Abstract
Imaging technologies developed in the early 20th century achieved contrast solely by relying on macroscopic and morphological differences between the tissues of interest and the surrounding tissues. Since then, there has been a movement toward imaging at the cellular and molecular level in order to visualize biological processes. This rapidly growing field is known as molecular imaging. In the last decade, many methodologies for imaging proteins have emerged. However, most of these approaches cannot be extended to imaging beyond the proteome. Here, we highlight some of the recently developed technologies that enable imaging of non-proteinaceous molecules in the cell: lipids, signalling molecules, inorganic ions, glycans, nucleic acids, small-molecule metabolites, and protein post-translational modifications such as phosphorylation and methylation.
Introduction
Molecular imaging is a powerful tool that has enabled the visualisation of biomolecules as they function in their native setting.1 The ability to monitor biological events in real time at the subcellular level has furthered our understanding of many physiological processes, including protein trafficking, protein localisation, and protein–protein interactions.2 Arguably, the most widely used set of tools in molecular imaging are fluorescent proteins. The discovery and development of the green fluorescent protein (GFP) by Shimomura, Chalfie, and Tsien, who were awarded the 2008 Nobel Prize in Chemistry for their efforts, has enabled the tagging and imaging of many proteins of interest.3
Although imaging of target proteins using fluorescent protein fusions has revolutionised many areas of biology, extension of this strategy to other components of the cell such as glycans, lipids, and nucleic acids has remained challenging. While proteins comprise the largest fraction of biological molecules in the cell, non-proteinaceous biomolecules also play important roles in cell biology.4 Thus, the ability to directly visualise all the components of the cell would allow for a more comprehensive understanding of cellular biochemistry. Here, we discuss the development of emerging technologies that enable imaging beyond the proteome, namely protein post-translational modifications such as glycosylation, phosphorylation, methylation, and lipidation, as well as other classes of biomolecules, including lipids, glycans, nucleic acids, small-molecule metabolites, and inorganic ions.
1. Fluorescent proteins
Traditionally, fluorescent proteins have been genetically fused to the gene that encodes the protein of interest in order to produce a chimeric protein that contains the fluorescent protein at its N- or C-terminus.5 Beyond the now routine use of this strategy to image proteins, several groups have applied fluorescent proteins to image lipids, signalling molecules, and post-translational modifications in the cell. Through protein-small molecule interactions, certain proteins can faithfully report on the location of non-proteinaceous biomolecules in the cell, perhaps most notably many classes of lipids.
1.1 Genetically-encoded probes for imaging membrane lipids
Membrane lipids, such as diacylglycerol, phosphoinositides, and phosphatidylserine, are important regulators of cellular homeostasis and many signal transduction pathways.6 These lipids are predominantly found in the inner leaflet of the plasma membrane and on the cytosolic face of organelle membranes and are responsible for recruiting a wide variety of proteins and initiating a variety of signalling pathways.7 Fluorescent protein fusions to the lipid-binding domains of these recruited proteins have been used to study lipid dynamics in live cells using fluorescence microscopy.8,9 For example, the pleckstrin homology domains of phospholipase C δ and Akt have been used to image the phosphoinositides PtdIns(4,5)P2 and PtdIns(3,4,5)P3, respectively.7 Fluorescent protein fusions of other lipid-binding domains have enabled imaging of other phosphoinositides and membrane lipids as well.7 This methodology has been instrumental in determining the localisation of these important lipids during various biological processes.
1.2 Genetically-encoded and Förster resonance energy transfer (FRET)-based sensors of signalling molecules
In addition to their contributions toward developing the fluorescent protein toolkit, Tsien and co-workers have also developed a FRET-based reporter system for imaging secondary messengers.3 They were initially interested in visualising cyclic adenosine 3′,5′-monophosphate (cAMP), a small molecule that is involved in intracellular signal transduction. Their first FRET reporter consisted of fluorescein-labeled catalytic subunits of cAMP-dependent protein kinase (PKA) and rhodamine-labeled regulatory subunits of PKA.10 Upon binding of cAMP, the regulatory subunit of PKA dissociates from the catalytic subunit, thus causing a loss in FRET between the two labels. Despite the utility of this probe, Tsien hoped to replace the small-molecule organic fluorophores with fluorescent proteins in order to enable imaging of other metabolites as well as proteins through genetic fusion.3 Using mutagenesis, they were able to develop GFP variants with enhanced fluorescence as well as fluorescent proteins of different colors. They discovered that the yellow fluorescent protein (YFP) is a good FRET acceptor for the cyan fluorescent protein (CFP). Following this development, Pozzan and co-workers teamed up with the Tsien laboratory to replace the two fluorophores with CFP and YFP.11 In their work, they demonstrated that this fluorescent protein FRET-based reporter can be used to image cAMP in stimulated rat cardiac myocytes.12 More recently, Dyachok et al. have developed a FRET-based sensor to measure cAMP dynamics in pancreatic beta cells using ratiometric evanescent wave microscopy.13,14 Other applications of cAMP imaging probes have been comprehensively reviewed elsewhere.15
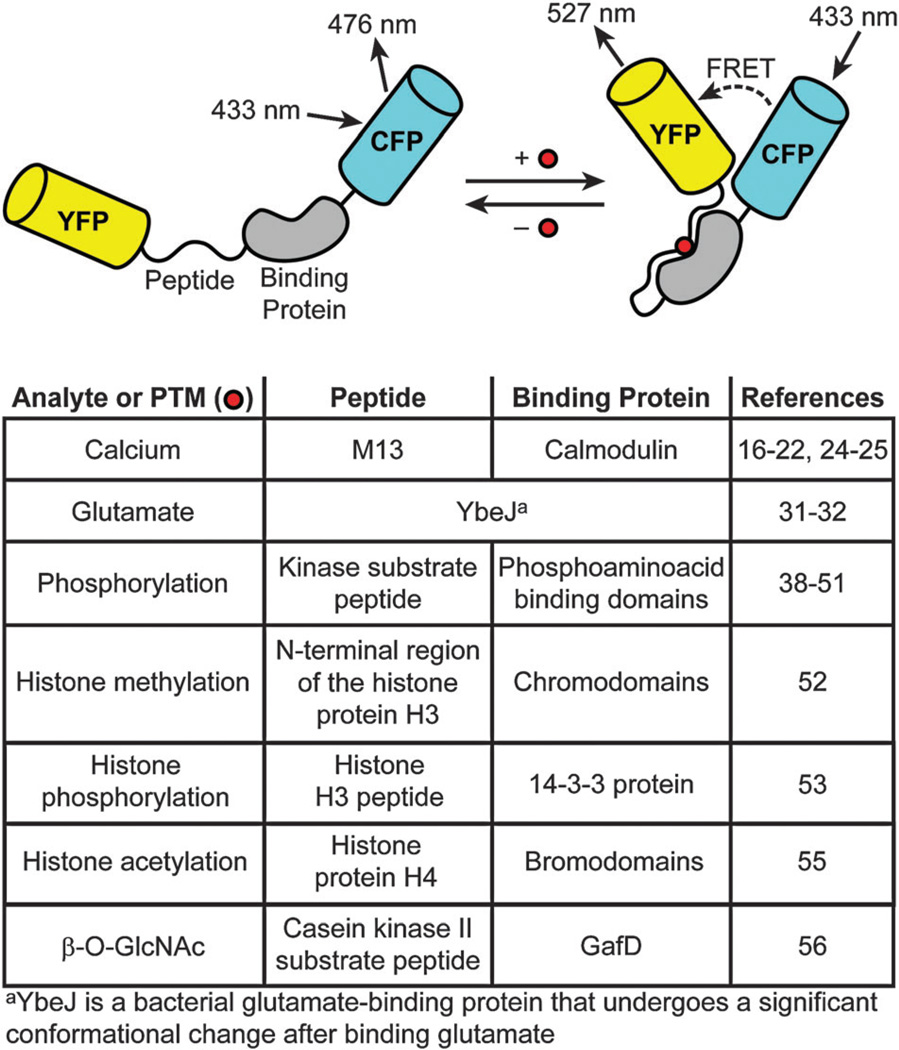
Tsien and co-workers also developed fluorescent protein FRET-based sensors of Ca2+, one of the most important secondary messengers in cell biology.16 Dubbed ‘cameleons,’ these indicators consisted of fusions between CFP, the Ca2+-binding protein calmodulin (CaM), the CaM-binding peptide M13, and YFP (Fig. 1).17–20 Increased levels of Ca2+ causes CaM to bind to M13, thus changing the distance between the two fluorescent proteins and causing an increase in FRET. More recently, an improved CaM-based FRET indicator was used to detect single action potentials in neurons from brain slices and in vivo, a feat which was not possible with previously existing Ca2+ sensors.21,22 The success of these probes, however, is limited by both sensitivity and impaired targeting efficiency due to the large size of the chimeric probe.23 Thus, alternative fluorescent protein-based Ca2+ sensors have been developed. One class of second-generation probes, named ‘camgaroos,’ is based on a single YFP which is pH-sensitive.19 Another second-generation Ca2+ sensor, named ‘pericam’ because it is a circularly permuted version of YFP, enables ratiometric measurements, or the simultaneous recording of two distinct fluorescent signals that can allow for quantitative readouts.24 A more recent improvement to this probe known as yellow cameleon-nano was used to image subtle changes in Ca2+ dynamics.25 Imoto and co-workers have developed a high-affinity Ca2+ probe composed of a single GFP fused to CaM and M13 known as GCaMP.26 Others have developed improved versions of GCaMP termed GCaMP2 and GCaMP3. GCaMP2 has been used to image Ca2+ in the murine heart27 and to measure synaptic activity in zebrafish.28 GCaMP3, a more recent improvement to GCaMP2, is brighter, has a greater dynamic range, and has a higher affinity for Ca2+.29 Recently, the palette of genetically-encoded sensors for Ca2+ was expanded to include improved green and red fluorescent protein indicators.30
Fig. 1.
Genetically-encoded fluorescent protein FRET reporters.
Fluorescent protein-based FRET reporters for sensing glutamate were also developed by Tsien and co-workers as well as Frommer and co-workers. Glutamate is the major excitatory neurotransmitter in the brain, and monitoring its levels could provide insight into many neurological processes. Okumoto et al. have developed a FRET-based sensor consisting of CFP fused to both a clamshell-shaped bacterial glutamate periplasmic binding protein (YbeJ) and YFP (Fig. 1).31 Hires et al. have improved this sensor by optimizing the linker sequences and glutamate affinities to enhance its signal-to-noise ratio.32,33
A genetically-encoded fluorescent indicator of intracellular H2O2 was developed by Lukyanov and co-workers.34 H2O2 is a reactive oxygen species (ROS) that is speculated to be involved in paracrine signalling.35 In their design, they fused a bacterial H2O2-sensitive transcription factor (OxyR) to a circularly permuted YFP. Cysteine oxidation of the OxyR portion induces a conformational change that changes the emission profile of the fluorescent protein. This probe has been used to image peroxide production during wound healing in zebrafish.36
1.3 Genetically-encoded and FRET-based sensors of protein post-translational modifications
Post-translational modifications of proteins are important regulators of protein function.37 Genetically-encoded FRET probes based on the initial design pioneered by Tsien and co-workers have been constructed for monitoring post-translational modifications such as phosphorylation, methylation, acetylation, and glycosylation (Fig. 1).38,39
Protein phosphorylation by protein kinases is key for the activation of numerous signal transduction pathways.40 FRET-based probes for phosphorylation, which sense kinase activities, consist of CFP and YFP fused to a consensus substrate for the relevant kinase and a binding domain.38,41 These sensors have been used to detect protein kinase A,42 protein tyrosine kinase,43 Akt/protein kinase B,44,45 and protein kinase C46 activities. Improved versions of these sensors with better specificity and reversibility have enabled visualisation of protein phosphorylation by the tyrosine kinase SrcA during cell mechanotransduction as well as monitoring of protein kinase A activity during insulin signalling.47,48 More recently, Kapoor and co-workers have examined the dynamics of protein phosphorylation by aurora B kinase, a key mitotic regulator, using FRET-based sensors in live cells.49 There have also been reports of genetically-encoded sensors with enhanced sensitivity and optimized signal-to-noise ratios for fluorescence lifetime imaging.50,51
Histone modifications play important roles in regulating gene expression. Ting and co-workers have developed CFP/YFP reporters of histone phosphorylation and methylation, which are important modifications that regulate transcription of nearby genes.52–54 More recently, changes in histone acetylation were monitored with a FRET-based reporter.55 Similarly, Mahal and co-workers have developed a FRET-based sensor of protein O-GlcNAcylation, a reversible form of glycosylation of intracellular proteins akin to phosphorylation.56,57 Development of this probe has allowed for dynamic monitoring of O-GlcNAc modifications in live cells.
2. Small-molecule sensors
Small-molecule indicators can be advantageous relative to genetically-encoded ones because they often exhibit greater dynamic ranges and increased sensitivity.58 They also tend to exhibit faster response kinetics. However, unlike genetically-encoded sensors, with few exceptions they cannot be localised or specifically targeted to a particular organelle in the cell by fusion to the protein of interest.16
Small-molecule fluorophores have been developed for sensing metal ions such as calcium, zinc, copper, and iron. Metal ions are interesting targets for imaging because they play critical roles in cell biology.59 The design of small-molecule sensors for metal ions is challenging because the probe must be selective for the specified metal over other biologically abundant cations, including those that exist at much higher cellular concentrations such as Na+, K+, and Mg2+. Thus, the design of metal-selective probes requires the successful application of principles of coordination chemistry.
