Abstract
Accurate determination of the amount of a given RNA within a cell is necessary to gain a full understanding of the RNA's function and regulation. Typically, the abundance of RNA is measured by quantitative polymerase chain reaction (qPCR). With qPCR, however, absolute quantification is not possible unless an adequate reference standard curve is generated. The method is not well suited for detecting low copy number templates and values vary depending on the specific primers used. To overcome these drawbacks, digital PCR (dPCR) has been developed to obtain exact values for RNA copies in a sample. Here we report the characterization of droplet digital PCR (ddPCR). We used ddPCR to quantify long noncoding RNAs from various subcellular compartments within human cells and found that results obtained using ddPCR parallel those from qPCR. Mutant huntingtin (HTT) protein is the cause of Huntington's Disease, and we show that we can quantify human HTT messenger RNA and discriminate between the mutant and wild-type HTT alleles using ddPCR. These results reveal insights into the design of experiments using ddPCR and show that ddPCR can be a robust tool for identifying the number of RNA species inside of cells.
Introduction
Accurate RNA measurement is one of the foundations of modern experimental biology and an essential part of any serious effort to develop nucleic acid therapeutics. Typically, RNA measurements may involve initial characterization of a transcript by northern analysis or polymerase chain reaction. Quantitative PCR (qPCR) is often used to gain a quantitative understanding of how a transcript's levels vary with respect to a given treatment. Missing in almost all of these analyses, however, is a recognition of the actual number of transcripts that exist in a sample. This variable, while critical for obtaining a complete understanding of RNA function and susceptibility to designed antagonists, is almost universally ignored.
Quantitative PCR, which relies on an increase in fluorescence signal that is proportional to the polymerase reaction product, or amplicon produced, uses the cycle threshold (CT) as a metric. This value is defined as the number of thermal cycles required for the fluorescence signal to rise above background noise but is often taken to be the point at which amplification enters log phase. It is therefore a rate-based measurement and a powerful guide to relative concentration.
Typically, CT values for specific genes are referenced to well-known housekeeping genes, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or ribosomal protein L30 (RPL30), across samples and used for normalization. This can become problematic as expression levels of housekeeping genes and their transcripts vary between cell populations, cellular compartments, and tissue types and samples (Warren et al., 2006; vide infra). These problems can be partially avoided through the use of an exogenous “spike-in” control. This method, however, does not account for any template-specific effect or bias introduced through primer design. Minute variations in technique, like pipetting, can also introduce significant errors in quantitation from spike-in controls. To estimate the absolute number of a given RNA species, data must be compared to a previously generated standard curve of the same template with identical primers and conditions. While straightforward in theory, the additional manipulations are cumbersome and extreme care must be taken when measuring the reference sample and comparing the reference and experimental standard curves.
Digital PCR (dPCR) methods aim to provide a more direct measurement of RNA copy number. The earliest dPCR strategies involved dilution of template to isolate individual molecules in a single reaction, followed by PCR detection (Kalinina et al., 1997; Vogelstein and Kinzler, 1999). Subsequent methods have refined dPCR by (1) increasing the number of reactions to enable more powerful statistical analysis; (2) minimizing reaction volume to reach a saturating concentration of PCR product with fewer starting templates while reducing the amount of sample necessary to obtain robust results; and (3) improving hardware and software for automation and detection to increase throughput and decrease cost.
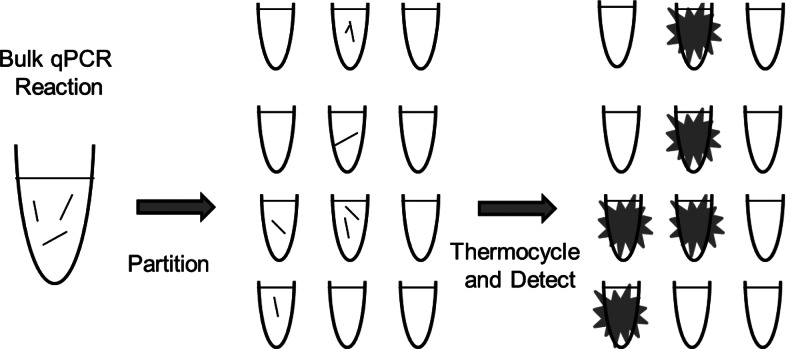
Several dPCR methods that rely on similar principles are currently available. A bulk sample, containing the sequence(s) of interest, is partitioned into a distinct environment so that no further mixing may occur. Partitioning is calibrated so that almost all partitions contain zero or one template molecule. These partitioned solutions are then subjected to standard thermal cycling and PCR conditions. Any partition containing at least one template will act as a mini-reaction vessel for amplification that results in an observable signal (Fig. 1). The ratio of positive to negative reactions can then be interpreted as an absolute copy/well number using statistical methods.
FIG. 1.
