Abstract
The Wnt signal transduction pathway is dysregulated in many highly prevalent diseases, including cancer. Unfortunately, drug discovery efforts have been hampered by the paucity of targets and drug-like lead molecules amenable to drug discovery. Recently, we reported the FDA-approved anthelmintic drug niclosamide inhibits Wnt/β-catenin signaling by a unique mechanism, though the target responsible remains unknown. We interrogated the mechanism and structure-activity relationships to understand drivers of potency and to assist target identification efforts. We found inhibition of Wnt signaling by niclosamide appears unique among the structurally-related anthelmintic agents tested and found the potency and functional response was dependent on small changes in the chemical structure of niclosamide. Overall, these findings support efforts to identify the target of niclosamide inhibition of Wnt/β-catenin signaling and the discovery of potent and selective modulators to treat human disease.
Keywords: Wnt signaling inhibitor, Wnt signaling activator, Small molecule, Mechanism of action, Target identification, Niclosamide
The Wnt signaling pathway is a key signal transduction pathway involved in tissue development and homeostasis, stem cell maintenance and renewal, and is dysregulated in many cancers 1-3. The pathway is of immense importance in biology and holds promise in the treatment of a number of highly prevalent diseases 4, 5. For example, in colorectal cancer (CRC) more than 80% of all sporadic and hereditary cancers show hyperactivation of the pathway due to mutations in the adenomatous polyposis coli (APC) or the β-catenin gene 1, 2. As a result, inhibitors of the pathway are highly sought-after as the basis of a new generation of targeted therapeutic agents.
A mechanistic understanding of the canonical Wnt/β-catenin signaling pathway has evolved over the past decade6. Briefly, Wnt proteins are secreted glycoproteins that bind and activate the seven transmembrane receptor Frizzled. Wnt binding to Frizzled results in activation of cytosolic proteins called Dishevelled, leading to internalization of the Frizzled receptor7, 8. Downstream signaling events resulting from Wnt binding include the stabilization and translocation of cytosolic β-catenin proteins into the nucleus, activation of the transcription factor LEF/TCF and transcription of Wnt/β-catenin target genes. Overall, the Wnt signal transduction pathway consists of many protein-protein interactions that traditionally have been difficult to target with small drug-like molecules 1, 9. The barrier to progress in the field stems from the lack of targets in the pathway amenable to the design of small drug-like molecules, good chemical starting points, and a gap in our ability to design drugs that penetrate cells and target intracellular protein-protein interactions. Recent publications, including our own, have begun to provide insights into mechanisms and chemical starting points 1, 10-30 though no approved drugs specifically targeting the pathway have been identified 1, 2, 30.
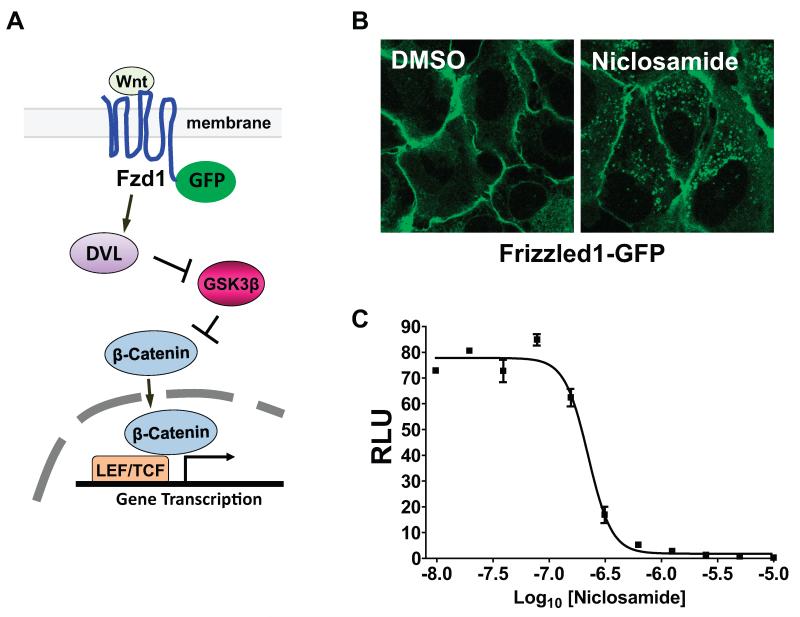
Reasoning plasma membrane proteins play critical roles in signal transduction and offer good targets for therapeutic intervention to treat human disease, we developed a novel cell-based high-throughput screening assay capable of detecting inhibitors or activators of Wnt signaling using a chimeric Frizzled1–GFP receptor (Figure 1) 7, 10. In this assay, called the Frizzled internalization assay, a U2OS cell line stably expressing Frizzled1-GFP (Fzd1GFP-U2OS) localizes Frizzled1-GFP predominantly to the plasma membrane with almost no internalized vesicles present until stimulated with Wnt ligands. Upon stimulation and visualization with confocal microscopy, a punctate pattern is observed in cells