Abstract
The Pharmacogenomics Knowledgebase (PharmGKB) is a resource that collects, curates, and disseminates information about the impact of human genetic variation on drug responses. It provides clinically relevant information, including dosing guidelines, annotated drug labels, and potentially actionable gene–drug associations and genotype–phenotype relationships. Curators assign levels of evidence to variant–drug associations using well-defined criteria based on careful literature review. Thus, PharmGKB is a useful source of high-quality information supporting personalized medicine–implementation projects.
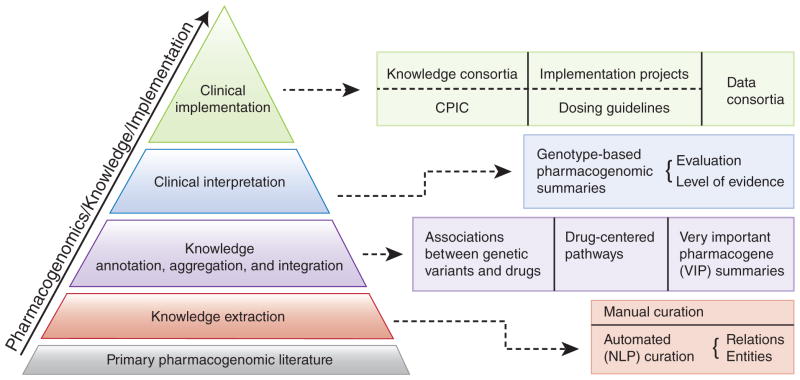
The PharmGKB (http://www.pharmgkb.org) is a publicly available Web-based knowledge base whose aim is to aid researchers in understanding how genetic variation among individuals contributes to differences in reactions to drugs. A visual summary of the data available and how these various types of information are integrated within the PharmGKB is provided in the PharmGKB Knowledge Pyramid (see Figure 1).
Figure 1.
The PharmGKB Knowledge Pyramid. CPIC, Clinical Pharmacogenetics Implementation Consortium; NLP, natural-language processing. From PharmGKB with the permission of PharmGKB and Stanford University. Copyright PharmGKB.
The foundation of the knowledge base is the primary pharmacogenetics and pharmacogenomics literature that is annotated, aggregated, and integrated in the form of (i) gene variant annotations, (ii) drug-centered pathways, and (iii) very important pharmacogene (VIP) summaries. Gene–drug–disease relationships are extracted from the literature using manual curation and natural language–processing techniques. Variant annotations are the core component of the knowledge in PharmGKB. Each variant annotation is based on a published article and describes the reported association between a single variant (single-nucleotide polymorphism or haplotype) and a drug phenotype. Multiple variant annotations may be created for a single publication if it reports multiple associations between variants and drugs. Key study parameters such as study size, population ethnicity, and statistics (e.g., P value and odds ratio) are recorded within each annotation.1 Diagrams and descriptions of drug-centered pathways depict the genes involved in the pharmacokinetics or pharmacodynamics of a particular drug and are based on published evidence. VIP gene summaries provide a concise overview of critical genes involved in drug response, with PharmGKB links to the literature, important variant details, noteworthy haplotypes, and relevant drugs.
Building on variant annotations, “clinical annotations” combine multiple-variant annotations into a single summary of the relevant variant–drug–phenotype association. For example, many studies have reported the relationship between the TPMT*3B variant (rs1800460) and adverse reactions to purine analogs. After gathering the relevant individual-variant annotations within the PharmGKB, we combine and summarize the associations from each publication to produce a single clinical annotation. Thus, each clinical annotation is linked to several PubMed identifiers that support the variant annotations and contain a summary for each genotype. In the case of TPMT*3B, the AA genotype contains two copies of the *3B variant and is associated with a significantly increased risk of side effects due to decreased enzyme levels; the AG genotype contains one copy of the *3B variant and is associated with a slightly increased risk of side effects due to moderately decreased enzyme levels; and the GG genotype contains no copies of the *3B variant and is not associated with increased risk of side effects (see Table 1).
Table 1.