Diagram displaying the principle of digital polymerase chain reaction (PCR). A bulk PCR reaction is partitioned into small volume aliquots that may or may not lack template. After thermal cycling, aliquots that contained at least 1 template molecule will show a positive signal. The ratio of positive to negative reactions is then analyzed using Poisson statistics to predict the absolute copy number in the original bulk sample.
Two basic categories of instruments are available. The first—and most well established—category uses microfluidic wells to separate individual reactions. Platforms such as the BioMark HD system from Fluidigm and Applied Biosystem's OpenArray rely on microfluidic dynamic arrays with charge-coupled device camera detection of fluorescence readout. The number of partitions available for statistical analysis limits these systems. Balanced against this limitation, each individual well may be monitored as an independent qPCR reaction (and thus provide a CT value) and end point melt analyses can be performed. These platforms are often applied as a high-throughput qPCR technology rather than for absolute quantitation of RNA. MicroRNA expression analysis (Petriv et al., 2010), single cell gene expression analysis (Citri et al., 2011), and targeted resequencing (Grossmann et al., 2011) are examples of these instruments being used as a high-throughput qPCR system.
The second category operates by emulsion PCR, which partitions a bulk sample into thousands of uniform, small volume aqueous droplets in oil. In these systems, the same principle of end point PCR and application of Poisson statistics to the ratio of positive to negative droplets may be applied. Examples include the Bio-Rad QX100 droplet digital PCR (ddPCR) system (Hindson et al., 2011; Pinheiro et al., 2011) or RainDance Technologies' RainStorm system (Kiss et al., 2008). The benefit of this approach is that the number of partitions can be increased from hundreds to many thousands. This yields more accurate quantitation and facilitates quantitation of low abundance nucleic acids. Emulsion PCR has been used to detect viruses (Henrich et al., 2012; Hayden et al., 2013), determine the expression of rare disease-related transcripts in tissue samples (Heredia et al., 2013), and determine genetic copy number variation among populations (Hindson et al., 2011). One drawback to this technique is that individual droplets are not monitored during amplification and only the endpoint is detected therefore an accompanying CT value may not be extracted from the data.
In this report we explore the application of droplet digital PCR using a Bio-Rad QX100 Droplet Digital PCR (ddPCR) system. Approximately 20,000 droplets per reaction are generated and each reaction is transferred to an individual well on a standard 96-well plate. After thermocycling, droplets containing a template will register a fluorescence signal upon amplification whereas a droplet lacking template will not. Each well containing 20,000 droplets is then “read” by passing the droplets through a fluorescence detector. The ratio of positive to negative signals effectively counts the RNA molecules in the starting sample. We provide insights into how to set up ddPCR experiments and develop several applications relevant to the design and evaluation of nucleic acid therapeutics.
Materials and Methods
Tissue culture
All reagents were obtained from Sigma unless otherwise noted. All culture media were supplemented with 10% (v/v) fetal bovine solution. HeLa cells (American Type Culture Collection) were maintained in RPMI-1640, A549 cells were maintained in F12-K media (ATCC). Huntington's disease patient-derived fibroblast cell line GM04281 (69 CAG repeats) and GM04795 wild-type fibroblasts were obtained from the Coriell Institute and maintained in Eagle's minimum essential medium.
Cellular fractionation and RNA isolation
Cells were scraped from plates with Dulbecco's phosphate buffered saline or detached using 1×trypsin ethylenediaminetetraacetic acid solution and pelleted at 500 g for 5 minutes at 4°C. Cells were counted using either a Beckman Coulter Z1 particle counter or a hemocytometer. Cells were divided into aliquots containing 5 million cells and processed as follows. The cytoplasmic membrane was lysed by the addition of hypotonic lysis buffer [10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.35% Triton X-100 containing RNasein RNase inhibitor (1.25 μL/mL)], complete protease inhibitor (Roche; 20 μL per mL) and dithiothreitol (1 μL/mL of 1 M stock). Resuspended cells were incubated on ice for 10 minutes then centrifuged at 800 g for 5 minutes, 4°C. The supernatant was then decanted and used as cytoplasmic fraction. Crude nuclei were washed 4 more times with hypotonic lysis buffer, centrifuging at 500 g for 5 minutes and washed once more centrifuging at 200 g for 5 minutes. For total nuclear fractions, the pure nuclei were dissolved in 1 mL TRI Reagent. For nucleoplasmic and chromatin fractions, nuclei were lysed in chromatin isolation buffer (0.3 M NaCl, 1 M urea, 1% NP-40) on ice with occasional mixing for 20 minutes and then centrifuged at 400 g. The supernatant was set aside as the nucleoplasmic fraction and the precipitated chromatin was further washed 2 more times with chromatin isolation buffer. Cytoplasmic and nucleoplasmic fractions were precipitated with 70% ethanol and 0.3 M sodium acetate then dissolved in 1 mL TRI reagent. The chromatin fraction was dissolved in 1 mL TRI reagent. To all samples dissolved in 1 mL of TRI reagent, 200 μL of chloroform was added. After vigorous vortexing and centrifugation, RNA was precipitated from the aqueous layer with 1 volume of isopropanol (500 μL). Isolated RNA was treated with 2U of DNase 1 (Worthington) in a total volume of 50 μL at 37°C for 30 minutes followed by heat inactivation at 80°C for 5 minutes.