indicative of receptor translocation. Upon screening in 384-well format, we discovered the FDA-approved anthelmintic drug Niclosamide inhibits Wnt/β-catenin signaling 10, a finding since confirmed by others 31, 32. Niclosamide produces robust internalization of the receptor (Figure 1B), and despite expectations that internalization of Frizzled receptor by Niclosamide would activate the pathway, Niclosamide inhibits the pathway by decreasing levels of cytosolic dishevelled2 and β-catenin, and decreasing Wnt/β-catenin gene transcription (see Figure 1C) 10. Though the specific molecular target of Niclosamide remains unknown, a number of studies now indicate the novel mechanism of Niclosamide-mediated inhibition of Wnt/β-catenin signaling can be exploited to treat CRC 31-33. For example, preclinical studies using well-known human colon cancer cell lines and patient derived colorectal cancer cells isolated from surgically resected metastatic disease, as well as studies using patient derived tumor explants in immunodeficient mice all indicate Niclosamide decreases dishevelled and β-catenin levels and inhibits Wnt/β-catenin signaling even in the presence of APC mutations; findings which strongly support its use to clinically evaluate its efficacy to treat CRC 33.
Figure 1. Niclosamide promotes Frizzled1-GFP internalization and inhibits Wnt3A-induced Wnt/β-catenin signaling.
(A) Illustration of Frizzled-GFP internalization assay with canonical Wnt signaling. (B) Confocal image of U2OS cells harboring Frizzled1-GFP treated with DMSO or Niclosamide (12.5 μM) for 6 hrs. Punctuate structure is the internalized vesicles of Frizzled1-GFP. (C) Niclosamide inhibits the activity of Wnt3A-induced TOPflash reporter assay. RLU: relative luciferase unit.
Niclosamide is a member of the salicylanilide class of anthelmintic agents and is currently listed on the World Health Organization’s list of essential medicines 34. It has been used as an anthelmintic agent in livestock since the early 1960s and was approved by the FDA for use in humans to treat tapeworm infections in 1982 35, 36. Niclosamide exerts its anthelmintic effect via a mechanism involving uncoupling of oxidative phosphorylation, though no specific target has been identified and the mechanism has not been well-delineated 37, 38. In the years since its discovery, additional biological activities have been reported 31, 32, 39-51. However, in nearly every case, the interaction of Niclosamide with a specific molecular target was not identified.
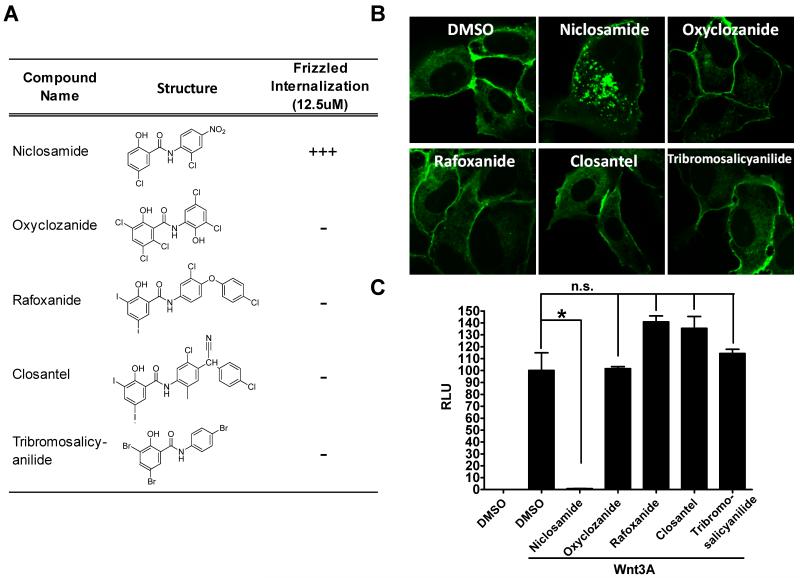
Given its intriguing mechanism and promise in treating CRC 10, 33, we initiated pilot studies to interrogate the mechanism and structure-activity relationships of the chemotype with the goal of enabling target identification studies and assessing the potential to optimize the chemical series for CRC clinical studies. To better understand the mechanism of Wnt inhibition, we acquired commercially available anthelmintic compounds structurally-related to Niclosamide - all in the same salicylanilide drug class and all with reported mechanisms of action involving uncoupling of oxidative phosphorylation 36, 38. We then tested these anthelmintic agents side-by-side in the Frizzled internalization assay and in the Wnt/β-catenin gene transcription assay (TOPflash). With the exception of Niclosamide, none of the compounds tested produced a robust punctate pattern indicative of Frizzled internalization nor did they inhibit Wnt3A-stimulated gene transcription in the Wnt/β-catenin TOPflash gene transcription assay (see Figure 2). Given these compounds are all anthelmintic agents and share a common mechanism of action, these findings suggest the mechanism responsible for Niclosamide inhibition of Wnt signaling appears different from its mechanism of anthelmintic activity.