2.1 Fluorescent Ca2+ indicators
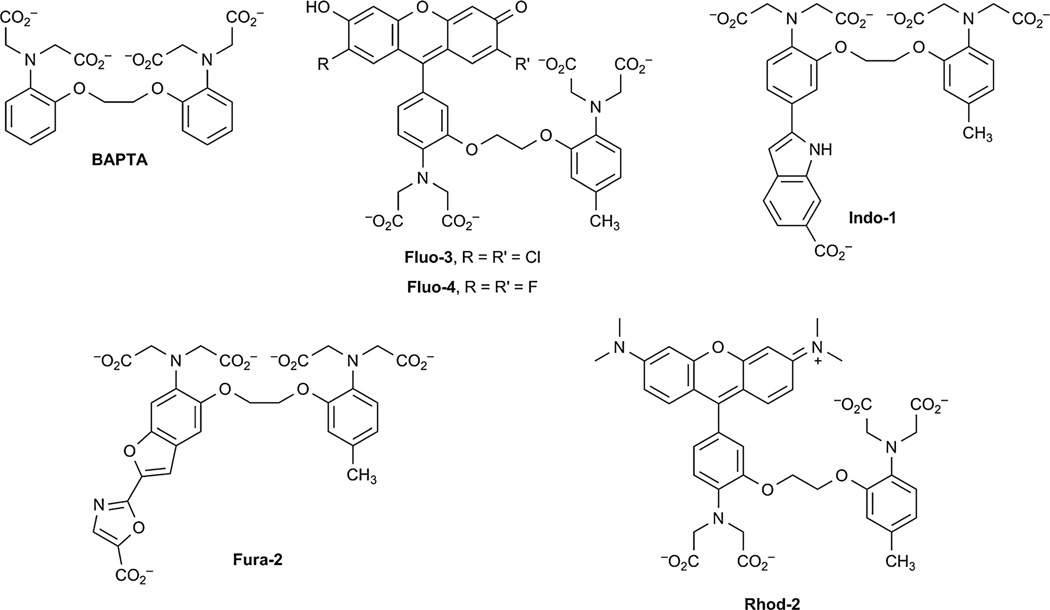
The first small-molecule fluorescent Ca2+ sensor was developed by Tsien.60 In this pioneering work, he replaced the two methylene groups of a well-known Ca2+ chelator with two benzene rings that enable the molecule to function as a chromophore (BAPTA, Fig. 2). Since then, many low- and high-affinity dyes for Ca2+-sensing have been developed that emit UV and visible light. Some of these probes include the fluorescein derivatives Fluo-3, developed by Tsien and co-workers, and Fluo-4, a brighter, more photostable derivative of Fluo-3, developed by Molecular Probes (Eugene, OR) (Fig. 2).58 Indo-1 and Fura-2 are ratiometric dyes that are widely considered the standard for quantitative intracellular Ca2+ measurements (Fig. 2).61 Red-shifted Ca2+ indicators based on the rhodamine scaffold, including Rhod-2, have also been developed to minimize autofluorescence from biological samples (Fig. 2).61,62 Tour et al. have combined the concept of a small-molecule sensor of Ca2+ and the localisation that the tetracysteine motif and FlAsH/ReAsH reagents provide by developing a biarsenical Ca2+ indicator to probe Ca2+ levels around specific proteins.63
Fig. 2.
Small-molecule fluorescent Ca2+ sensors.
2.2 Small-molecule Zn2+ indicators
In the past century, Zn2+ has emerged as an important regulator of protein function and signalling.64–66 It has also been implicated in the pathophysiology of several neurodegenerative disorders, including Alzheimer’s disease.67 Thus, the ability to image Zn2+ using small-molecule sensors could elucidate metal ion homeostasis in relation to disease.
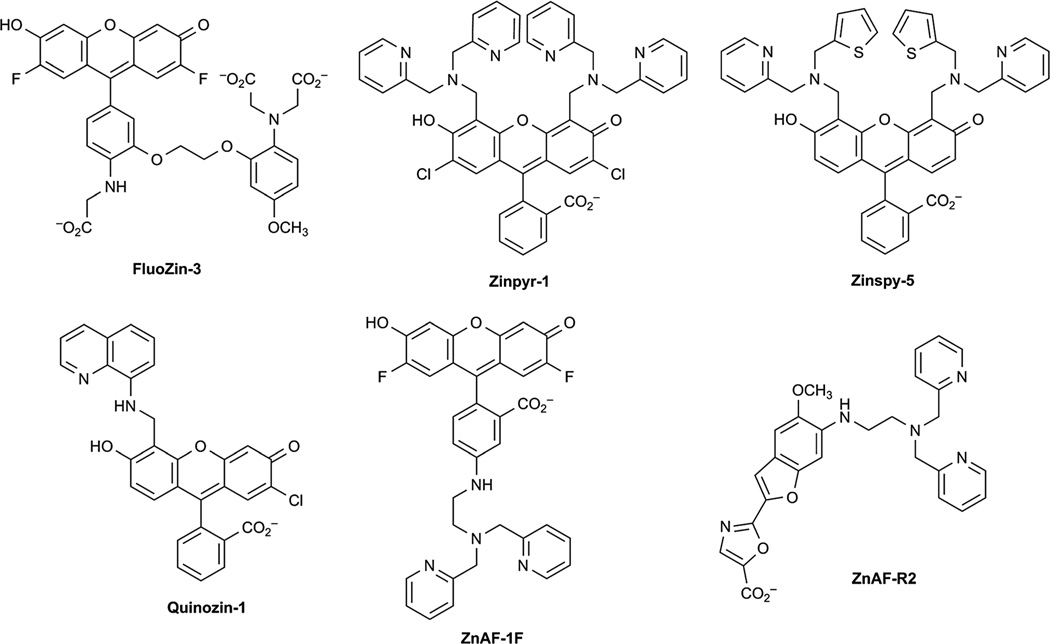
Several groups have developed Zn2+ sensors that were inspired by analogous probes for Ca2+ detection. Gee et al. have removed a single acetate arm from the Ca2+ chelator of Fluo-3 to create FluoZin-3, which maintains high affinity for Zn2+ (Fig. 3).68 Lippard and co-workers have developed several classes of fluorescein-based sensors for Zn2+ that are modified with a Zn2+ chelator on the xanthene core. These include the Zinpyr,69–73 Zinspy,74 and Quinozin75 series of probes (Fig. 3). An improvement to the Zinpyr family of dyes, known as ZPP1, exhibits lower background fluorescence and higher fluorescence turn-on when complexed to zinc.76 This probe was recently used to image high levels of Zn2+ in a mouse model of prostate cancer.77 Nagano and co-workers have developed fluorescein-based Zn2+ sensors that are functionalised at the phenyl ring of the fluorophore-scaffold.78 These include the ZnAF series (Fig. 3). In addition to fluorescein-based reagents, BODIPY,79 cyanine,80,81 and coumarin82,83 probes for sensing Zn2+ in biological systems have also been developed.
Fig. 3.
Small-molecule fluorescent Zn2+ sensors.
Several ratiometric sensors of Zn2+ that capitalize on different mechanisms have been synthesized for quantitative measurements of Zn2+ concentrations in vivo. FuraZin and IndoZin are ratiometric Zn2+ sensors derived from the Ca2+ probes Fura and Indo, which operate via an internal charge transfer mechanism.84 Nagano and co-workers have also developed the ratiometric Zn2+ sensors ZnAF–R1 and –R2, which are based on the Fura scaffold (Fig. 3).85 Taki et al. have developed ratiometric sensors known as the Zinbo series that exploit excited-state intramolecular proton transfer processes in benzoxazole scaffolds.86 Lippard and co-workers have also developed ratiometric Zn2+-sensors that rely on a two-fluorophore system in which ester-mediated hydrolysis liberates coumarin as an internal standard.87 In addition to fluorescent sensors, Zn2+ sensors that rely on alternative imaging modalities such as magnetic resonance imaging (MRI) have been developed.88–90
2.3 Small-molecule Cu+ and Fe2+/Fe3+ indicators
Oxidative stress is thought to cause neuronal cell damage that can eventually lead to pathologies such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis.91 Redox-active metals such as Cu+ and Fe2+ have been implicated in causing oxidative stress by producing ROS such as H2O2, superoxide (O2−•), and hydroxyl radicals (•OH).92 Therefore, there has been much interest in imaging not only the ROS themselves (see sections 1.2 and 2.4) but the metal ions as well.
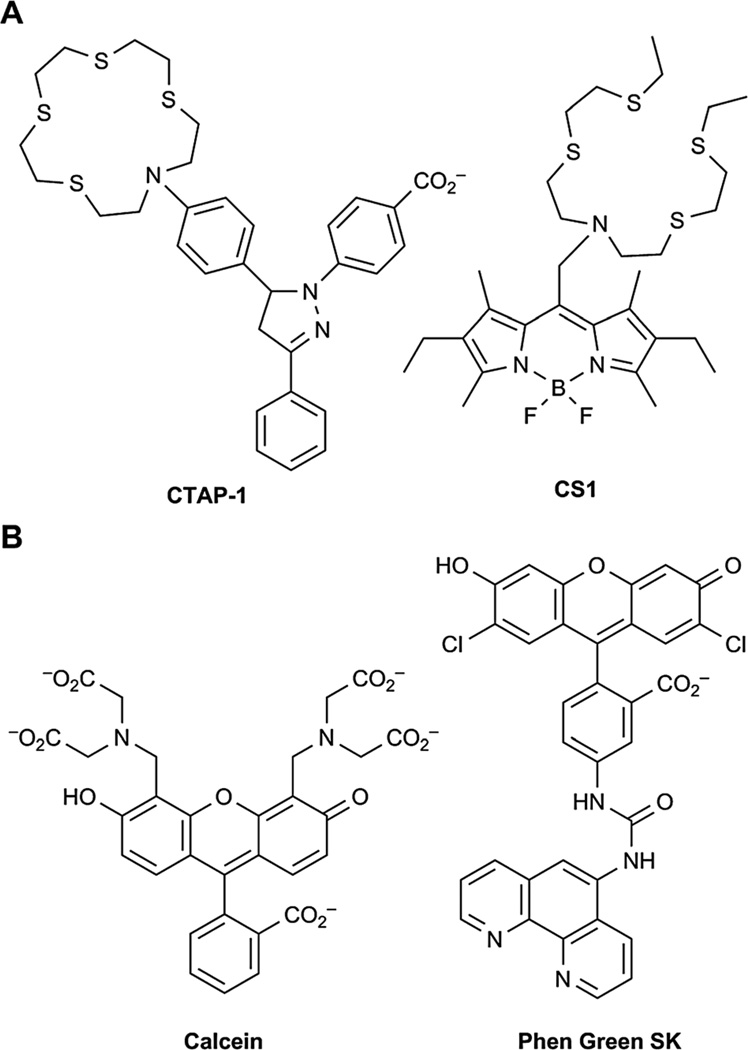
Fluorescence detection of copper using small-molecule sensors is difficult because Cu+ is the major active species within the reducing environment of the cytosol; however, this ion can readily disproportionate to Cu2+ and Cu0. In addition, both Cu+ and Cu2+ are capable of quenching the fluorescence of many fluorophores.64 Currently, there are a few reports of fluorescent Cu+ sensors that have been successfully used for visualizing Cu+ in live cells. Yang et al. have developed a sensor known as CTAP-1 (Fig. 4A), which comprises a pyrazoline dye platform appended to an azatetrathiacrown receptor for metal coordination.93 A more water-soluble version of this probe, CTAP-2, was recently described.94 Chang and co-workers have developed a Cu+ indicator known as CS1 (Fig. 4A) that combines a BODIPY dye and a thioether metal chelator.95,96 Since then, a ratiometric fluorescent sensor based on this dye has been described, as well as a mitochondrial targeted version that allows for imaging of mitochondrial pools of copper.97,98 A second-generation probe with different substituents on the boron center of the BODIPY dye known as CS3 was used in combination with X-ray fluorescence microscopy to image labile copper pools in neurons.99 Recently, a near-infrared copper sensor, CS790, was reported for imaging copper in a murine model of Wilson disease.100 Chang and co-workers have also developed an MRI-based sensor for copper.101,102
Fig. 4.
Small-molecule fluorescent Cu+ and Fe2+/Fe3+ sensors.
Like copper, the major forms of iron in the cell, Fe2+ and Fe3+, also exhibit significant quenching capabilities, and designing fluorogenic sensors of iron is thus difficult as well. As a result, all of the sensors for detecting Fe2+/Fe3+ in cellular systems are turn-off probes. For example, calcein (Fig. 4B) is a commercially available fluorophore that uses a metal chelator for turn-off Fe2+ detection.61 Phen Green SK (Fig. 4B) is another commercial probe that can detect both Fe2+ and Fe3+.61 One drawback of these probes, however, is that they are only partially selective for iron and can detect other metal ions as well.103 The most specific sensors for Fe3+ rely on receptors derived from siderophores, which are small, high-affinity iron chelating compounds secreted by bacteria, that can be conjugated directly onto fluorophores.61,104,105 A few turn-on sensors for Fe3+ have been reported; however, their utility for imaging in cells has not yet been established.106,107
2.4 Small-molecule ROS and RNS indicators
As discussed in Section 2.3, there is mounting evidence that ROS are linked to several neurodegenerative diseases.91 Reactive nitrogen species (RNS) are also thought to cause neuropathological disorders.108 More recently, ROS and RNS have been implicated as signalling molecules during normal physiological processes.35,109 The canonical ROS and RNS involved in these processes are H2O2 and NO, respectively. Due to space limitation, herein we will discuss only small-molecule fluorescence indicators of H2O2 and NO because in-depth reviews on sensors for other ROS and RNS as well as metal-based fluorogenic probes for NO have been written.110–113
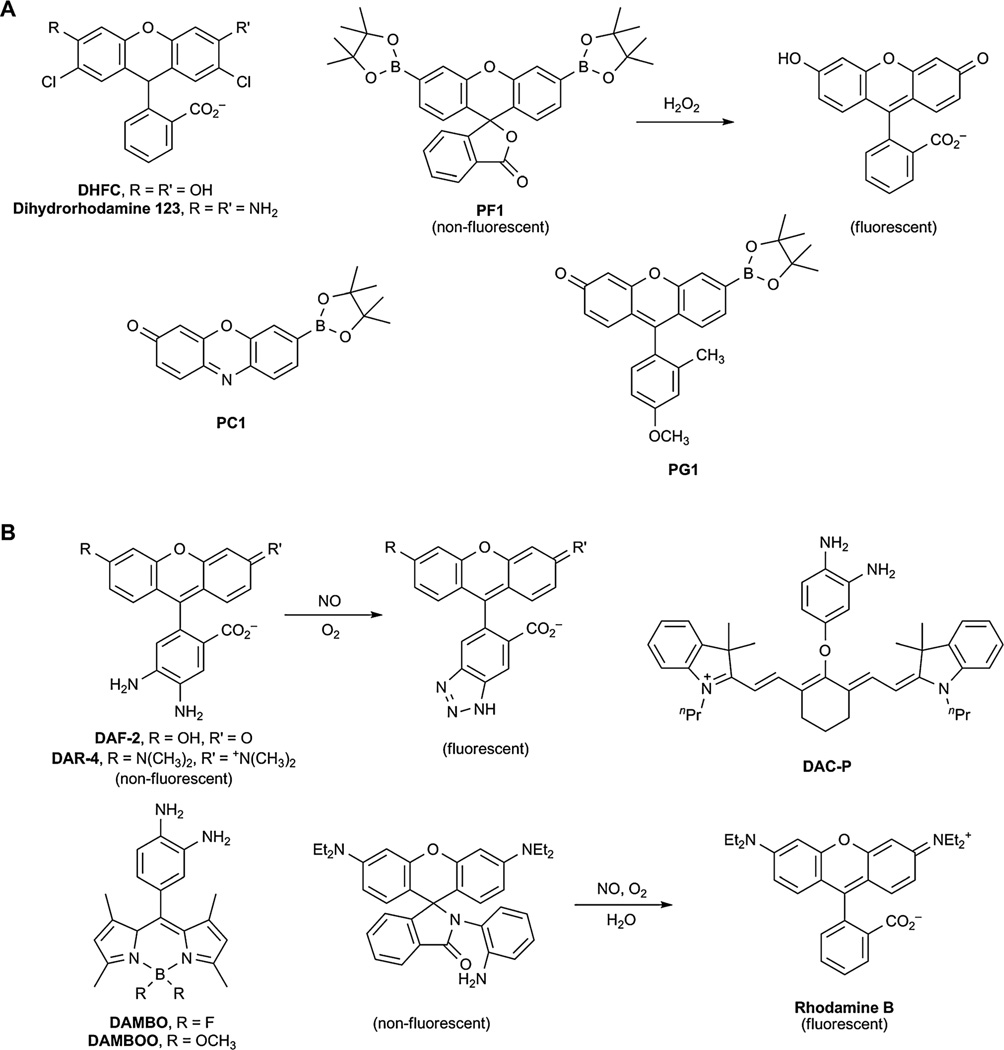
The traditional indicators for sensing H2O2 include dihydrofluorescein (DHFC, Fig. 5A)114 or dihydrorhodamine (Fig. 5A);115 however, these dyes suffer from auto-oxidation by the excitation light as well as reactivity with other ROS.116,117 Chang et al. have developed a class of boronate-based fluorescent probes for sensing H2O2, which includes their first generation probe Peroxy-fluor-1 (PF1, Fig. 5A).118 In the presence of H2O2, the boronic esters are chemoselectively deprotected to yield a fluorescent molecule. This concept has also been applied to blue- and red-shifted analogs, Peroxyxanthone-1 (PX1) and Peroxyresorufin-1 (PR1).119 Second-generation probes that are more sensitive and are capable of detecting H2O2 at physiologically relevant concentrations have also been developed. These include Peroxycrimson-1 (PC1) and Peroxygreen-1 (PG1) (Fig. 5A).120 Further, sensors of additional colors and FRET-based ratiometric probes for sensing H2O2 have been recently described.121,122 Hydrogen peroxide sensors that can be targeted to specific organelles for visualizing ROS within subcellular locations have also been developed. These include probes for visualizing H2O2 in the mitochondria, nucleus, and endoplasmic reticulum.123–125 Finally, new hydrogen peroxide sensors have been used to elucidate that redox signalling through Nox2 is essential for the normal growth and proliferation of neural stem cells.126 These sensors were also used to show that H2O2 uptake is mediated by the water channel aquaporin to modulate downstream signalling events.127
Fig. 5.