PharmGKB criteria for levels of evidence
| Level | Criteria | Example |
|---|---|---|
| 1A | Annotation for a variant– drug combination in a CPIC- or medical society–endorsed pharmacogenomics guideline, or implemented at a PGRN site, or in another major health system | rs1800460 in TPMT (TPMT*3B)a and thiopurines: This association is published as a CPIC guideline and used in multiple clinics Drugs: azathioprine, mercaptopurine, purine analogues, thioguanine CC: Patients with the CC genotype may have a decreased, but not absent, risk for toxicity with thiopurine drugs and purine analogues as compared to patients with the CT or TT genotype. Patients with the CC genotype may still be at risk for toxicity when taking thiopurine drugs and purine analogues based on their genotype. Other genetic and clinical factors may also influence a patient’s risk for toxicity CT: Patients with the CT genotype may have an increased risk for toxicity with thiopurine drugs and purine analogues as compared to patients with the CC genotype. Other genetic and clinical factors may also influence a patient’s risk for toxicity TT: Patients with the TT genotype may have an increased risk for toxicity with thiopurine drugs and purine analogues as compared to patients with a CC genotype. Other genetic and clinical factors may also influence a patient’s risk for toxicityb |
| 1B | Annotation for a variant–drug combination in which the preponderance of evidence shows an association. The association must be replicated in more than one cohort with significant P values, and, preferably with a strong effect size | rs1801133 in mthfr and methotrexate: PharmGKB has multiple articles for this association with significant P values and several with high odds ratios Drug: methotrexate AA: Patients with the AA genotype with leukemia or lymphoma who are treated with methotrexate regimens may have an increased risk and increased severity of mucositis, as compared to patients with the GA or GG genotype. Other genetic and clinical factors may also influence a patient’s risk of oral mucositis AG: Patients with the AG genotype with leukemia or lymphoma who are treated with methotrexate regimens may have a decreased risk and decreased severity of mucositis as compared to patients with the AA genotype. Other genetic and clinical factors may also influence a patient’s risk of oral mucositis GG: Patients with the GG genotype with leukemia or lymphoma who are treated with methotrexate regimens may have a decreased risk and decreased severity of mucositis as compared to patients with the AA genotype. Other genetic and clinical factors may also influence a patient’s risk of oral mucositis |
| 2A | Annotation for a variant–drug combination that qualifies for level 2B, in which the variant is within a VIP as defined by PharmGKB where their functional significance is more likely known | rs12248560 in CYP2C19 and omeprazole: PharmGKB has multiple articles for this association, and it qualifies for level 2 and is in a pharmacogene Drug: omeprazole CC: Adult patients with the CC genotype who are treated with omeprazole may require a decreased dose as compared to patients with the TT genotype. Other genetic factors, including other CYP2C19 alleles *17 rs12248560, *2 rs4244285, *3 rs4986893, and clinical factors may also influence a patient’s required dose and should be taken into consideration. May not be applicable to pediatric patients CT: Patients with this genotype were not studied TT: Adult patients with the TT genotype who are treated with omeprazole may require an increased dose as compared to patients with the CC genotype. Other genetic factors, including other CYP2C19 alleles *17 rs12248560, *2 rs4244285,*3 rs4986893, and clinical factors may also influence a patient’s required dose and should be taken into consideration. May not be applicable to pediatric patientsd |
| 2B | Annotation for a variant–drug combination with moderate evidence of an association. The association must be replicated, but there may be some studies that do not show statistical significance, and/or the effect size may be small | rs2234922 in EPHX1 and carbamazepine: PharmGKB contains two articles with significant P values, one with no association reported; population sizes ranging from 70 to 234; no odds ratios reported Drug: carbamazepine AA: Patients with the AA genotype may require a decreased dose of carbamazepine as compared with patients with the AG or GG genotype. Other genetic and clinical factors may also influence dose of carbamazepine AG: Patients with the AG genotype may require an increased dose of carbamazepine as compared with patients with the AA genotype. Other genetic and clinical factors may also influence dose of carbamazepine GG: Patients with the GG genotype may require an increased dose of carbamazepine as compared with patients with the AA genotype. Other genetic and clinical factors may also influence dose of carbamazepinee |
| 3 | Annotation for a variant–drug combination based on a single significant (not yet replicated) study or annotation for a variant–drug combination evaluated in multiple studies but lacking clear evidence of an association | rs993648 in CERKL and iloperidone: PharmGKB contains a genome-wide association study article reporting a statistically significant association, but it is not replicated Drug: iloperidone CC: Patients with the CC genotype who are treated with iloperidone may have an increased risk for adverse cardiovascular events as compared with patients with the CT genotype. Other genetic and clinical factors may also influence a patient’s response CT: Patients with the CT genotype who are treated with iloperidone may have a decreased, but not absent, risk for adverse cardiovascular events as compared with patients with the CC or TT genotype. It is unclear at this time why the heterozygous genotype would confer a phenotype different from either homozygous genotype TT: Patients with the TT genotype who are treated with iloperidone may have an increased risk for adverse cardiovascular events as compared with patients with the CT genotype. Other genetic and clinical factors may also influence a patient’s responsef |
| 4 | Annotation based on a case report, nonsignificant study, or in vitro, molecular, or functional assay evidence only | rs61750900 in UGT2B10 and nicotine: PharmGKB contains two in vitro studies for this association Drug: nicotine GG: The GG genotype is not associated with changes in nicotine metabolism/clearance in human liver microsomes from subjects with the GG genotype and HEK293 overexpressing UGT2B10 GT: The GT genotype is significantly associated with a decrease in nicotine metabolism/clearance in human liver microsomes from subjects with the GT genotype as compared with subjects with the GG genotype TT: The TT genotype is significantly associated with a decrease in nicotine metabolism/clearance in human liver microsomes from subjects with the TT genotype as compared with subjects with the GG genotype and HEK293 overexpressing UGT2B10 variant constructg |
CPIC, Clinical Pharmacogenetics Implementation Consortium; PGRN, Pharmacogenomics Research Network; PharmGKB, the Pharmacogenomics Knowledgebase; VIP, very important pharmacogene.