Figure 2. Structures of anthelmintic derivatives tested and their activity in the Frizzled1-GFP internalization and the Wnt3A-stimulated TOPflash β-catenin reporter assay.
(A) Chemical structures of salicylanilide anthelmintics tested and amount of punctate observed after 6 hours with different anti-helminthic compounds at 12.5 μM in the Frizzled1-GFP internalization assay. Punctate observed in confocal images of Frizzled1-GFP USOS cells were scored visually and with Metamorph software (Molecular Devices, LLC). 10 to 20 cells per sample were analyzed to reach statistical significance; (B) Confocal images of Frizzled1-GFP U2OS stable cells treated with different anti-helminthic compounds at 12.5 μM for 6 hours; and (C) Wnt-3a stimulated TOPflash reporter activity of salicylanilide anthelmintics. HEK293 cells harboring TOPflash reporter were treated with DMSO (control) or with Wnt3A- conditional medium and different anthelmintic compounds at 12.5 μM for 8 hours. RLU: relative luciferase unit. n.s., not significant (P > 0.05); *, P < 0.05 (t tests).
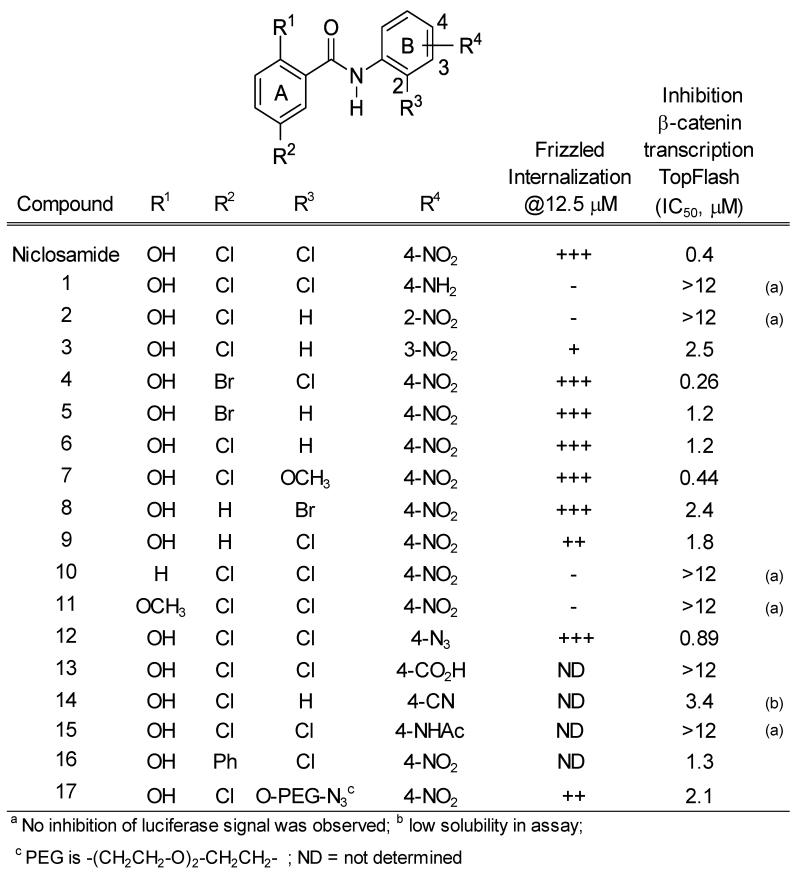
Intrigued that a mechanism with a novel target may exist, we purchased and synthesized focused libraries of compounds to conduct pilot SAR studies that could enable target identification and lead optimization efforts (Figure 3) 52 (see also supplemental information). In all cases studied, we found a good correlation between Frizzled1-GFP receptor internalization and inhibition of Wnt3A-stimulated inhibition in the TOPflash assay (See Supplemental Fig.1 for confocal images). Given this correlation, we focused on the TOPflash assay to develop the SAR. In the A-ring, methylation of the hydroxyl group (R1) (compound 11) or substitution of the hydroxyl group with an H-atom (compound 10) yielded a loss of activity in Wnt/β-catenin signaling. Substituting the 4-chloro substituent (R2) with a hydrogen atom (compound 8 and 9) generally resulted in a slight decrease in activity, while substitution by bromine produced a compound with slightly greater potency (compound 4). In the B-ring, substitution of the 2-chloro group (R3) with a hydrogen atom at the B-2 position produced an active compound with a slight loss in activity (compound 6), while replacement by a methoxy group produced a compound of similar potency (compound 7). The overall SAR at this position indicated inhibitory potency was fairly insensitive to substitution and suggested to us that substitution at this position with a peg-ether tethering group may provide access to affinity reagents to help identify the target. In the event, we were pleased to find that a peg-substituted derivative useful to prepare affinity reagents for target identification studies resulted in an active inhibitor (compound 17).