Small-molecule fluorescent ROS/RNS sensors. (A) Fluorogenic H2O2 probes. (B) Fluorogenic NO probes.
Most sensors for NO utilize an o-phenylenediamine scaffold that is oxidized to the corresponding aryl triazole in the presence of NO and air (Fig. 5B).109 In this design, the electron-rich diamine quenches the fluorescence of the dye by photoinduced electron transfer. Reaction with NO and O2 generates an electron-poor triazole that triggers the fluorescence turn-on of the dye. Nagano and co-workers have developed a wide variety of probes based on this design that contain fluorescein (DAF-2),128 rhodamine (DAR-4),129 BODIPY (DAMBO and DAMBOO),130,131 and cyanine (DAC-P, Fig. 5B).132 This group has also developed fluorescent sensors of NO with increased sensitivity due to longer intracellular retention times.133 Recently, a variation of the o-phenylenediamine scaffold was developed that is based on a quenched rhodamine B spirolactam (Fig. 5B).134
3. Chemical reporter strategy
For biomolecules such as glycans, certain protein post-translational modifications, and nucleic acids, detection via genetically-encoded reporters such as fluorescent proteins or small-molecule sensors has largely not been possible. Thus, alternative strategies have been developed.
3.1 Lectins and antibodies for imaging glycans
Glycans are interesting targets for imaging because they play key roles in many dynamic biological processes. For example, cell-surface glycans are known to mediate cell–cell adhesion and communication, as well as host-pathogen interactions.135 Other examples include embryonic development,136 leukocyte homing,137 and cancer cell metastasis.138 Traditionally, glycans have been detected using antibodies or carbohydrate-binding proteins known as lectins.139 Though lectins have been widely used for the detection and enrichment of glycoconjugates,140,141 these glycan-binding proteins generally have low affinities for their targets and require multivalency for high-avidity binding.142 Other drawbacks include toxicity and tissue impermeability.143 For these reasons, lectins have limited capabilities for in vivo imaging; however, they have been used routinely for imaging glycans ex vivo on cultured cells and on tissue samples.144,145
Like lectins, antibodies have limited use for in vivo imaging; however, there is one report of their use for glycan-specific imaging in mice. In this study, Licha et al. imaged a peripheral lymph node endothelial glycan termed sulfoadhesin using the MECA-79 antibody.146 Though there are a number of monoclonal antibodies against distinct epitopes, it is difficult to generate antibodies against certain epitopes because the synthesis of many glycan structures can be very cumbersome.147 Thus, applications of antibodies toward in vivo imaging are also limited. Furthermore, like lectins, antibodies also suffer from poor tissue access.148
3.2 Bioorthogonal chemical reporter strategy for imaging glycans
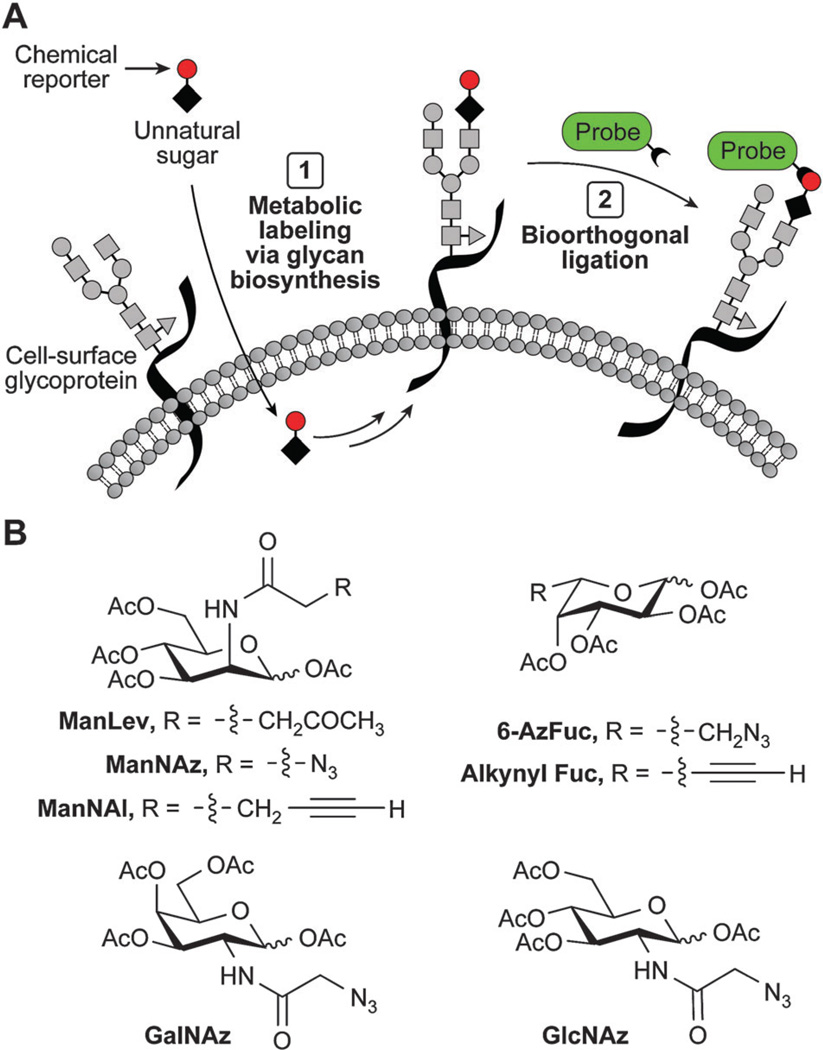
Though lectins and antibodies have been used to image glycans, they are not ideal for imaging dynamic changes in the glycome, the ensemble of glycans displayed on the cell surface. We have developed an approach for imaging the glycome that enables the visualization of glycan dynamics in vivo.149 Our method is a two-step strategy in which the first step involves the metabolic incorporation of an unnatural monosaccharide into an organism’s glycome.150 This substrate analog contains a reactive group known as a “chemical reporter.’ In the second step, the chemical reporter is labelled with an imaging probe via a chemoselective reaction.
The requirements for such a chemical reaction are quite rigorous. First, the chemical reporter and its reaction partner must react in a physiological environment (pH 6–8, 37 °C), while simultaneously remaining inert to all of the surrounding functionality in the biological environment. This task is challenging because there are many nucleophilic and electrophilic functional groups found in cells and living organisms. Second, the reaction must not produce any toxic byproducts or cause harm to the biological sample. Finally, the reporter and its complementary probe must have good bioavailability and form a stable bond. Chemical reactions that meet this collection of criteria are known as “bioorthogonal.”151
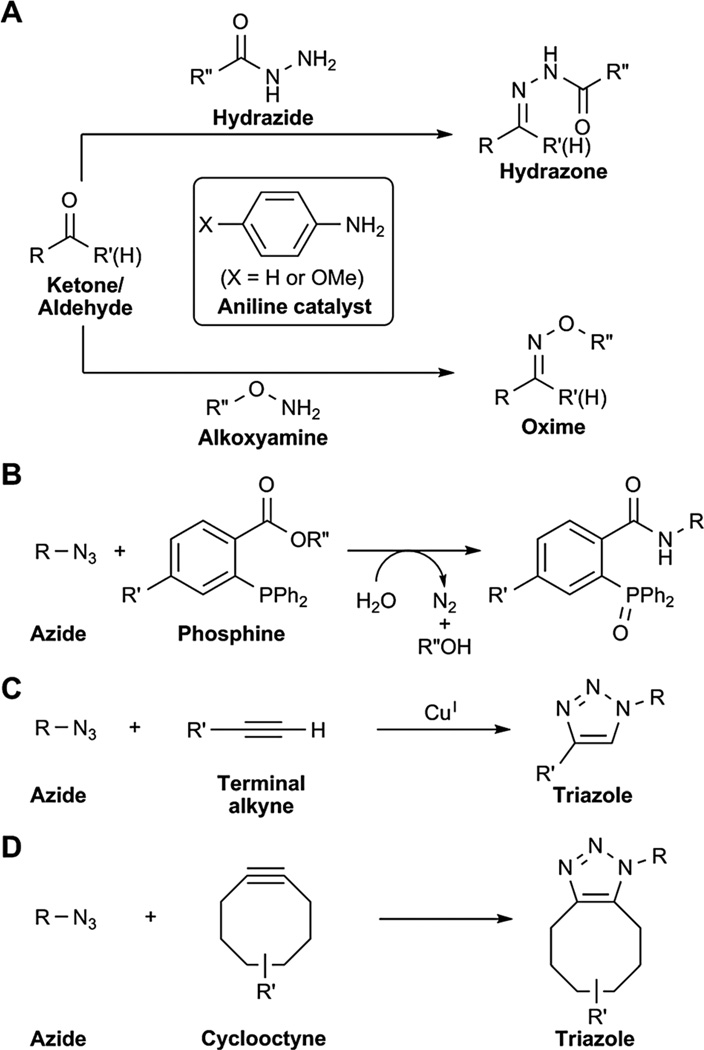
Currently, only a handful of reactions possess the quality of bioorthogonality. These reactions include the condensation of ketones and aldehydes with hydrazide or alkoxyamine probes, the Staudinger ligation of triarylphosphines and azides, and Cu-free click chemistry between cyclooctynes and azides (Fig. 6A–B, D). These reactions have been used to label not only proteins but glycans and lipids as well.
Fig. 6.
Bioorthogonal chemical reactions. (A) Ketone and aldehyde condensation with hydrazides or alkoxyamines. (B) Staudinger ligation between azides and triarylphosphines. (C) Copper-catalysed azidealkyne 1,3-dipolar cycloaddition (CuAAC). (D) Cu-free click chemistry.
Ketones and aldehydes are not truly bioorthogonal because keto and aldehydic metabolites are abundant within cells and in biological fluids. In addition, the pH optima of hydrazone and oxime formation with hydrazide and alkoxyamine groups is 3–4, which precludes their use in vivo.152 Ketones and aldehydes have been utilised to label proteins and glycans on cell surfaces and in the extracellular environment. These mild electrophiles can form reversible Schiff bases with primary amines such as lysine side chains; however, in water, the equilibrium favors the carbonyl. In contrast, the formation of hydrazones and oximes using hydrazide and aminooxy groups, respectively, are favored in physiological conditions and are quite stable (Fig. 6A). Dawson and co-workers have greatly accelerated both oxime and hydrazone formation via the use of a nucleophilic aniline catalyst.153,154
We have recognized the utility of ketones and aldehydes as chemical reporters and have used them to label glycans.155 In this work, Mahal et al. reported that an unnatural keto analog of N-acetylmannosamine (ManNAc), N-levulinoylmannosamine (ManLev), can be metabolized by mammalian cells to the corresponding keto sialic acid (SiaLev) and incorporated within cell-surface glycans (Fig. 7A–B).155 Sadamoto et al. have introduced ketones into bacterial cell walls and labeled the chemical reporters with a hydrazide fluorophore probe.156
Fig. 7.
Bioorthogonal chemical reporter strategy. (A) Metabolic oligosaccharide engineering. (B) Unnatural sugars.