PharmGKB reports alleles on the positive chromosomal strand.
Adapted from PharmGKB with the permission of PharmGKB and Stanford University. Copyright PharmGKB.
The level of risk for any given genotype is reported in a relative fashion as compared with other genotypes. For example, the AA genotype is associated with an increased risk of side effects as compared with the AG and GG genotypes—but not necessarily at an increased risk of side effects for patients on the drug in general, as this would depend on a detailed examination of the target-population allele frequencies and the populations on which the original US Food and Drug Administration approval is based. Thus, we report risk relative to other genotypes because the incidence of efficacy or adverse events for any given drug has usually not been quantified. Also, the distribution of genotypes for any given population is often not available. Although many groups use HapMap frequencies to calculate population major alleles or typical genotypes, the HapMap populations are small and ethnically very specific. Their frequencies do not necessarily represent larger population frequencies (nor were they meant to). Therefore, we do not report the risk of a particular drug response as compared with “normal” because normal is typically not well defined. We report the risk as compared with other possible genotypes.
Each clinical annotation is assigned a “level of evidence” score that is a measure of confidence in the association as determined by the PharmGKB curators. This score is based on several criteria, including replication of the association, P value (after correction for multiple-hypothesis testing, if applicable), and odds ratio, if available. Table 1 describes the four levels of evidence and the criteria for each, with an example from the knowledge base. Levels 1 and 2 are divided into A and B subtypes. Level 1 annotations involve a variant–drug combination in which the preponderance of evidence shows an association. The association must be replicated in more than one cohort with significant P values and, preferably, with a strong effect size. Level 1A annotations are associations for which PharmGKB staff is also aware of clinical implementation tests or deployments. Level 2 annotations are for variant–drug combinations with moderate evidence of an association. The association for level 2 annotations must be replicated but may include negative studies as well. Level 2A annotations are those that involve PharmGKB VIP genes, and thus are particularly well documented.
Level 3 annotations are based on a single significant (not yet replicated) study or annotation for a variant–drug combination evaluated in multiple studies but lacking clear evidence of an association. Level 4 annotations are based on a case report; on a study that did not achieve significance but is biologically plausible; or on in vitro, molecular, or functional assay evidence. In cases in which the only literature evidence available is that there is no association, no clinical annotation is written. The lack of evidence for an association can be important in a research setting, and so although variant annotations are created in these situations, they are not summarized as clinical annotations because they would have no clinical utility to a doctor, pharmacist, patient, or direct-to-consumer genotyping customer. Curators use their experience and judgment when assigning evidence levels. They may use their discretion to move the level up or down for a particular annotation, and the change is typically discussed as a team and recorded in the annotation. The PharmGKB welcomes input from the scientific and clinical community (e.g., via feedback@pharmgkb.org) regarding an assigned level of evidence for a specific clinical annotation.
All annotations are available in tab-delimited files sent from PharmGKB after execution of the Data Usage Agreement. The supporting information for variant annotations (e.g., P value, study size, and odds ratio) is included. All approved users can then reevaluate the PharmGKB clinical annotations using their own criteria and rankings. This is critical because various implementation research programs, such as the 1200 Patients Project2 and the University of Florida and Shands Personalized Medicine programs3 may use the PharmGKB clinical annotations in different detailed ways in making decisions regarding pharmacogenomics variants to use.
The PharmGKB is focused on pharmacogenomics knowledge and implementation. The clinical implementation of pharmacogenomics represents the top level of the PharmGKB knowledge pyramid (Figure 1). PharmGKB supports several clinically relevant projects, including the data-sharing consortia and several implementation projects. For example, the Clinical Pharmacogenetics Implementation Consortium (CPIC) provides drug-dosing guidelines based on an individual’s genotype if genetic information is already available.4 CPIC guidelines help clinicians understand how available genetic test results can be used to optimize drug therapy, rather than whether tests should be ordered. Key assumptions underlying the CPIC guidelines are that clinical high-throughput and preemptive (pre-prescription) genotyping will become more widespread, and that clinicians will be faced with having patients’ genotypes available even if they have not explicitly ordered a test with a specific drug in mind.
Pharmacogenetics and pharmacogenomics are at a critical juncture. As the field moves from the bench to clinical implementation, it requires a high-quality and reliable source of up-to-date information about human genetic variation and its impact on drug response. The PharmGKB is the preeminent resource for enabling clinicians and translational researchers to implement pharmacogenomic knowledge in the context of personalized medicine.
Acknowledgments
We thank the members of the PharmGKB Scientific Advisory Board and Peter O’Donnell (University of Chicago) for useful feedback. In particular, we acknowledge in-depth discussions with Julie Johnson (University of Florida). The PharmGKB is supported by National Institutes of Health grant R24 GM061374.
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark Med. 2011;5:795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donnell PH, et al. The 1200 Patients Project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–449. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012;92:437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]