Figure 3. Structure-Activity Relationships in the Frizzled1-GFP internalization and the TOPflash β-catenin reporter assays.
Punctate produced by derivatives in Frizzled1-GFP U2OS stable cell line were evaluated by confocal imaging and visually scored by the amount of punctate observed: Comparable to control (−); trace (+); moderate (++); strong (+++); or ND (Not Determined).
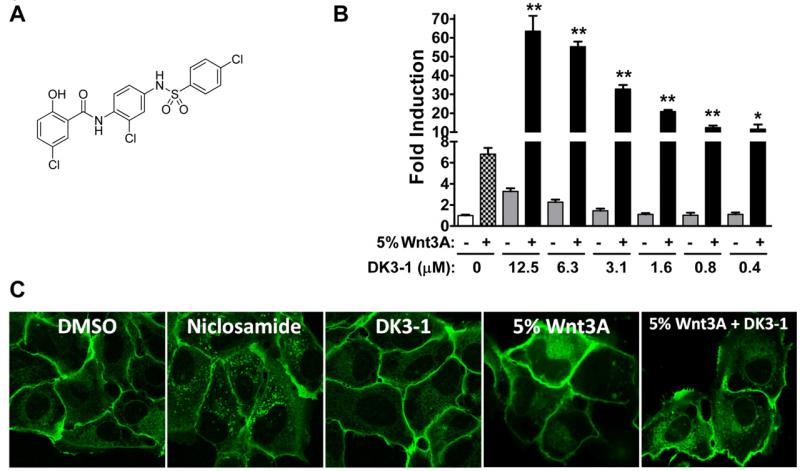
The SAR at the para position in the B-ring was particular interesting (Figure 3). Here, a substituent change within the chemotype not only effected the potency of inhibition but could also change the inhibitory functional response to Wnt3A stimulation to an activation functional response. A dramatic loss of inhibitory potency resulted when the nitro group at B-4 (R4) in the anilide ring was substituted by an amine (compound 1) or an acetamide (compound 15). Substitution with cyano (compound 14) resulted in comparable activity to the similarly substituted nitro derivative (compound 6). To our delight, replacement of the nitro group with an azide group that could provide access to photo-affinity reagents and assist target and binding site identification produced an active compound (compound 12). Moving the nitro group to the ortho or meta position resulted in reduced activity (compound 2 and 3, respectively). Remarkably, substitution of the nitro group with an aryl sulfonamide (DK3-1) produced the opposite functional response in the TOPflash assay and increased β-catenin transcriptional activity (Figure 4). This change in functional response was interesting. Upon further evaluation, we found that DK3-1 synergistically increased β-catenin gene transcription in a dose dependent manner in the presence of Wnt3A-ligand (Figure 4B)53. Compound DK3-1 alone does not appear to increase the punctate pattern of Frizzled1-GFP in the Frizzled internalization assay. However, it does produce a small dose-dependent increase in activity in the TOPflash assay (Figure 4C). Co-treatment of DK3-1 and 5% Wnt3A-conditioned media produces a visible punctate phenotype and a very significant increase in functional response. Overall, the SAR findings indicate that small variation in structure can change not only the Wnt/β-catenin inhibitory potency, but also change the functional response. This dynamic SAR supports a hypothesis that the Wnt-modulating activity in the niclosamide chemotype may allow optimization.