More recently, the azide has emerged as the chemical reporter of choice. This functional group is unique in that it is truly bioorthogonal to the biological milieu. Only one naturally occurring azido-metabolite, which was isolated from unialgal cultures, has been reported to date.157 Moreover, azides can undergo highly selective reactions such as the Staudinger ligation with triarylphosphines, the copper-catalysed azide-alkyne 1,3-dipolar cycloaddition (CuAAC), and Cu-free click chemistry with strained alkynes (Fig. 6B–D).158
The Staudinger ligation is based on the classic Staudinger reduction, which was developed by Hermann Staudinger in 1919. In this work, Staudinger reported that azides react with triarylphosphines under mild conditions to yield aza-ylide intermediates.159 In the presence of water or an appropriate electrophile, this intermediate can be hydrolyzed or trapped to yield an amine and the corresponding phosphine oxide.160 We modified the classic Staudinger reaction by introducing an electrophilic trap onto the phosphine.161 Termed the Staudinger ligation, this reaction ultimately results in the formation of a covalent amide bond between the azide and phosphine along with oxidation of the phosphine (Fig. 6B). This reaction also proceeds at physiological pH with no apparent toxicity to live cells and whole organisms.162
We have shown that an azide-containing analog of ManNAc, termed N-azidoacetylmannosamine (ManNAz), can also be metabolically incorporated into cell-surface glycans as the corresponding azido sialic acid (SiaNAz) (Fig. 7).161 The Staudinger ligation has been used to image these labelled glycans on live cells using phosphine fluorophore conjugates163 as well as bioluminescent and fluorogenic phosphines.164,165 Furthermore, we and others have shown that this reaction can be used to label glycans within living animals.162,166,167
Paulson and co-workers have shown that natural sialic acids can be oxidized to the corresponding aldehyde using sodium periodate and then subsequently imaged using oxime chemistry with alkoxyamine probes.168 In addition to metabolic labelling of sialic acid-containing glycans,169,170 other sectors of the glycome can be metabolically labelled using azido analogs of N-acetylglucosamine (β-O-GlcNAcylated proteins),171 N-acetylgalactosamine (mucin-type O-linked glycans and β-O-GlcNAcylated proteins),172,173 and fucose (fucosylated glycans) (Fig. 7B).174,175 Extending the scope of this methodology, which has thus far focused on labelling individual monosaccharides within the larger glycan structure, a recent report described the targeting of the disaccharide LacNAc (Galβ-1,4-GlcNAc).176
Azides can also be exploited as 1,3-dipoles in a [3+2] cycloaddition with alkynes to form triazole products (Fig. 6C).158 Sharpless and co-workers and Meldal and co-workers independently showed that the normally sluggish reaction between azides and acetylenes can be greatly accelerated with the addition of a copper catalyst.177,178 The copper-catalysed azide-alkyne 1,3-dipolar cycloaddition (CuAAC) is one of the most popular reactions from a family of “click reactions,” defined by their highly selective and modular approach to forming chemical bonds.179 CuAAC is much faster than the Staudinger ligation; however, the necessary copper catalyst is cytotoxic, although new biocompatible Cu(i) catalysts have been described to reduce toxicity to live cells.150,180–183 CuAAC has been used for imaging glycans using alkynyl ManNAc analogs as well as azido and alkynyl fucose analogs in fixed cells (Fig. 7B).175,184,185 In addition, CuAAC was recently used to image lipopolysacccharide (LPS) in bacterial membranes by targeting 3-deoxy-d-mannooctulosonic acid, a specific and essential component of the inner core of LPS.186
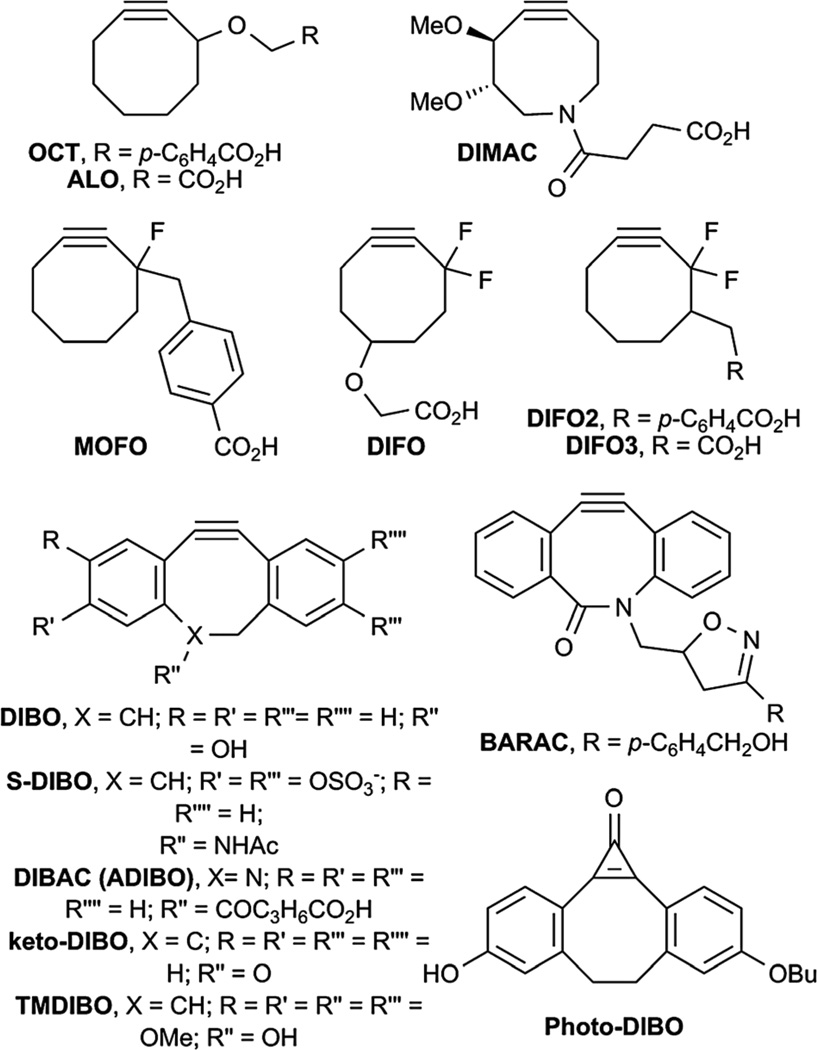
We have eliminated the need for a toxic copper catalyst by activation of the alkyne via an alternative mechanism—ring strain. This reaction is accordingly known as Cu-free click chemistry. Agard et al. first demonstrated this concept using a cyclooctyne reagent (OCT) whose kinetics were on par with the Staudinger ligation (Fig. 6D and 8).187 In order to accelerate the strain-promoted cycloaddition, Baskin et al. introduced fluorine atoms in an attempt to lower the LUMO energy of the alkyne (MOFO, DIFO, DIFO2, DIFO3; Fig. 8).188,189 This reaction has been used to image glycans on live cells,188 the nematode C. elegans,190 and zebrafish embryos.191–194 We have also shown that Cu-free click chemistry can be used to label glycans within mice.195 Second-generation cyclooctynes have been developed, including more water-soluble probes (ALO and DIMAC, Fig. 8)189,196 as well as faster reagents based on a dibenzocyclooctynyl scaffold that have been utilized for imaging glycans on cells (DIBO, S-DIBO, DIBAC, BARAC, Photo-DIBO, TMDIBO, keto-DIBO; Fig. 8),197–203 a coumarin-based fluorogenic cyclooctyne,204 and thiacycloalkynes with improved stability.205 Several theoretical studies have been published to explain the enhanced reactivity of a number of these cyclooctynes.206–208
Fig. 8.
Cyclooctyne reagents for Cu-free click chemistry.
In addition to Cu-free click chemistry using strained alkynes, several new chemistries using strained alkenes have emerged for labelling proteins in live cells and animals.209 These include reactions of strained alkenes with azides,210 tetrazines,211,212 nitrile oxides,213 and nitrile imines,214 as well as photoinducible tetrazole–alkene cycloadditions, known as photo-click chemistry.215 Recently, a new bioorthogonal reaction between highly strained quadricyclanes and Ni(bis)thiolenes was described that is selective and proceeds readily in aqueous environments.216 Although these reactions have only been described for targeting proteins thus far, their selectivity and versatility will hopefully encourage their use in imaging beyond the proteome. Applications using multiple bioorthogonal chemistries would ultimately enable simultaneous visualisation of different classes of biomolecules within the same cell.
3.3 Bioorthogonal chemical reporter strategy for imaging lipids
As secondary metabolites and protein post-translational modifications, lipids, like glycans, are not amenable to imaging via genetically-encoded reporters. Lipids are attractive targets for imaging because they play important roles in cellular processes such as signal transduction and membrane fusion.217 In addition, lipidation of proteins regulates the signalling properties, subcellular localisation, trafficking, and activity of many proteins.218 One approach to imaging lipids in live cells has been the application of the bioorthogonal chemical reporter strategy.219
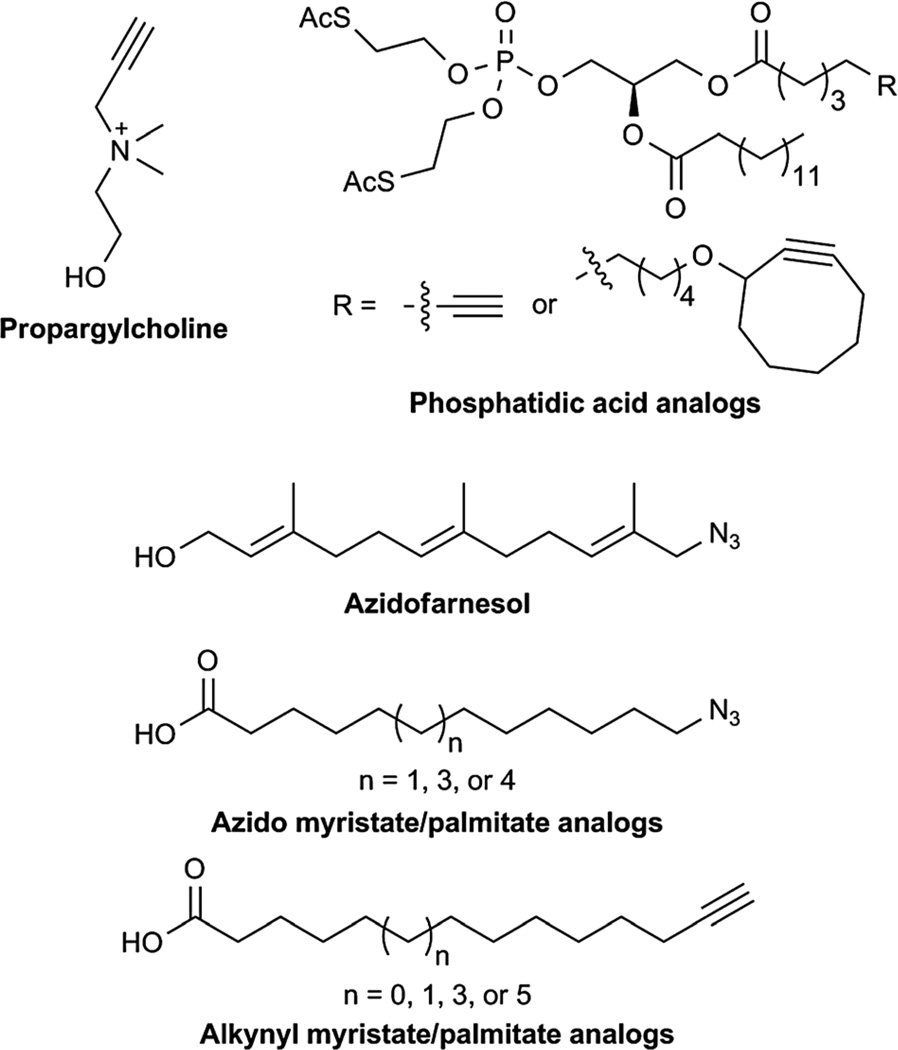
Phospholipids are the major constituents of cell membranes. Those containing the head group choline are important structural components of membranes and play critical roles in cell signalling.220 Despite the crucial roles of these lipids, little is known about their localisation and trafficking.221 Thus, Jao et al. have utilized the chemical reporter strategy in order to image their dynamics in cells and tissues. In order to achieve this goal, they synthesized a choline analog that contains a terminal alkyne moiety, which can be detected by using CuAAC (Fig. 9). Using this technology, they were able to examine the kinetics of phospholipid turnover and the distribution of the lipids in cells and in mice.221 Similarly, Schultz and co-workers have shown that phosphatidic acid analogs that contain either terminal alkynes or cyclooctynes within the lipid tail can be used to image these lipids in cells (Fig. 9).222
Fig. 9.
Chemical reporters for lipids.
