronic sequence or delete exonic sequence, and a large tandem insertion. We posit that these mutant alleles produce shortened titin with abnormal properties that cause dilated cardiomyopathy.
The frequency of TTN truncating variants identified by next-generation sequencing of samples from subjects with hypertrophic cardiomyopathy and controls was similar (1% and 3%, respectively; P = 0.34). In addition, each subject with hypertrophic cardiomyopathy who had a TTN variant also had a pathogenic mutation in an established hypertrophic cardiomyopathy gene, suggesting that TTN truncations rarely, if ever, cause hypertrophic cardiomyopathy.
TTN truncating mutations were more common in subjects with dilated cardiomyopathy than in subjects with hypertrophic cardiomyopathy (P = 3×10−16) or controls (P = 9×10−14). Among the subjects with dilated cardiomyopathy, more TTN truncations were identified in those studied with the use of next-generation sequencing (groups A and B) than dideoxy sequencing (group C; P<0.001). Despite clinical differences (including more familial disease in group A and more severe dilated cardiomyopathy in group B) (Table 2, and Fig. 1 in the Supplementary Appendix), the frequency of TTN truncating mutations was similar in groups A and B (28% and 24%, respectively; P = 0.74). The reduced frequency of mutations identified in the subjects in group C raises the possibility that the detection of mutations was better with the next-generation sequencing strategy than with dideoxy sequencing.
TTN truncating mutations and dilated cardiomyopathy were coinherited in families (combined lod score, 11.1) (Fig. 2 and Table 13 in the Supplementary Appendix). Segregation analyses of frame-shift or nonsense mutations in 17 families (lod score, 9.3) and splice-site mutations in 3 families (lod score, 1.8) confirmed the coinheritance of each type of truncating mutation. Family studies also showed that the penetrance of TTN truncating mutations was more than 95% for the subjects who were more than 40 years of age (Fig. 2 and Table 12 in the Supplementary Appendix).
Among the subjects with dilated cardiomyopathy, those with and those without TTN truncating mutations had similar clinical manifestations and similar morbidity and mortality, but men with TTN mutations had adverse events at significantly earlier ages than did women (P = 4×10−5). Sex is reported to influence outcomes in heart failure of various causes.36 However, the suggestion that sex would substantially influence an autosomal monogenic cause of heart failure is unexpected and warrants further study.
Mutations that substantially disrupt the structure of full-length titin might cause dilated cardiomyopathy by means of several mechanisms. RNA- and protein-surveillance pathways most likely degrade some truncated titin peptides.37 Decreased titin levels could limit sarcomere formation and might cause cardiac dysfunction and remodeling. Yet this is not the case for previously reported TTN mutations that delete only part of the M-band portion of titin14 (Fig. 1, and Table 14 in the Supplementary Appendix); immunohisto-chemical studies showed that some of these carboxy-terminal truncated titin proteins are integrated into the sarcomere and cause recessive, early-onset skeletal and cardiac myopathy, not dominant dilated cardiomyopathy. In addition, if more proximal TTN truncating mutations caused dilated cardiomyopathy through haploinsufficiency, the distribution of such mutations would be rather uniform across the susceptible portion of titin. In contrast, we observed a skewed mutation distribution in subjects with dilated cardiomyopathy, which was distinct from that observed in subjects without the disease (Fig. 1). This unequal mutation distribution may indicate that truncated titin proteins found in subjects with dilated cardiomyopathy, like previously studied carboxy-terminal titin truncations, are integrated into the sarcomere and cause dilated cardiomyopathy by means of a dominant negative mechanism.
If truncated titin proteins in subjects with dilated cardiomyopathy were incorporated into the sarcomere, they would most likely be anchored at the Z-line and would interact with the full complement of Z-disk factors (Fig. 1). However, these truncated titin proteins would not include the M-band residues that anchor titin to the middle of the sarcomere through myomesin, encode a kinase domain, and interact with many proteins.38 The M-band portion of titin is implicated in sensing and modulating sarcomeric force.12,39–41 We suggest that the loss of these interactions could lead to dilated cardiomyopathy through the disturbance of the normal regulation of sarcomeric force.