Figure 4. The niclosamide derivative DK3-1 synergistically activates TOPflash reporter activity in presence of Wnt-3A ligand.
(A) chemical structure of DK3-1. (B) DK3-1 synergizes with Wnt3A and activates TOPflash reporter activity. HEK293 cells harboring TOPflash reporter were treated without or with Wnt3A conditional medium and DK3-1 at the concentration indicated for 8 hours. *: p<0.05, **: p<0.001, compared to 5% Wnt3A only sample by t test. (C) Confocal images of Frizzled1-GFP U2OS stable cells treated with DMSO, Niclosamide (12.5 μM), DK3-1 (12.5 μM), 5% Wnt3A conditioned Medium, and combination of 5%Wnt3A and DK3-1 (12.5 μM).
Our findings to date suggest the mechanism of action by which Niclosamide inhibits Wnt signaling is distinct from the mechanism of anthelmintic activity within the salicylanilide class of anthelmintic agents. Our pilot SAR studies indicate that a good correlation exists between the Frizzled internalization assay and the TOPflash β-catenin transcription assay and that a dynamic SAR exists within the chemotype. Additional SAR, target ID and mechanism studies are underway. These studies offer the opportunity to identify small molecule modulators of Wnt/β-catenin signaling that enhance our understanding of Wnt/β-catenin pathway regulation while at the same time providing therapeutic agents to treat diseases such as colon cancer that are mediated via dysregulation of Wnt signaling.
Supplementary Material
Acknowledgements
This work was funded in part by 5R01 CA113656-03 (WC), the Pediatric Brain Tumor Foundation (WC) and a Clinical Oncology Research Center Development Grant 5K12-CA100639-08 (RAM, MC). Wei Chen is a V foundation Scholar and an American Cancer Society Research Scholar. NMR instrumentation in the Duke NMR Spectroscopy Center was funded by the NIH, NSF, NC Biotechnology Center and Duke University. The authors gratefully acknowledge this support and the support of Professor Eric Toone and the Duke Small Molecule Synthesis Facility.
Abbreviations
- APC
Adenomatous Polyposis Coli
- CRC
colorectal cancer
- Dvl
dishevelled
- LEF/TCF
Lymphoid enhancer factor/T cell factor
- Fzd1
Frizzled1
- GFP
green fluorescent protein
- GSK3
glycogen synthase kinase 3
- SAR
structure-activity relationships.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
None declared
References and notes
- 1.Barker N, Clevers H. Nature Reviews Drug Discovery. 2006;5:997. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 2.Coombs GS, Covey TM, Virshup DM. Current Drug Targets. 2008;9:513. doi: 10.2174/138945008784911796. [DOI] [PubMed] [Google Scholar]
- 3.Besancon R, Valsesia-Wittmann S, Puisieux A, de Fromentel CC, Maguer-Satta V. Current Medicinal Chemistry. 2009;16:394. doi: 10.2174/092986709787315531. [DOI] [PubMed] [Google Scholar]
- 4.Moon RT, Kohn AD, Ferrari GVD, Kaykas A. Nat Rev Genet. 2004;5:691. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H. Cell. 2006;127:469. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.MacDonald BT, Tamai K, He X. Developmental Cell. 2009;17:9. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. Science. 2003;301:1391. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 8.Sato A, Yamamoto H, Sakane H, Koyama H, Kikuchi A. The EMBO Journal. 2010;29:41. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meireles LMC, Mustata G. Curr. Top. Med. Chem. 2011;11:248. doi: 10.2174/156802611794072632. [DOI] [PubMed] [Google Scholar]