Lipidation of proteins via fatty-acylation and prenylation has been known to greatly affect the specificity and efficiency of signal transduction as well as protein–protein interactions.218 Several groups have applied the bioorthogonal chemical reporter strategy to image these protein post-translational modifications.223–225 Zhao and co-workers have used azido analogs of farnesylated proteins in order to profile proteins that are post-translationally modified (Fig. 9).226,227 Alkynyl farnesol reporters have also been described.228,229 Several groups have demonstrated that fatty-acylation of eukaryotic proteins by attachment of S-palmitoyl groups and N-myristoyl to cysteine and N-terminal glycine residues, respectively, can be imaged or profiled by employing azido and alkynyl analogs of these lipids and the Staudinger ligation or CuAAC (Fig. 9).230–236 More recently, a chemical reporter of cholesterylated proteins has also been described.237
3.4 Bioorthogonal chemical reporter strategy for imaging nucleic acids
Nucleic acids are traditionally detected by intercalating dyes (e.g., 4′6-diamidino-2-phenylindole), in situ hybridization techniques, and metabolic incorporation of nucleoside analogs that are either radiolabelled or detected via antibodies. Hybridization approaches include molecular beacons, which have been used for visualizing both DNA and RNA.238,239 Development of these fluorescent oligonucleotide probes requires the incorporation of fluorescent nucleotides, which can be synthesized using different chemistries.240 For example, Ju and co-workers have synthesized fluorescent oligonucleotides via both CuAAC and the Staudinger ligation for the purposes of DNA sequencing using fluorescence detection.241,242 Excess fluorophore conjugates of oligonucleotide probes must be washed away to eliminate background signal, so fluorogenic approaches are highly sought after as well. Toward this end, Cai et al. have developed a fluorogenic peptide nucleic acid-based probe that is activated by the Staudinger ligation upon DNA hybridization.243
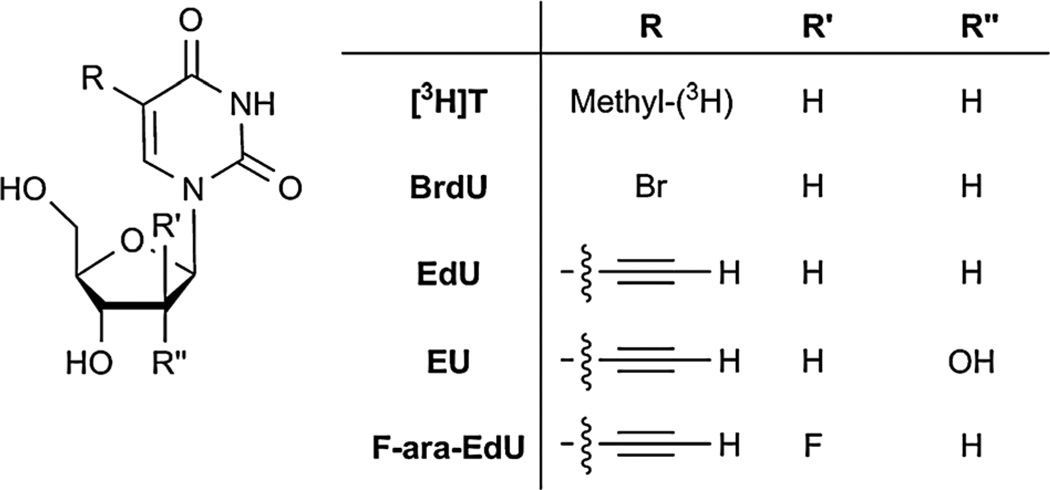
Other techniques include metabolic incorporation of deoxynucleoside analogs such as [3H]thymidine ([3H]T) and 5-bromo-2′-deoxyuridine (BrdU) (Fig. 10). [3H]T and BrdU are detected by autoradiography and immunohistochemistry, respectively. Though these analogs have been useful for probing DNA synthesis, both of these techniques are not ideal. Autoradiography can be cumbersome, and BrdU immunostaining can compromise the integrity of the tissue samples.244 Mitchison and co-workers have synthesized an alkynyl analog of thymidine, 5-ethynyl-2′-deoxyuridine (EdU, Fig. 10), which is readily incorporated into DNA during DNA replication.245 This chemical reporter can then be detected with CuAAC and has enabled visualization of DNA synthesis in cells and in tissue sections from mice. In addition, this methodology causes much less degradation of biological specimens relative to traditional BrdU immunostaining. Arabinosylated EdU analogs (F-ara-EdU, Fig. 10) and alkynyl cytosine analogs have been reported that were used for imaging DNA in cells and zebrafish.246,247 More recently, an alkynyl analog of uridine, 5-ethynyluridine (EU, Fig. 10) has been used to image RNA synthesis in vivo using an analogous strategy.248
Fig. 10.
Metabolic labels for nucleic acids.
4. Label-free imaging methods
4.1 Imaging mass spectrometry
All of the technologies discussed thus far require labels or contrast agents in order to visualise the biological molecules of interest. Imaging mass spectrometry (IMS) is an emerging label-free technique that allows for the visualisation of endogenous proteins, lipids, and small molecules.249 IMS can be used to create 2D and 3D images of the proteomic or small-molecule content of cells or tissues by combining the measurement capability of mass spectrometers with a surface sampling process.250 Currently, while there exist many different modes of ionisation, two popular ionisation methods are used to generate images.251 These techniques include matrix-assisted laser desorption ionisation (MALDI) and secondary ion mass spectrometry (SIMS) IMS. Though IMS has been used for imaging both proteins and other biological molecules, for the purposes of this review, we will focus exclusively on imaging small molecules.
Of these two IMS techniques, MALDI IMS has seen more widespread use for analysing biological samples due to its large mass-to-charge ratio (m/z) range (up to 100 kDa), which is useful for analysing proteomes.252 Imaging with MALDI mass spectrometry, however, is only capable of achieving spatial resolutions of about 20 µm while SIMS can achieve better than 100 nm with specialised instruments.253 On the other hand, SIMS has been documented to detect molecules of masses up to only 1 kDa.250 While many groups have taken advantage of these imaging technologies, a number of laboratories continue to pursue the development of new and improved methodologies, including those that can achieve ionisation under ambient conditions.254,255
For analysis by MALDI IMS, biological samples are prepared in a similar manner to protein or peptide samples. Typically, a thin cryosection of tissue is covered with an organic matrix. The sample is then scanned by the mass spectrometer, and spectra are collected over a predefined area. The resulting dataset contains an array of spectra including different signals of measured mass and intensity. The intensity of signal, or combinations of signals, can then be plotted to generate an image over the sample surface.256 This technique has been used to image many different molecules, including phospholipids.256 As discussed in Section 3.3, these lipids are involved in cell signalling pathways and have been implicated in diseases such as lipid storage disorders as well as Alzheimer’s disease and Down syndrome.257 McLean et al., Jackson et al., and Rujoi et al. have used MALDI IMS to visualise phospholipids in rat brains and mammalian lens tissue.258–261 A growing number of researchers are applying IMS to the diagnosis of diseases such as cancer by looking at differences in lipid profiles between diseased and healthy tissue.262–265 In addition to lipids, Dorrestein and co-workers have applied MALDI IMS to imaging secondary metabolites produced by marine cyanobacteria and sponges as well as Bacillus subtilis and Streptomyces coelicolor.266–268
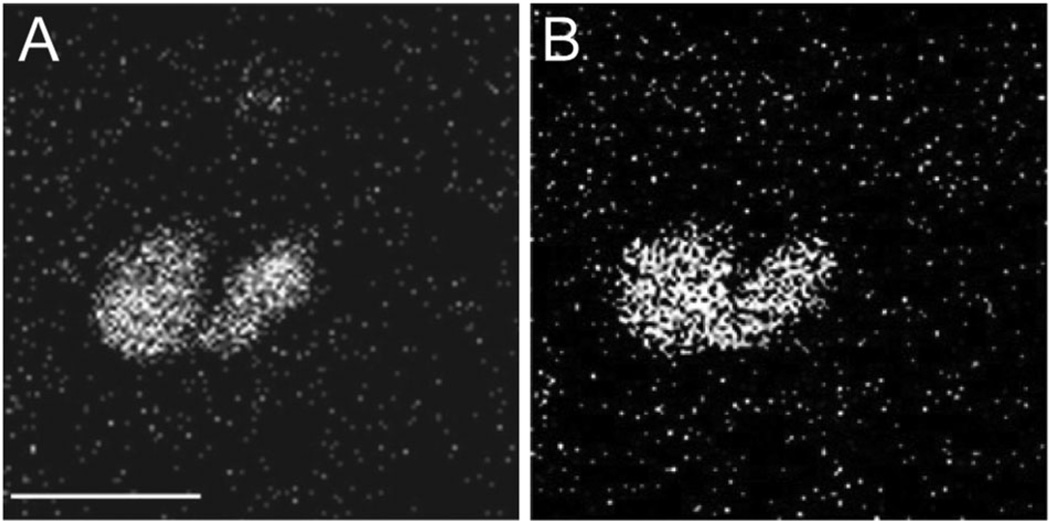
SIMS uses an accelerated primary ion beam to bombard the sample, a process that generates secondary ions that are then detected by a mass spectrometer.253 Based on instrument design, there are two different approaches to SIMS. The first method, time-of-flight (TOF) SIMS, uses a primary ion beam that is pulsed, and the resulting secondary ions are detected by a TOF mass spectrometer. The second approach, termed dynamic SIMS, uses a continuous primary ion beam, and preselected ions are detected by a magnetic sector mass spectrometer. Specialised instruments have been developed to improve both the sensitivity and spatial resolution of SIMS (vide infra).253 TOF-SIMS has been used to image lipids in cells and tissues.269–271 TOF-SIMS has also been used to image highly curved membranes during Tetrahymena mating (Fig. 11).272 In this study, Ostrowski et al. demonstrated that the fusion region contains elevated amounts of phosphatidylethanolamine, a high-curvature lipid.
Fig. 11.
Tetrahymena thermophila mating results in lipid heterogeneity at membrane fusion sites. (A) SIMS image for m/z 184, representative of the phosphocholine headgroup, demonstrating the absence of phosphatidylcholine at the conjugation junction. (B) SIMS image for m/z 126, representative of the phosphoethanolamine headgroup, demonstrating the presence of phosphatidylethanolamine at the conjugation junction. SIMS images were acquired using a 200 nm beam spot size (B250 nm lateral resolution). Scale bar = 50 µm. From Science 2004, 305, 71. Reprinted with permission from AAAS.
In the last decade, advancements in instrumentation have greatly improved the utility of SIMS for imaging biological samples. These developments include nanoSIMS, which enables image resolution of a few tens of nanometers.253 This technique has been applied in tracer studies examining 13C-labeled free fatty acid transport across cell membranes.273 Boxer and co-workers have used nanoSIMS to image lipid domains within supported lipid bilayers with a lateral resolution of 100 nm.274 NanoSIMS has also been used to demonstrate that deep-sea anaerobic methane-oxidizing archaea fix nitrogen and cyanide and share the products with sulfate-reducing bacterial symbionts.275 Lechene et al. have developed multiple-isotope imaging mass spectrometry (MIMS), in which multiple isotopes are detected simultaneously, enabling both imaging and quantification of labeled molecules within subcellular compartments.276 This technique was utilised to test the idea that stem cells retain the older template DNA to insure lifetime genetic stability, a concept known as the immortal strand hypothesis.277 In this study, the authors showed that the DNA strands are randomly segregated during cell proliferation. Another technical advance includes cluster time-of-flight (TOF)-SIMS, which has been shown to improve the yield of secondary ions produced.278 This technique utilises heavier primary ions that have improved efficiency for production of secondary ions, thus allowing easier identification of biomolecules. Many groups have demonstrated that cluster TOF-SIMS imaging can be used to image lipids in rodent brains and tissues.279–283 Finally, Heeren and co-workers have developed matrix-enhanced SIMS (ME-SIMS), which combines the high spatial resolution of SIMS with the sample preparation of MALDI IMS.284 Like cluster TOF-SIMS, ME-SIMS also increases the molecular ion yield.
4.2 Raman and coherent anti-Stokes Raman scattering (CARS) microscopy
Raman and coherent anti-Stokes Raman scattering (CARS) microscopy are emerging label-free methods for cellular imaging.285 These two techniques rely on optical signals and have been used to image proteins, lipids, and nucleic acids.286,287 Xie and co-workers have recently demonstrated that stimulated emission can also be used as a contrast mechanism for microscopy.288 They have used this technique to image chromoproteins, small-molecule drug distributions in vivo, and haemoglobin. For the purposes of this review, we will focus exclusively on Raman and CARS applications for imaging small molecules.
Raman microscopy relies on a phenomenon known as Raman scattering. This effect occurs when the wavelength of scattered light shifts slightly from the original wavelength of light due to excitation of the light-scattering molecules to a vibrationally excited state.286 Because the shift in wavelength depends strongly on the molecule’s structure and chemical environment, the resulting spectra can be used to identify specific molecules in the sample. In Raman microscopy, spectra are obtained from each position in the sample to construct a 2D or 3D image of molecular distribution based on signal intensities. This imaging technique has been used to visualize nucleic acids289,290 and lipid bodies291 in live cells. Raman microscopy has also been used to discern malignant tissue from healthy tissue for cancer diagnostic purposes.292–294 Raman microscopy, however, suffers from several limitations.295 First, the Raman effect is very weak, and consequently, data acquisition times are long. Second, creation of images requires high-powered lasers and long integration times per pixel, impeding video-rate microscopy. Xie and co-workers have greatly improved the sensitivity of Raman microscopy by developing a new technique known as stimulated Raman scattering (SRS) microscopy, which has been used to visualize lipids in live cells and tissues from mice.296 Furthermore, in vivo imaging was enabled by enhancement of the signal and by increasing the imaging speed to video-rate.297 Recently, Freudiger et al. have improved the spectral selectivity of SRS, thus allowing the molecular distinction between related molecules by using spectrally tailored excitation.298
Stronger vibrational signals can be obtained using CARS microscopy. In CARS, light beams with two different optical frequencies interact with the sample. A strong CARS signal is produced when the difference of the two frequencies matches the vibrational frequency of the molecules. Because CARS is orders of magnitude more sensitive than Raman microscopy, it allows video-rate vibrational imaging. The Reintjes group was the first to use CARS as a contrast mechanism for microsopy.299 The technique was popularized by Xie and co-workers almost two decades later when they developed a greatly improved method that allowed for higher sensitivity, higher spatial resolution, and three-dimensional sectioning capabilities.300
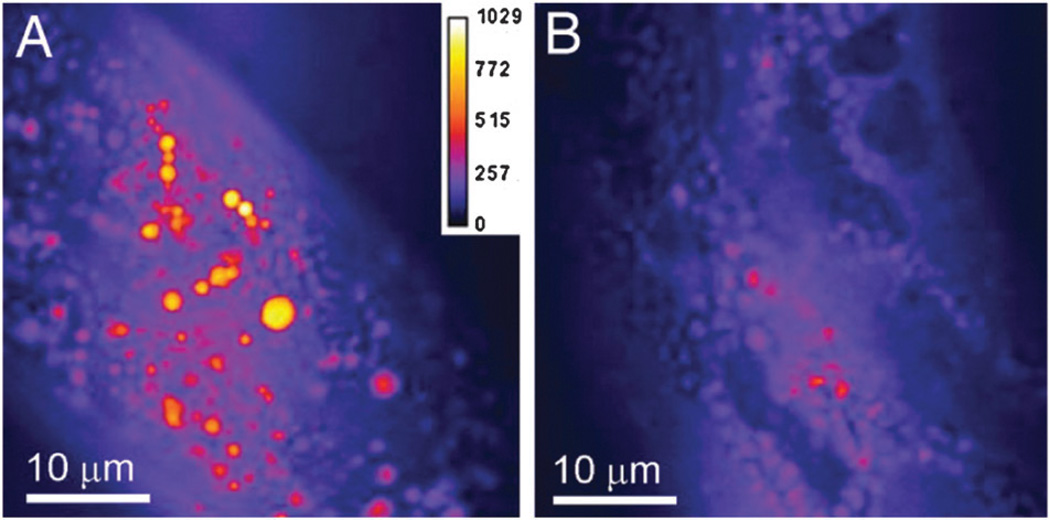
With these improvements, CARS has been used extensively for many in vitro and in vivo imaging applications. Wurpel et al. and Potma et al. have used CARS to image lipid vesicles and lipids on supported bilayers.301,302 Xie and co-workers have also used CARS to image lipids in live cells.303–305 Others have imaged lipids in infection models,306,307 mouse brains,308 and whole organisms such as C. elegans (Fig. 12).309–311 Improvements to CARS include video-rate microscopy applications of imaging lipids in the skin of live mice312 and frequency modulation CARS, which significantly increases the detection sensitivity of CARS by decreasing the nonresonant background.313
Fig. 12.