We conclude that TTN truncating mutations are the most common known genetic cause of dilated cardiomyopathy. Ongoing analyses of other classes of TTN variation (e.g., missense variants) may expand our understanding of the importance of TTN in the pathogenesis of dilated cardiomyopathy. Incorporation of next-generation sequencing analyses of TTN into clinical genetic screens should increase the detection of dilated cardiomyopathy mutations by approximately 50%, permitting earlier diagnosis and interventions to prevent disease progression. Further study of the functional consequences of TTN truncating mutations on myocardial physiological features and myocyte signaling is warranted.
Supplementary Material
Acknowledgments
Supported by grants from the Howard Hughes Medical Institute (to Dr. C.E. Seidman); the National Institutes of Health (to Drs. Herman, Taylor, Graw, Ackerman, Mitchell, Murry, Ho, Mestroni, and J.G. Seidman); the Leducq Foundation (to Drs. Cook, J.G. Seidman, and C.E. Seidman); the American Heart Association and the Muscular Dystrophy Association (to Drs. Taylor and Mestroni); the U.K. National Institute for Health Research Cardiovascular Biomedical Research Unit (Royal Brompton and Harefield National Health Service Foundation Trust and Imperial College), the British Heart Foundation, and the Medical Research Council U.K. (to Drs. Banner, Pennell, Cook, and Barton); and the J. Ira and Nicki Harris Family Research Award (to Dr. Ho). Sequencing in subjects with dilated cardiomyopathy in group C was funded by a contract from the National Heart, Lung, and Blood Institute (N01-HV-48194).
We thank Lynne Stevenson, Michael Givertz, and Eldrin Lewis (Brigham and Women’s Hospital); Joshua Gorham, Tatjana Levi, Steven McCarroll, Lee Rodman, Adam Saltzman, and Mei Zhu (Harvard Medical School); John Smith (Harefield Hospital Transplantation Tissue Typing Laboratory); Paula Rogers (Brompton and Harefield Biomedical Research Unit); and G. Kees Hovingh (Department of Vascular Medicine, Academic Medical Center, Amsterdam).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Morita H, Rehm HL, Menesses A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. doi: 10.4065/83.6.630. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 4.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardio-myopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 6.Baig MK, Goldman JH, Caforio AL, Coonar AS, Keeling PJ, McKenna WJ. Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J Am Coll Cardiol. 1998;31:195–201. doi: 10.1016/s0735-1097(97)00433-6. [DOI] [PubMed] [Google Scholar]
- 7.Mestroni L, Rocco C, Gregori D, et al. Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity. J Am Coll Cardiol. 1999;34:181–190. doi: 10.1016/s0735-1097(99)00172-2. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman RS, Cox S, Lakdawala NK, et al. A novel custom resequencing array for dilated cardiomyopathy. Genet Med. 2010;12:268–278. doi: 10.1097/GIM.0b013e3181d6f7c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siu BL, Niimura H, Osborne JA, et al. Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation. 1999;99:1022–1026. doi: 10.1161/01.cir.99.8.1022. [DOI] [PubMed] [Google Scholar]
- 10.Gerull B, Atherton J, Geupel A, et al. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J Mol Med. 2006;84:478–483. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- 11.Gerull B, Gramlich M, Atherton J, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 12.Lange S, Xiang F, Yakovenko A, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 13.Hackman P, Vihola A, Haravuori H, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmignac V, Salih MA, Quijano-Roy S, et al. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann Neurol. 2007;61:340–351. doi: 10.1002/ana.21089. [DOI] [PubMed] [Google Scholar]
- 15.Hackman P, Marchand S, Sarparanta J, et al. Truncating mutations in C-terminal titin may cause more severe tibial muscular dystrophy (TMD) Neuromuscul Disord. 2008;18:922–928. doi: 10.1016/j.nmd.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Trinick J, Knight P, Whiting A. Purification and properties of native titin. J Mol Biol. 1984;180:331–356. doi: 10.1016/s0022-2836(84)80007-8. [DOI] [PubMed] [Google Scholar]