- 10.Chen MY, Wang JB, Lu JY, Bond MC, Ren XR, Lyerly HK, Barak LS, Chen W. Biochemistry. 2009;48:10267. doi: 10.1021/bi9009677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BZ, Dodge ME, Tang W, Lu JM, Ma ZQ, Fan CW, Wei SG, Hao WN, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L. Nature Chemical Biology. 2009;5:100. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang SMA, Mishina YM, Liu SM, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi XY, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao WL, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Nature. 2009;461:614. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 13.Lu JM, Ma ZQ, Hsieh JC, Fan CW, Chen BZ, Longgood JC, Williams NS, Amatruda JF, Lum L, Chen C. Bioorganic & Medicinal Chemistry Letters. 2009;19:3825. doi: 10.1016/j.bmcl.2009.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii N, You L, Xu ZD, Uematsu K, Shan JF, He B, Mikami I, Edmondson LR, Neale G, Zheng J, Guy RK, Jablons DM. Cancer Research. 2007;67:573. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Gwak J, Cho MJ, Song T, Won J, Kim DE, Shin JG, Oh S. Molecular Pharmacology. 2006;70:960. doi: 10.1124/mol.106.024729. [DOI] [PubMed] [Google Scholar]
- 16.Grandy D, Shan JF, Zhang XX, Rao S, Akunuru S, Li HY, Zhang YH, Alpatov I, Zhang XA, Lang RA, Shi DL, Zheng JJ. Journal of Biological Chemistry. 2009;284:16256. doi: 10.1074/jbc.M109.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You L, Xu ZD, Punchihewa C, Jablons DM, Fujii N. Molecular Cancer Therapeutics. 2008;7:1633. doi: 10.1158/1535-7163.MCT-08-0155. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YN, Appleton BA, Wiesmann C, Lau T, Costa M, Hannoush RN, Sidhu SS. Nature Chemical Biology. 2009;5:217. doi: 10.1038/nchembio.152. [DOI] [PubMed] [Google Scholar]
- 19.Gwak J, Song T, Song JY, Yun YS, Choi IW, Jeong Y, Shin JG, Oh S. Biochemical and Biophysical Research Communications. 2009;387:444. doi: 10.1016/j.bbrc.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Liu H, Wang S, Hao X, Li L. Cell Research. 2011;21:730. doi: 10.1038/cr.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepourcelet M, Chen YN, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Cancer Cell. 2004;5:91. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 22.Cheng D, Zhang G, Han D, Gao W, Pan S. WO2010101849A1. 2010:205pp. [Google Scholar]
- 23.Handeli S, Simon JA. Molecular Cancer Therapeutics. 2008;7:521. doi: 10.1158/1535-7163.MCT-07-2063. [DOI] [PubMed] [Google Scholar]
- 24.Gong Y-D, Dong M-S, Lee S-B, Kim N, Bae M-S, Kang N-S. Bioorganic & Medicinal Chemistry. 2011;19:5639. doi: 10.1016/j.bmc.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 25.Waaler J, Machon O, von Kries JP, Wilson SR, Lundenes E, Wedlich D, Gradl D, Paulsen JE, Machonova O, Dembinski JL, Dinh H, Krauss S. Cancer Research. 2011;71:197. doi: 10.1158/0008-5472.CAN-10-1282. [DOI] [PubMed] [Google Scholar]
- 26.Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, Nagourney R, Cardozo T, Brown AM, DasGupta R. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5954. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, Kim K, Melancon B, Ghidu VP, Sulikowski GA, LaFleur B, Salic A, Lee LA, Miller DM, Lee E. Nature Chemical Biology. 2010;6:829. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni TT, Rellinger EJ, Mukherjee A, Xie S, Stephens L, Thorne CA, Kim K, Hu J, Lee E, Marnett L, Hatzopoulos AK, Zhong TP. Chemistry & Biology. 2011;18:1658. doi: 10.1016/j.chembiol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Oh SW, Kim HY, Moon SH, Ha JR, Kahn M. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16707. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiselyov AS, Tkachenko SE, Balakin KV, Ivachtchenko AV. Expert Opinion on Therapeutic Targets. 2007;11:1087. doi: 10.1517/14728222.11.8.1087. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Lin C, Roberts MJ, Waud WR, Piazza GA, Li Y. PloS one. 2011;6:e29290. doi: 10.1371/journal.pone.0029290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, Fichtner I, Schlag Peter M, Shoemaker Robert H, Stein U. Journal of the National Cancer Institute. 2011;103:1018. doi: 10.1093/jnci/djr190. [DOI] [PubMed] [Google Scholar]