CARS imaging of lipid storage in C. elegans. CARS microscopy reveals a dramatic difference in lipid droplet densities from wild-type C. elegans (A) and a feeding-defective mutant, pha-3 (B). From Proc Natl Acad Sci 2007, 104, 14658. Reprinted with permission from National Academy of Sciences, U.S.A.
Conclusions
Recent advances in molecular imaging techniques have been made possible by fundamental advances in disparate areas of chemistry, spanning from metal-coordination chemistry to biochemistry to spectroscopy. These new methods have enabled the visualisation of virtually every major class of molecule inside living cells. As these methods are further improved and gain wider usage, it is our hope that they will continue to shed light on many biological processes.
Acknowledgements
P.V.C. was supported by the National Science Foundation and American Chemical Society Division of Medicinal Chemistry predoctoral fellowships. We thank Jeremy Baskin and Kimberly Beatty for critical reading of the manuscript.
Biographies

Pamela V. Chang
Pamela V. Chang received a BS in Chemistry from the Massachusetts Institute of Technology (MIT) in 2004. At MIT she worked in the laboratory of Professor Stephen J. Lippard. Upon graduating from MIT, she pursued graduate studies at the University of California, Berkeley, with Professor Carolyn R. Bertozzi, and completed her PhD in Chemistry in 2010. As a graduate student, Pamela’s research interests focused on developing chemical tools for imaging glycans in vivo. Pamela is currently a Jane Coffin Childs postdoctoral fellow at Yale University in the Department of Immunobiology, under the guidance of Professor Ruslan Medzhitov.

Carolyn R. Bertozzi
Carolyn R. Bertozzi is the T.Z. and Irmgard Chu Distinguished Professor of Chemistry and Professor of Molecular and Cell Biology at University of California, Berkeley and an Investigator of the Howard Hughes Medical Institute. She completed her undergraduate degree in Chemistry from Harvard University in 1988 and her PhD in Chemistry from UC Berkeley in 1993. After postdoctoral work at the University of California, San Francisco, in the field of cellular immunology, she joined the UC Berkeley faculty in 1996. Prof. Bertozzi’s research interests span the disciplines of chemistry and biology with an emphasis on glycobiology, microbiology, and nanomaterials.
References
- 1.Massoud TF, Gambhir SS. Genes Dev. 2003;17:545. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Science. 2002:296, 503. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 3.Tsien RY. Angew. Chem., Int. Ed. 2009;48:5612. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 4.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2008. [Google Scholar]
- 5.Zhang J, Campbell RE, Ting AY, Tsien RY. Nat. Rev. Mol. Cell Biol. 2002;3:906. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 6.Martin TF. Annu. Rev. Cell Dev. Biol. 1998;14:231. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- 7.Lemmon MA. Nat. Rev. Mol. Cell Biol. 2008;9:99. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 8.Halet G. Biol. Cell. 2005;97:501. doi: 10.1042/BC20040080. [DOI] [PubMed] [Google Scholar]
- 9.Varnai P, Balla T. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2006;1761:957. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY. Nature. 1991;349:694. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 11.Zaccolo M, De Giorgi F, Cho CY, Feng L, Knapp T, Negulescu PA, Taylor SS, Tsien RY, Pozzan T. Nat. Cell Biol. 1999;2:25. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 12.Zaccolo M, Pozzan T. Science. 2002;295:1711. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 13.Dyachok O, Isakov Y, Sagetorp J, Tengholm A. Nature. 2006;439:349. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- 14.Dipilato LM, Zhang J. Curr. Opin. Chem. Biol. 2010;14:37. doi: 10.1016/j.cbpa.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willoughby D, Cooper DM. Nat. Methods. 2007;5:29. doi: 10.1038/nmeth1135. [DOI] [PubMed] [Google Scholar]
- 16.McCombs JE, Palmer AE. Methods. 2008;46:152. doi: 10.1016/j.ymeth.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 18.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. J. Biol. Chem. 2001;276:29188. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 19.Baird GS, Zacharias DA, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11241. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. Nat. Biotechnol. 2002;20:87. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 21.Wallace DJ, Meyer zum Alten Borgloh S, Astori S, Yang Y, Bausen M, Kugler S, Palmer AE, Tsien RY, Sprengel R, Kerr JN, Denk W, Hasan MT. Nat. Methods. 2008;5:797. doi: 10.1038/nmeth.1242. [DOI] [PubMed] [Google Scholar]
- 22.Palmer AE, Tsien RY. Nat. Protoc. 2006;1:1057. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 23.Rudolf R, Mongillo M, Rizzuto R, Pozzan T. Nat. Rev. Mol. Cell Biol. 2003;4:579. doi: 10.1038/nrm1153. [DOI] [PubMed] [Google Scholar]
- 24.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10554. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horikawa K, Yamada Y, Matsuda T, Kobayashi K, Hashimoto M, Matsu-ura T, Miyawaki A, Michikawa T, Mikoshiba K, Nagai T. Nat. Methods. 2010;7:729. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 26.Nakai J, Ohkura M, Imoto K. Nat. Biotechnol. 2001;19:137. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 27.Tallini YN, Ohkura M, Choi BR, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin HB, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, Kotlikoff MI. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4753. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreosti E, Odermatt B, Dorostkar MM, Lagnado L. Nat. Methods. 2009;6:883. doi: 10.1038/nmeth.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Nat. Methods. 2009;6:875. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. Science. 2011;333:1888. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8740. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hires SA, Zhu Y, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4411. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsien RY. FEBS Lett. 2005;579:927. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Nat. Methods. 2006;3:281. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 35.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Curr. Opin. Cell Biol. 2005;17:183. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Niethammer P, Grabher C, Look AT, Mitchison TJ. Nature. 2009;459:996. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wold F. Annu. Rev. Biochem. 1981;50:783. doi: 10.1146/annurev.bi.50.070181.004031. [DOI] [PubMed] [Google Scholar]
- 38.Aye-Han NN, Ni Q, Zhang J. Curr. Opin. Chem. Biol. 2009;13:392. doi: 10.1016/j.cbpa.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura H, Hayashi-Takanaka Y, Yamagata K. Curr. Opin. Cell Biol. 2010;22:412. doi: 10.1016/j.ceb.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Tarrant MK, Cole PA. Annu. Rev. Biochem. 2009;78:797. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Allen MD. Mol. BioSyst. 2007;3:759. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Ma Y, Taylor SS, Tsien RY. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14997. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurokawa K, Mochizuki N, Ohba Y, Mizuno H, Miyawaki A, Matsuda M. J. Biol. Chem. 2001;276:31305. doi: 10.1074/jbc.M104341200. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki K, Sato M, Umezawa Y. J. Biol. Chem. 2003;278:30945. doi: 10.1074/jbc.M212167200. [DOI] [PubMed] [Google Scholar]
- 45.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. J. Biol. Chem. 2004;280:5581. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Violin JD, Zhang J, Tsien RY, Newton AC. J. Cell Biol. 2003;161:899. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Nature. 2005;434:1040. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Nature. 2005;437:569. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 49.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Nature. 2008;453:1132. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ouyang M, Sun J, Chien S, Wang Y. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14353. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19264. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CW, Jao CY, Ting AY. J. Am. Chem. Soc. 2004;126:5982. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- 53.Lin CW, Ting AY. Angew. Chem., Int. Ed. 2004;43:2940. doi: 10.1002/anie.200353375. [DOI] [PubMed] [Google Scholar]
- 54.Felsenfeld G, Groudine M. Nature. 2003;421:448. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 55.Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16257. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carrillo LD, Krishnamoorthy L, Mahal LK. J. Am. Chem. Soc. 2006;128:14768. doi: 10.1021/ja065835+. [DOI] [PubMed] [Google Scholar]
- 57.Hart GW, Housley MP, Slawson C. Nature. 2007;446:1017. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 58.Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD. Methods. 2008;46:143. doi: 10.1016/j.ymeth.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. Mill Valley: University Science Books; 1994. [Google Scholar]
- 60.Tsien RY. Biochemistry. 1980;19:2396. doi: 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]
- 61.Haugland RP. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. Eugene, OR: Molecule Probes, Inc.; 2005. [Google Scholar]
- 62.Minta A, Kao JP, Tsien RY. J. Biol. Chem. 1989;264:8171. [PubMed] [Google Scholar]
- 63.Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. Nat. Chem. Biol. 2007;3:423. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:1517. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 65.Domaille DW, Que EL, Chang CJ. Nat. Chem. Biol. 2008;4:168. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- 66.Frederickson C. Science’s STKE. 2003;2003:1. doi: 10.1126/stke.2003.182.pe18. [DOI] [PubMed] [Google Scholar]
- 67.Frederickson CJ, Koh JY, Bush AI. Nat. Rev. Neurosci. 2005;6:449. doi: 10.1038/nrn1671. [DOI] [PubMed] [Google Scholar]
- 68.Gee KR, Zhou ZL, Qian WJ, Kennedy R. J. Am. Chem. Soc. 2002;124:776. doi: 10.1021/ja011774y. [DOI] [PubMed] [Google Scholar]
- 69.Chang CJ, Nolan EM, Jaworski J, Burdette SC, Sheng M, Lippard SJ. Chem. Biol. 2004;11:203. doi: 10.1016/j.chembiol.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 70.Woodroofe CC, Masalha R, Barnes KR, Frederickson CJ, Lippard SJ. Chem. Biol. 2004;11:1659. doi: 10.1016/j.chembiol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 71.Burdette SC, Frederickson CJ, Bu W, Lippard SJ. J. Am. Chem. Soc. 2003;125:1778. doi: 10.1021/ja0287377. [DOI] [PubMed] [Google Scholar]
- 72.Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ. J. Am. Chem. Soc. 2001;123:7831. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 73.Walkup GK, Burdette SC, Lippard SJ, Tsien RY. J. Am. Chem. Soc. 2000;122:5644. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 74.Nolan EM, Ryu JW, Jaworski J, Feazell RP, Sheng M, Lippard SJ. J. Am. Chem. Soc. 2006;128:15517. doi: 10.1021/ja065759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nolan EM, Jaworski J, Okamoto K, Hayashi Y, Sheng M, Lippard SJ. J. Am. Chem. Soc. 2005;127:16812. doi: 10.1021/ja052184t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang XA, Hayes D, Smith SJ, Friedle S, Lippard SJ. J. Am. Chem. Soc. 2008;130:15788. doi: 10.1021/ja807156b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh SK, Kim P, Zhang XA, Yun SH, Moore A, Lippard SJ, Medarova Z. Cancer Res. 2010;70:6119. doi: 10.1158/0008-5472.CAN-10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirano T, Kikuchi K, Urano Y, Higuchi T, Nagano T. J. Am. Chem. Soc. 2000;122:12399. [Google Scholar]
- 79.Wu Y, Peng X, Guo B, Fan J, Zhang Z, Wang J, Cui A, Gao Y. Org. Biomol. Chem. 2005;3:1387. doi: 10.1039/b501795e. [DOI] [PubMed] [Google Scholar]
- 80.Tang B, Huang H, Xu KH, Tong LL, Yang GW, Liu X, An LG. Chem. Commun. 2006:3609. doi: 10.1039/b606809j. [DOI] [PubMed] [Google Scholar]
- 81.Kiyose K, Kojima H, Urano Y, Nagano T. J. Am. Chem. Soc. 2006;128:6548. doi: 10.1021/ja060399c. [DOI] [PubMed] [Google Scholar]
- 82.Lim NC, Schuster JV, Porto MC, Tanudra MA, Yao L, Freake HC, Bruckner C. Inorg. Chem. 2005;44:2018. doi: 10.1021/ic048905r. [DOI] [PubMed] [Google Scholar]
- 83.Lim NC, Bruckner C. Chem. Commun. 2004:1094. doi: 10.1039/b403448a. [DOI] [PubMed] [Google Scholar]
- 84.Gee KR, Zhou ZL, Ton-That D, Sensi SL, Weiss JH. Cell Calcium. 2002;31:245. doi: 10.1016/S0143-4160(02)00053-2. [DOI] [PubMed] [Google Scholar]
- 85.Maruyama S, Kikuchi K, Hirano T, Urano Y, Nagano T. J. Am. Chem. Soc. 2002;124:10650. doi: 10.1021/ja026442n. [DOI] [PubMed] [Google Scholar]
- 86.Taki M, Wolford JL, O’Halloran TV. J. Am. Chem. Soc. 2004;126:712. doi: 10.1021/ja039073j. [DOI] [PubMed] [Google Scholar]
- 87.Woodroofe CC, Lippard SJ. J. Am. Chem. Soc. 2003;125:11458. doi: 10.1021/ja0364930. [DOI] [PubMed] [Google Scholar]
- 88.Zhang XA, Lovejoy KS, Jasanoff A, Lippard SJ. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10780. doi: 10.1073/pnas.0702393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanaoka K, Kikuchi K, Urano Y, Narazaki M, Yokawa T, Sakamoto S, Yamaguchi K, Nagano T. Chem. Biol. 2002;9:1027. doi: 10.1016/s1074-5521(02)00216-8. [DOI] [PubMed] [Google Scholar]
- 90.Hanaoka K, Kikuchi K, Urano Y, Nagano T. J. Chem. Soc., Perkin Trans. 2. 2001:1840. [Google Scholar]
- 91.Barnham KJ, Masters CL, Bush AI. Nat. Rev. Drug Discovery. 2004;3:205. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 92.Halliwell B, Gutteridge J. Free Radicals in Biology and Medicine. Oxford: Oxford University Press; 1999. [Google Scholar]