- 17.Granzier HL, Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- 18.Liversage AD, Holmes D, Knight PJ, Tskhovrebova L, Trinick J. Titin and the sarcomere symmetry paradox. J Mol Biol. 2001;305:401–409. doi: 10.1006/jmbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 19.van der Ven PF, Bartsch JW, Gautel M, Jockusch H, Furst DO. A functional knockout of titin results in defective myofibril assembly. J Cell Sci. 2000;113:1405–1414. doi: 10.1242/jcs.113.8.1405. [DOI] [PubMed] [Google Scholar]
- 20.Musa H, Meek S, Gautel M, Peddie D, Smith AJ, Peckham M. Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J Cell Sci. 2006;119:4322–4331. doi: 10.1242/jcs.03198. [DOI] [PubMed] [Google Scholar]
- 21.Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys J. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowits R, Kempner ES, Bisher ME, Podolsky RJ. A physiological role for titin and nebulin in skeletal muscle. Nature. 1986;323:160–164. doi: 10.1038/323160a0. [DOI] [PubMed] [Google Scholar]
- 23.Muhle-Goll C, Habeck M, Cazorla O, Nilges M, Labeit S, Granzier H. Structural and functional studies of titin’s fn3 modules reveal conserved surface patterns and binding to myosin S1 — a possible role in the Frank-Starling mechanism of the heart. J Mol Biol. 2001;313:431–447. doi: 10.1006/jmbi.2001.5017. [DOI] [PubMed] [Google Scholar]
- 24.Cazorla O, Wu Y, Irving TC, Granzier H. Titin-based modulation of calcium sensitivity of active tension in mouse skinned cardiac myocytes. Circ Res. 2001;88:1028–1035. doi: 10.1161/hh1001.090876. [DOI] [PubMed] [Google Scholar]
- 25.Guo W, Bharmal SJ, Esbona K, Greaser ML. Titin diversity — alternative splicing gone wild. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/753675. 753675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bang ML, Centner T, Fornoff F, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 27.Herman DS, Hovingh GK, Iartchouk O, et al. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6:507–510. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mestroni L, Maisch B, McKenna WJ, et al. Guidelines for the study of familial dilated cardiomyopathies: Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 29.Maron BJ, McKenna WJ, Danielson GK, et al. American College of Cardiology/European Society of Cardiology clinical expert consensus document on hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the European Society of Cardiology Committee for Practice Guidelines. J Am Coll Cardiol. 2003;42:1687–1713. doi: 10.1016/s0735-1097(03)00941-0. [DOI] [PubMed] [Google Scholar]
- 30.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 33.Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol. 2011 doi: 10.1002/0471142727.mb0412s94. Chapter 4:Unit4 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dec GW, Fuster V. Idiopathic dilated cardiomyopathy. N Engl J Med. 1994;331:1564–1575. doi: 10.1056/NEJM199412083312307. [DOI] [PubMed] [Google Scholar]
- 35.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 36.Jones RC, Francis GS, Lauer MS. Predictors of mortality in patients with heart failure and preserved systolic function in the Digitalis Investigation Group trial. J Am Coll Cardiol. 2004;44:1025–1029. doi: 10.1016/j.jacc.2004.05.077. [DOI] [PubMed] [Google Scholar]
- 37.Gramlich M, Michely B, Krohne C, et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009;47:352–358. doi: 10.1016/j.yjmcc.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gräter F, Shen J, Jiang H, Gautel M, Grubmüller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J, Raddatz K, Molkentin JD, et al. Cardiac hypertrophy and reduced contractility in hearts deficient in the titin kinase region. Circulation. 2007;115:743–751. doi: 10.1161/CIRCULATIONAHA.106.645499. [DOI] [PubMed] [Google Scholar]
- 41.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res. 2007;100:456–459. doi: 10.1161/01.RES.0000259559.48597.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.