- 33.Osada T, Chen MY, Yang XY, Spasojevic I, Vandeusen JB, Hsu D, Clary BM, Clay TM, Chen W, Morse MA, Lyerly HK. Cancer Research. 2011;71:4172. doi: 10.1158/0008-5472.CAN-10-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO, editor. The Selection and Use of Essential Medicines. World Health Organization; Geneva: 2007. [Google Scholar]
- 35.Pearson RD, Hewlett EL. Annals of Internal Medicine. 1985;102:550. doi: 10.7326/0003-4819-102-4-550. [DOI] [PubMed] [Google Scholar]
- 36.Swan GE. J. S. Afr. Vet. Assoc. 1999;70:61. doi: 10.4102/jsava.v70i2.756. [DOI] [PubMed] [Google Scholar]
- 37.Weinbach EC, Garbus J. Nature. 1969;221:1016. doi: 10.1038/2211016a0. [DOI] [PubMed] [Google Scholar]
- 38.Terada H. Environmental Health Perspectives. 1990;87:213. doi: 10.1289/ehp.9087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yo YT, Lin YW, Wang YC, Balch C, Huang RL, Chan MW, Sytwu HK, Chen CK, Chang CC, Nephew KP, Huang T, Yu MH, Lai HC. Mol Cancer Ther. 2012;11:1703. doi: 10.1158/1535-7163.MCT-12-0002. [DOI] [PubMed] [Google Scholar]
- 40.Wu CJ, Jan JT, Chen CM, Hsieh HP, Hwang DR, Liu HW, Liu CY, Huang HW, Chen SC, Hong CF, Lin RK, Chao YS, Hsu JTA. Antimicrobial Agents and Chemotherapy. 2004;48:2693. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang AM, Ku HH, Liang YC, Chen YC, Hwu YM, Yeh TS. Journal of Cellular Biochemistry. 2009;106:682. doi: 10.1002/jcb.22065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, Pan J, Pei D, Ding K. ACS Medicinal Chemistry Letters. 2010;1:454. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SJ, Shin JH, Kang H, Hwang JJ, Cho DH. BMB reports. 2011;44:517. doi: 10.5483/bmbrep.2011.44.8.517. [DOI] [PubMed] [Google Scholar]
- 44.Khanim FL, Merrick BAME, Giles HV, Jankute M, Jackson JB, Giles LJ, Birtwistle J, Bunce CM, Drayson MT. Blood Cancer Journal. 2011;1:e39. doi: 10.1038/bcj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, Sun X, Wu Y, Zhou J, Pan J. Cancer Research. 2010;70:2516. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]
- 46.Gies E, Wilde I, Winget JM, Brack M, Rotblat B, Novoa CA, Balgi AD, Sorensen PH, Roberge M, Mayor T. PloS One. 2010;5:e14410. doi: 10.1371/journal.pone.0014410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang ZD, Huang ZP, Yu H, Dias J, Minami T, Michnick SW, Westwick JK. Nature Chemical Biology. 2006;2:329. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 48.Balgi AD, Fonseca BD, Donohue E, Tsang TCF, Lajoie P, Proud CG, Nabi IR, Roberge M. PloS One. 2009:4. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merschiohann K, Steverding D. Experimental Parasitology. 2008;118:637. doi: 10.1016/j.exppara.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Navab M, Ruchala P, Waring AJ, Lehrer RI, Hama S, Hough G, Palgunachari MN, Anantharamaiah GM, Fogelman AM. Journal of Lipid Research. 2009;50:1538. doi: 10.1194/jlr.M800539-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fonseca BD, Diering GH, Bidinosti MA, Dalal K, Alain T, Balgi AD, Forestieri R, Nodwell M, Rajadurai CV, Gunaratnam C, Tee AR, Duong F, Andersen RJ, Orlowski J, Numata M, Sonenberg N, Roberge M. J. Biol. Chem. 2012;287:17530. doi: 10.1074/jbc.M112.359638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubey SK, Singh AK, Singh H, Sharma S, Iyer RN, Katiyar JC, Goel P, Sen AB. J. Med. Chem. 1978;21:1178. doi: 10.1021/jm00209a020. [DOI] [PubMed] [Google Scholar]
- 53.The mechanism responsible for pathway activation requires further study. Whereas it is possible an agonist or antagonist response may result from small structural changes in a ligand binding to the same biochemical target, it is also possible different derivatives in the chemotype bind to different targets that produce an opposite functional response. Our studies to date do not provide insights into the target or mechanism driving pathway activation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.