- 93.Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11179. doi: 10.1073/pnas.0406547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morgan MT, Bagchi P, Fahrni CJ. J. Am. Chem. Soc. 2011;133:15906. doi: 10.1021/ja207004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng L, Miller EW, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2006;128:10. doi: 10.1021/ja055064u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller EW, Zeng L, Domaille DW, Chang CJ. Nat. Protoc. 2006;1:824. doi: 10.1038/nprot.2006.140. [DOI] [PubMed] [Google Scholar]
- 97.Domaille DW, Zeng L, Chang CJ. J. Am. Chem. Soc. 2010;132:1194. doi: 10.1021/ja907778b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dodani SC, Leary SC, Cobine PA, Winge DR, Chang CJ. J. Am. Chem. Soc. 2011;133:8606. doi: 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dodani SC, Domaille DW, Nam CI, Miller EW, Finney LA, Vogt S, Chang CJ. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5980. doi: 10.1073/pnas.1009932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirayama T, Van de Bittner GC, Gray LW, Lutsenko S, Chang CJ. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2228. doi: 10.1073/pnas.1113729109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Que EL, Gianolio E, Baker SL, Wong AP, Aime S, Chang CJ. J. Am. Chem. Soc. 2009;131:8527. doi: 10.1021/ja900884j. [DOI] [PubMed] [Google Scholar]
- 102.Que EL, Chang CJ. J. Am. Chem. Soc. 2006;128:15942. doi: 10.1021/ja065264l. [DOI] [PubMed] [Google Scholar]
- 103.Petrat F, Rauen U, de Groot H. Hepatology. 1999;29:1171. doi: 10.1002/hep.510290435. [DOI] [PubMed] [Google Scholar]
- 104.Ma Y, Luo W, Camplo M, Liu Z, Hider RC. Bioorg. Med. Chem. Lett. 2005;15:3450. doi: 10.1016/j.bmcl.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 105.Lytton SD, Mester B, Libman J, Shanzer A, Cabantchik ZI. Anal. Biochem. 1992;205:326. doi: 10.1016/0003-2697(92)90443-b. [DOI] [PubMed] [Google Scholar]
- 106.Xiang Y, Tong A. Org. Lett. 2006;8:1549. doi: 10.1021/ol060001h. [DOI] [PubMed] [Google Scholar]
- 107.Xiang Y, Tong A, Jin P, Ju Y. Org. Lett. 2006;8:2863. doi: 10.1021/ol0610340. [DOI] [PubMed] [Google Scholar]
- 108.Ignarro LJ. Nitric Oxide Biology and Pathobiology. San Diego: Academic Press; 2000. [Google Scholar]
- 109.Miller EW, Chang CJ. Curr. Opin. Chem. Biol. 2007;11:620. doi: 10.1016/j.cbpa.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McQuade LE, Lippard SJ. Curr. Opin. Chem. Biol. 2009;2010 doi: 10.1016/j.cbpa.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 111.Gomes A, Fernandes E, Lima JL. J. Fluoresc. 2006;16:119. doi: 10.1007/s10895-005-0030-3. [DOI] [PubMed] [Google Scholar]
- 112.Soh N. Anal. Bioanal. Chem. 2006;386:532. doi: 10.1007/s00216-006-0366-9. [DOI] [PubMed] [Google Scholar]
- 113.Lim MH, Lippard SJ. Acc. Chem. Res. 2007;40:41. doi: 10.1021/ar950149t. [DOI] [PubMed] [Google Scholar]
- 114.LeBel CP, Ischiropoulos H, Bondy SC. Chem. Res. Toxicol. 1992;5:227. doi: 10.1021/tx00026a012. [DOI] [PubMed] [Google Scholar]
- 115.Henderson LM, Chappell JB. Eur. J. Biochem. 1993;217:973. doi: 10.1111/j.1432-1033.1993.tb18328.x. [DOI] [PubMed] [Google Scholar]
- 116.Marchesi E, Rota C, Fann YC, Chignell CF, Mason RP. Free Radical Biol. Med. 1999;26:148. doi: 10.1016/s0891-5849(98)00174-9. [DOI] [PubMed] [Google Scholar]
- 117.Afzal M, Matsugo S, Sasai M, Xu B, Aoyama K, Takeuchi T. Biochem. Biophys. Res. Commun. 2003;304:619. doi: 10.1016/s0006-291x(03)00641-7. [DOI] [PubMed] [Google Scholar]
- 118.Chang MC, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2004;126:15392. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller EW, Albers AE, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2005;127:16652. doi: 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Nat. Chem. Biol. 2007;3:263. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 121.Dickinson BC, Huynh C, Chang CJ. J. Am. Chem. Soc. 2010;132:5906. doi: 10.1021/ja1014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Albers AE, Okreglak VS, Chang CJ. J. Am. Chem. Soc. 2006;128:9640. doi: 10.1021/ja063308k. [DOI] [PubMed] [Google Scholar]
- 123.Dickinson BC, Chang CJ. J. Am. Chem. Soc. 2008;130:9638. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Srikun D, Albers AE, Nam CI, Iavarone AT, Chang CJ. J. Am. Chem. Soc. 2010;132:4455. doi: 10.1021/ja100117u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dickinson BC, Tang Y, Chang Z, Chang CJ. Chem. Biol. 2011;18:943. doi: 10.1016/j.chembiol.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dickinson BC, Peltier J, Stone D, Schaffer DV, Chang CJ. Nat. Chem. Biol. 2010;7:106. doi: 10.1038/nchembio.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller EW, Dickinson BC, Chang CJ. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15681. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Anal. Chem. 1998;70:2446. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 129.Kojima H, Hirotani M, Nakatsubo N, Kikuchi K, Urano Y, Higuchi T, Hirata Y, Nagano T. Anal. Chem. 2001;73:1967. doi: 10.1021/ac001136i. [DOI] [PubMed] [Google Scholar]
- 130.Gabe Y, Ueno T, Urano Y, Kojima H, Nagano T. Anal. Bioanal. Chem. 2006;386:621. doi: 10.1007/s00216-006-0587-y. [DOI] [PubMed] [Google Scholar]
- 131.Gabe Y, Urano Y, Kikuchi K, Kojima H, Nagano T. J. Am. Chem. Soc. 2004;126:3357. doi: 10.1021/ja037944j. [DOI] [PubMed] [Google Scholar]
- 132.Sasaki E, Kojima H, Nishimatsu H, Urano Y, Kikuchi K, Hirata Y, Nagano T. J. Am. Chem. Soc. 2005;127:3684. doi: 10.1021/ja042967z. [DOI] [PubMed] [Google Scholar]
- 133.Izumi S, Urano Y, Hanaoka K, Terai T, Nagano T. J. Am. Chem. Soc. 2009;131:10189. doi: 10.1021/ja902511p. [DOI] [PubMed] [Google Scholar]
- 134.Zheng H, Shang GQ, Yang SY, Gao X, Xu JG. Org. Lett. 2008;10:2357. doi: 10.1021/ol800206x. [DOI] [PubMed] [Google Scholar]
- 135.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press; 2008. [PubMed] [Google Scholar]
- 136.Haltiwanger RS, Lowe JB. Annu. Rev. Biochem. 2004;73:491. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 137.Rosen SD. Annu. Rev. Immunol. 2004;22:129. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 138.Fuster MM, Esko JD. Nat. Rev. Cancer. 2005;5:526. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 139.Sharon N. J. Biol. Chem. 2006;282:2753. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 140.Pilobello KT, Mahal LK. Methods Mol. Biol. 2007;385:193. doi: 10.1007/978-1-59745-426-1_14. [DOI] [PubMed] [Google Scholar]
- 141.Hirabayashi J. Glycoconjugate J. 2004;21:35. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- 142.Kiessling LL, Pohl NL. Chem. Biol. 1996;3:71. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- 143.Ohba H, Bakalova R. Cancer Chemother. Pharmacol. 2003;51:451. doi: 10.1007/s00280-003-0607-y. [DOI] [PubMed] [Google Scholar]
- 144.Carlsson S, Carlsson MC, Leffler H. Glycobiology. 2007;17:906. doi: 10.1093/glycob/cwm059. [DOI] [PubMed] [Google Scholar]
- 145.Paessens LC, Garcia-Vallejo JJ, Fernandes RJ, van Kooyk Y. Immunol. Lett. 2007;110:65. doi: 10.1016/j.imlet.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 146.Licha K, Debus N, Emig-Vollmer S, Hofmann B, Hasbach M, Stibenz D, Sydow S, Schirner M, Ebert B, Petzelt D, Buhrer C, Semmler W, Tauber R. J. Biomed. Opt. 2005;10:41205. doi: 10.1117/1.2007967. [DOI] [PubMed] [Google Scholar]
- 147.Hang HC, Bertozzi CR. Acc. Chem. Res. 2001;34:727. doi: 10.1021/ar9901570. [DOI] [PubMed] [Google Scholar]
- 148.Dvorak HF, Nagy JA, Dvorak AM. Cancer Cells. 1991;3:77. [PubMed] [Google Scholar]
- 149.Laughlin ST, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2008;106:12. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sletten EM, Bertozzi CR. Angew. Chem., Int. Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sletten EM, Bertozzi CR. Acc. Chem. Res. 2011;44:666. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jencks WP. J. Am. Chem. Soc. 1959;81:475. [Google Scholar]
- 153.Dirksen A, Dirksen S, Hackeng TM, Dawson PE. J. Am. Chem. Soc. 2006;128:15602. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 154.Dirksen A, Hackeng TM, Dawson PE. Angew. Chem., Int. Ed. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 155.Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 156.Sadamoto R, Niikura K, Ueda T, Monde K, Fukuhara N, Nishimura S. J. Am. Chem. Soc. 2004;126:3755. doi: 10.1021/ja039391i. [DOI] [PubMed] [Google Scholar]
- 157.Griffin RJ. Prog. Med. Chem. 1994;31:121. doi: 10.1016/s0079-6468(08)70020-1. [DOI] [PubMed] [Google Scholar]
- 158.Baskin JM, Bertozzi CR. QSAR Comb. Sci. 2007;26:1211. [Google Scholar]
- 159.Staudinger H, Meyer J. Helv. Chim. Acta. 1919;2:635. [Google Scholar]
- 160.Gololobov YG, Kasukhin LF. Tetrahedron. 1992;48:1353. [Google Scholar]
- 161.Saxon E, Bertozzi CR. Science. 2000;287:2007. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 162.Prescher JA, Dube DH, Bertozzi CR. Nature. 2004;430:873. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 163.Chang PV, Prescher JA, Hangauer MJ, Bertozzi CR. J. Am. Chem. Soc. 2007;129:8400. doi: 10.1021/ja070238o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hangauer MJ, Bertozzi CR. Angew. Chem., Int. Ed. 2008;47:2394. doi: 10.1002/anie.200704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cohen AS, Dubikovskaya EA, Rush JS, Bertozzi CR. J. Am. Chem. Soc. 2010;132:8563. doi: 10.1021/ja101766r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4819. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Neves AA, Stockmann H, Harmston RR, Pryor HJ, Alam IS, Ireland-Zecchini H, Lewis DY, Lyons SK, Leeper FJ, Brindle KM. FASEB J. 2011;25:2528. doi: 10.1096/fj.10-178590. [DOI] [PubMed] [Google Scholar]
- 168.Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. Nat. Methods. 2009;6:207. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moller H, Bohrsch V, Bentrop J, Bender J, Hinderlich S, Hackenberger CP. Angew. Chem., Int. Ed. Engl. 2012;51:5986. doi: 10.1002/anie.201108809. [DOI] [PubMed] [Google Scholar]
- 170.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VD, Yarema KJ. Glycobiology. 2009;19:1382. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9116. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14846. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3141. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR. J. Am. Chem. Soc. 2006;128:12078. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12371. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zheng T, Jiang H, Gros M, del Amo DS, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. Angew. Chem., Int. Ed. 2011;50:4113. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem., Int. Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 178.Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 179.Kolb HC, Finn MG, Sharpless KB. Angew. Chem., Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 180.Hong V, Steinmetz NF, Manchester M, Finn MG. Bioconjugate Chem. 2010;21:1912. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Soriano Del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J. Am. Chem. Soc. 2010;132:16893. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano Del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem., Int. Ed. 2011;50:8051. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, Gee KR, Ting AY. Angew. Chem., Int. Ed. Engl. 2012;51:5852. doi: 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew. Chem., Int. Ed. 2009;48:4030. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2614. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dumont A, Malleron A, Awwad M, Dukan S, Vauzeilles B. Angew. Chem., Int. Ed. 2012;51:3143. doi: 10.1002/anie.201108127. [DOI] [PubMed] [Google Scholar]
- 187.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 188.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16793. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem. Biol. 2006;1:644. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 190.Laughlin ST, Bertozzi CR. ACS Chem. Biol. 2009;4:1068. doi: 10.1021/cb900254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10360. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dehnert KW, Beahm BJ, Huynh TT, Baskin JM, Laughlin ST, Wang W, Wu P, Amacher SL, Bertozzi CR. ACS Chem. Biol. 2011;6:547. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. ChemBioChem. 2012;13:353. doi: 10.1002/cbic.201100649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Codelli J, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1821. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sletten EM, Bertozzi CR. Org. Lett. 2008;10:3097. doi: 10.1021/ol801141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jewett JC, Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2010;132:3688. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poloukhtine AA, Mbua NE, Wolfert MA, Boons GJ, Popik VV. J. Am. Chem. Soc. 2009;131:15769. doi: 10.1021/ja9054096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ning X, Guo J, Wolfert MA, Boons GJ. Angew. Chem., Int. Ed. 2008;47:2253. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Debets MF, van Berkel SS, Schoffelen S, Rutjes FP, van Hest JC, van Delft FL. J. Chem. Commun. 2010;46:97. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 201.Dommerholt J, Schmidt S, Temming R, Hendriks LJ, Rutjes FP, van Hest JC, Lefeber DJ, Friedl P, van Delft FL. J. Angew. Chem., Int. Ed. 2010;49:9422. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stockmann H, Neves AA, Stairs S, Ireland-Zecchini H, Brindle KM, Leeper FJ. Chem. Sci. 2011;2:932. doi: 10.1039/C0SC00631A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Friscourt F, Ledin PA, Mbua NE, Flanagan-Steet H, Wolfert MA, Steet R, Boons GJ. J. Am. Chem. Soc. 2012;134:5381. doi: 10.1021/ja3002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jewett JC, Bertozzi CR. Org. Lett. 2011;13:5937. doi: 10.1021/ol2025026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.de Almeida G, Sletten EM, Nakamura H, Palaniappan KK, Bertozzi CR. Angew. Chem., Int. Ed. 2012;51:2443. doi: 10.1002/anie.201106325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schoenebeck F, Ess DH, Jones GO, Houk KN. J. Am. Chem. Soc. 2009;131:8121. doi: 10.1021/ja9003624. [DOI] [PubMed] [Google Scholar]
- 207.Ess DH, Jones GO, Houk KN. Org. Lett. 2008;10:1633. doi: 10.1021/ol8003657. [DOI] [PubMed] [Google Scholar]
- 208.Gordon CG, Mackey JL, Jewett JC, Sletten EM, Houk KN, Bertozzi CR. J. Am. Chem. Soc. 2012;134:9199. doi: 10.1021/ja3000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Debets MF, van Berkel SS, Dommerholt J, Dirks AT, Rutjes FP, van Delft FL. Acc. Chem. Res. 2011;44:805. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 210.van Berkel SS, Dirks AT, Debets MF, van Delft FL, Cornelissen JJ, Nolte RJ, Rutjes FP. ChemBioChem. 2007;8:1504. doi: 10.1002/cbic.200700278. [DOI] [PubMed] [Google Scholar]
- 211.Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Devaraj NK, Weissleder R. Acc. Chem. Res. 2011;44:816. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wirges CT, Timper J, Fischler M, Sologubenko AS, Mayer J, Simon U, Carell T. Angew. Chem., Int. Ed. 2008;48:219. doi: 10.1002/anie.200803123. [DOI] [PubMed] [Google Scholar]
- 214.Kaya E, Vrabel M, Deiml C, Prill S, Fluxa VS, Carell T. Angew. Chem., Int. Ed. 2012;51:4466. doi: 10.1002/anie.201109252. [DOI] [PubMed] [Google Scholar]
- 215.Lim RK, Lin Q. Acc. Chem. Res. 2011;44:828. doi: 10.1021/ar200021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2011;133:17570. doi: 10.1021/ja2072934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Woscholski R. Signal Transduction. 2006;6:77. [Google Scholar]
- 218.Resh MD. Nat. Chem. Biol. 2006;2:584. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 219.Charron G, Wilson J, Hang HC. Curr. Opin. Chem. Biol. 2009;13:382. doi: 10.1016/j.cbpa.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 220.Vance JE, Vance DE. Biochem. Cell Biol. 2004;82:113. doi: 10.1139/o03-073. [DOI] [PubMed] [Google Scholar]
- 221.Jao CY, Roth M, Welti R, Salic A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15332. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Neef AB, Schultz C. Angew. Chem., Int. Ed. 2009;48:1498. doi: 10.1002/anie.200805507. [DOI] [PubMed] [Google Scholar]
- 223.Hannoush RN, Sun J. Nat. Chem. Biol. 2010;6:498. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 224.Hang HC, Wilson JP, Charron G. Acc. Chem. Res. 2011;44:699. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hang HC, Linder ME. Chem. Rev. 2011;111:6341. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12479. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Berry AF, Heal WP, Tarafder AK, Tolmachova T, Baron RA, Seabra MC, Tate EW. ChemBioChem. 2010;11:771. doi: 10.1002/cbic.201000087. [DOI] [PubMed] [Google Scholar]
- 228.Charron G, Tsou LK, Maguire W, Yount JS, Hang HC. Mol. BioSyst. 2011;7:67. doi: 10.1039/c0mb00183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.DeGraw AJ, Palsuledesai C, Ochocki JD, Dozier JK, Lenevich S, Rashidian M, Distefano MD. Chem. Biol. Drug Des. 2010;76:460. doi: 10.1111/j.1747-0285.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. J. Am. Chem. Soc. 2007;129:2744. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 231.Martin DD, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, Berthiaume LG. FASEB J. 2007;22:797. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. FASEB J. 2007;22:721. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yap MC, Kostiuk MA, Martin DD, Perinpanayagam MA, Hak PG, Siddam A, Majjigapu JR, Rajaiah G, Keller BO, Prescher JA, Wu P, Bertozzi CR, Falck JR, Berthiaume LG. J. Lipid Res. 2009;51:1566. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hannoush RN, Arenas-Ramirez N. ACS Chem. Biol. 2009;4:581. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 235.Heal WP, Wickramasinghe SR, Bowyer PW, Holder AA, Smith DF, Leatherbarrow RJ, Tate EW. Chem. Commun. 2008:480. doi: 10.1039/b716115h. [DOI] [PubMed] [Google Scholar]
- 236.Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. Org. Biomol. Chem. 2008;6:2308. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- 237.Heal WP, Jovanovic B, Bessin S, Wright MH, Magee AI, Tate EW. Chem. Commun. 2011;47:4081. doi: 10.1039/c0cc04710d. [DOI] [PubMed] [Google Scholar]
- 238.Dirks RW, Tanke HJ. BioTechniques. 2006;40:489. doi: 10.2144/000112121. [DOI] [PubMed] [Google Scholar]
- 239.Bao G, Rhee WJ, Tsourkas A. Annu. Rev. Biomed. Eng. 2009;11:25. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Gramlich PM, Wirges CT, Manetto A, Carell T. Angew. Chem., Int. Ed. 2008;47:8350. doi: 10.1002/anie.200802077. [DOI] [PubMed] [Google Scholar]
- 241.Wang CC, Seo TS, Li Z, Ruparel H, Ju J. Bioconjugate Chem. 2003;14:697. doi: 10.1021/bc0256392. [DOI] [PubMed] [Google Scholar]
- 242.Seo TS, Li Z, Ruparel H, Ju J. J. Org. Chem. 2003;68:609. doi: 10.1021/jo026615r. [DOI] [PubMed] [Google Scholar]
- 243.Cai J, Li X, Yue X, Taylor JS. J. Am. Chem. Soc. 2004;126:16324. doi: 10.1021/ja0452626. [DOI] [PubMed] [Google Scholar]
- 244.Rakic P. Nat. Rev. Neurosci. 2002;3:65. doi: 10.1038/nrn700. [DOI] [PubMed] [Google Scholar]
- 245.Salic A, Mitchison TJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Neef AB, Luedtke NW. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20404. doi: 10.1073/pnas.1101126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Guan L, van der Heijden GW, Bortvin A, Greenberg MM. ChemBioChem. 2011;12:2184. doi: 10.1002/cbic.201100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jao CY, Salic A. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15779. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nat. Methods. 2007;4:828. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 250.McDonnell LA, Heeren RM. Mass Spectrom. Rev. 2007;26:606. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 251.Seeley EH, Caprioli RM. Anal. Chem. 2012;84:2105. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Heeren RMA, McDonnell LA, Amstalden E, Luxembourg SL, Altelaar AFM, Piersma SR. Appl. Surf. Sci. 2006;252:6827. [Google Scholar]
- 253.Boxer SG, Kraft ML, Weber PK. Annu. Rev. Biophys. 2009;38:53. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 254.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 255.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 256.Francese S, Dani FR, Traldi P, Mastrobuoni G, Pieraccini G, Moneti G. Comb. Chem. High Throughput Screening. 2009;12:156. doi: 10.2174/138620709787315454. [DOI] [PubMed] [Google Scholar]
- 257.Woods AS, Jackson SN. AAPS J. 2006;8:E391. doi: 10.1007/BF02854910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.McLean JA, Ridenour WB, Caprioli RM. J. Mass Spectrom. 2007;42:1099. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 259.Jackson SN, Wang HY, Woods AS, Ugarov M, Egan T, Schultz JA. J. Am. Soc. Mass Spectrom. 2005;16:133. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 260.Rujoi M, Estrada R, Yappert MC. Anal. Chem. 2004;76:1657. doi: 10.1021/ac0349680. [DOI] [PubMed] [Google Scholar]
- 261.Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. Chem. Rev. 2011;111:6491. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Chughtai K, Heeren RM. Chem. Rev. 2010;110:3237. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NY. Cancer Res. 2011;72:645. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2007;855:98. doi: 10.1016/j.jchromb.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 265.Touboul D, Roy S, Germain DP, Chaminade P, Brunelle A, Laprevote O. Int. J. Mass Spectrom. 2007;260:158. [Google Scholar]
- 266.Esquenazi E, Coates C, Simmons L, Gonzalez D, Gerwick WH, Dorrestein PC. Mol. BioSyst. 2008;4:562. doi: 10.1039/b720018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Yang YL, Xu Y, Straight P, Dorrestein PC. Nat. Chem. Biol. 2009;5:885. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Watrous JD, Dorrestein PC. Nat. Rev. Microbiol. 2011;9:683. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fletcher JS. Analyst. 2009;134:2204. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 270.Johansson B. Surf. Interface Anal. 2006;38:1401. [Google Scholar]
- 271.Nygren H, Malmberg P, Kriegeskotte C, Arlinghaus HF. FEBS Lett. 2004;566:291. doi: 10.1016/j.febslet.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 272.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kleinfeld AM, Kampf JP, Lechene C. J. Am. Soc. Mass Spectrom. 2004;15:1572. doi: 10.1016/j.jasms.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 274.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Science. 2006;313:1948. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 275.Dekas AE, Poretsky RS, Orphan VJ. Science. 2009;326:422. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 276.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. J. Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, Lee RT, Lechene CP. Nature. 2012;481:516. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Mas S, Perez R, Martinez-Pinna R, Egido J, Vivanco F. Proteomics. 2008;8:3735. doi: 10.1002/pmic.200800115. [DOI] [PubMed] [Google Scholar]
- 279.Malmberg P, Borner K, Chen Y, Friberg P, Hagenhoff B, Mansson JE, Nygren H. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2007;1771:185. doi: 10.1016/j.bbalip.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 280.Amaya KR, Monroe EB, Sweedler JV, Clayton DF. Int. J. Mass Spectrom. 2007;260:121. [Google Scholar]
- 281.Nygren H, Borner K, Hagenhoff B, Malmberg P, Mansson JE. Biochim. Biophys. Acta. 2005;1737:102. doi: 10.1016/j.bbalip.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 282.Touboul D, Brunelle A, Halgand F, De La Porte S, Laprevote O. J. Lipid Res. 2005;46:1388. doi: 10.1194/jlr.M500058-JLR200. [DOI] [PubMed] [Google Scholar]
- 283.Sjovall P, Lausmaa J, Johansson B. Anal. Chem. 2004;76:4271. doi: 10.1021/ac049389p. [DOI] [PubMed] [Google Scholar]
- 284.McDonnell LA, Piersma SR, Altelaar AFM, Mize TH, Luxembourg SL, Verhaert PDEM, van Minnen J, Heeren RMA. J. Mass Spectrom. 2005;40:160. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 285.Pezacki JP, Blake JA, Danielson DC, Kennedy DC, Lyn RK, Singaravelu R. Nat. Chem. Biol. 2011;7:137. doi: 10.1038/nchembio.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Fujita K, Smith NI. Mol. Cell. 2008;26:530. [PubMed] [Google Scholar]
- 287.Min W, Freudiger CW, Lu S, Xie XS. Annu. Rev. Phys. Chem. 2011;62:507. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Min W, Lu S, Chong S, Roy R, Holtom GR, Xie XS. Nature. 2009;461:1105. doi: 10.1038/nature08438. [DOI] [PubMed] [Google Scholar]
- 289.Uzunbajakava N, Lenferink A, Kraan Y, Volokhina E, Vrensen G, Greve J, Otto C. Biophys. J. 2003;84:3968. doi: 10.1016/S0006-3495(03)75124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Puppels GJ, de Mul FF, Otto C, Greve J, Robert-Nicoud M, Arndt-Jovin DJ, Jovin TM. Nature. 1990;347:301. doi: 10.1038/347301a0. [DOI] [PubMed] [Google Scholar]
- 291.van Manen HJ, Kraan YM, Roos D, Otto C. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10159. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Huang Z, McWilliams A, Lui H, McLean DI, Lam S, Zeng H. Int. J. Cancer. 2003;107:1047. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 293.Shafer-Peltier KE, Haka AS, Fitzmaurice M, Crowe J, Myles J, Dasari RR, Feld MS. J. Raman Spectrosc. 2002;33:552. [Google Scholar]
- 294.Nijssen A, Schut TCB, Heule F, Caspers PJ, Hayes DP, Neumann MHA, Puppels GJ. J. Invest. Dermatol. 2002;119:64. doi: 10.1046/j.1523-1747.2002.01807.x. [DOI] [PubMed] [Google Scholar]
- 295.Evans CL, Xie XS. Annu. Rev. Anal. Chem. 2008;1:883. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 296.Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He CW, Tsai JC, Kang JX, Xie XS. Science. 2008;322:1857. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Science. 2010;330:1368. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Freudiger CW, Min W, Holtom GR, Xu B, Dantus M, Xie XS. Nat. Photonics. 2011;5:103. doi: 10.1038/nphoton.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Duncan MD, Reintjes J, Manuccia TJ. Opt. Lett. 1982;7:350. doi: 10.1364/ol.7.000350. [DOI] [PubMed] [Google Scholar]
- 300.Zumbusch A, Holtom GR, Xie XS. Phys. Rev. Lett. 1999;82:4142. [Google Scholar]
- 301.Wurpel GW, Rinia HA, Muller M. J. Microsc. 2005;218:37. doi: 10.1111/j.1365-2818.2005.01462.x. [DOI] [PubMed] [Google Scholar]
- 302.Potma EO, Evans C, Xie XS, Jones RJ, Ye J. Opt. Lett. 2003;28:1835. doi: 10.1364/ol.28.001835. [DOI] [PubMed] [Google Scholar]
- 303.Xie XS, Yu J, Yang WY. Science. 2006;312:228. doi: 10.1126/science.1127566. [DOI] [PubMed] [Google Scholar]
- 304.Nan X, Potma EO, Xie XS. Biophys. J. 2006;91:728. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Nan X, Cheng JX, Xie XS. J. Lipid Res. 2003;44:2202. doi: 10.1194/jlr.D300022-JLR200. [DOI] [PubMed] [Google Scholar]
- 306.Nan X, Tonary AM, Stolow A, Xie XS, Pezacki JP. ChemBioChem. 2006;7:1895. doi: 10.1002/cbic.200600330. [DOI] [PubMed] [Google Scholar]
- 307.Rakic B, Sagan SM, Noestheden M, Belanger S, Nan X, Evans CL, Xie XS, Pezacki JP. Chem. Biol. 2006;13:23. doi: 10.1016/j.chembiol.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 308.Evans CL, Xu XY, Kesari S, Xie XS, Wong STC, Young GS. Opt. Express. 2007;15:12076. doi: 10.1364/oe.15.012076. [DOI] [PubMed] [Google Scholar]
- 309.Hellerer T, Axang C, Brackmann C, Hillertz P, Pilon M, Enejder A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14658. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Le TT, Duren HM, Slipchenko MN, Hu CD, Cheng JX. J. Lipid Res. 2009;51:672. doi: 10.1194/jlr.D000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Folick A, Min W, Wang MC. Curr. Opin. Genet. Dev. 2011;21:585. doi: 10.1016/j.gde.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Evans CL, Potma EO, Puoris’haag M, Cote D, Lin CP, Xie XS. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16807. doi: 10.1073/pnas.0508282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ganikhanov F, Evans CL, Saar BG, Xie XS. Opt. Lett. 2006;31:1872. doi: 10.1364/ol.31.001872. [DOI] [PubMed] [Google Scholar]