Complementary DNA preparation, ddPCR, and qPCR
Total RNA from cell extracts was reverse transcribed in the same manner for qPCR and ddPCR experiments. Complementary DNA (cDNA) preparation was carried out using Applied Biosystems' high capacity reverse transcriptase kit with 5 μL of DNase-treated RNA (corresponding to RNA derived from 0.5 million cells) in a 20 μL reaction and subsequently diluted to 200 μL. Given the aforementioned dilutions, cDNA concentration was calculated to be at 2,500 cell equivalents of template per μL. The PCR reaction solution was reconstituted to a final volume of 20 μL using 1–5 μL of template and ddPCR Supermix (BioRad). Unless otherwise noted primer and probe concentrations were 0.5 μM and 0.025 μM respectively. Droplet formation was carried out using a QX100 droplet generator. To the bottom 8 wells of the droplet generation cartridge are added 70 μL of ddPCR droplet generation oil. The middle wells are loaded with 20 μL of reaction. A rubber gasket is placed over the cartridge and loaded into the droplet generator. Drawing the 2 liquids through microfluidic channels, an emulsion is created in the top well of the cartridge. The emulsion (∼40 μL in volume) is then slowly transferred using a multichannel pipette (great care must be taken to avoid droplet shearing) to a twin.tec semi-skirted 96-well plate (Eppendorf). The plate is then heat-sealed with foil and the emulsion was cycled to end point per the manufacturer's protocol. The samples were then read using a BioRad QX100 reader. Data from the droplet reader are given as copies per μL and were converted to copies per cell based on the known cell equivalents of input cDNA. Quanititative PCR was carried out on a BioRad CFX96 thermocycler in 20 μL reactions with 1–5 μL cDNA using iTaq supermix (BioRad). For qPCR and ddPCR, all primers were designed and synthesized as PrimeTime Assays by Integrated DNA Technologies. Hydrolysis probes contained a 5′-FAM fluorophore with an internal ZEN and a 3′-Iowa black quencher. Primers and probes for detection of GAPDH were designed and synthesized by Applied Biosystems. Primers and probes were designed to amplify and detect all known transcript isoforms. Primers and probes for the genes studied are indicated below. All sequences are given 5′→3′ (F, forward primer; R, reverse primer; P, probe):
COX-2 lncRNA (cyclooxygenase 2 long noncoding RNA): F, GCTCACTGCAAGTCGTATGA; R, CACATGGGCTTGGTTTTCAG; P, CAATTGGTCGCTAACCGAGAGAACCTT.
MALAT1 (metastasis associated lung adenocarcinoma transcript 1): F, ACCATCGTTACCTTGAAACCG; R, GATCTAGCACAGACCCTTCAC; P, CTCACCTCGATGCAGCCAGTAGC.
NEAT1 (nuclear enriched abundant transcript 1): F, TCTCTTCCTCCACCATTACCA; R, CCTCCCTTTAACTTATCCATTCAC; P, AACAATACCGACTCCAACAGCCACT.
U14 (small nucleoler RNA U14; SNORD14A): F, CACTGTGATGATGGTTTTCCAAC; R, AGGAAGGTTTACCCAACACTAAG; P, CGCAGTTTCCACCAGAAAGGTTTTCC.
HOTAIR (HOX antisense intergenic RNA): F, GCTTCTAAATCCGTTCCATTCC; R, GAGTTCCACAGACCAACACC; P, TCAATCAGAAAGGTCCTGCTCCGC.
RPL30: F, CACCAGTTTTAGCCAACATAGC; R, GATCAGACAAGGCAAAGCGA; P, CAACTGCCCAGCTTTGAGGAAATCT.
dPCR evaluation of huntingtin alleles
Huntington's disease patient-derived fibroblast cell line GM04281 (69 CAG repeats) and GM04795 wild-type fibroblasts were detached from plates using 1×trypsin, and counted with a hemocytometer. Forty thousand cells were then taken up in 1 mL Trizol, and RNA isolation and cDNA preparation were carried out as for previous examples. Primers for the quantification of HTT messenger RNA (mRNA) were overlapping the CAG repeat region with the probe immediately at the 3′ end of the repeat: F, ATGGCGACCCTGGAAAAG; R, GGCTGAGGAAGCTGAGGAG; P, TTCGAGTCCCTCAAGTCCTTCC. Standard primer and probe concentrations were used for the quantification of HTT mRNA (0.5 μM and 0.025 μM respectively). High primer and probe concentrations (1.6 μM and 0.5 μM respectively) were used to obtain allelic discrimination of huntingtin mRNA.
Results and Discussion
Quantitative PCR versus ddPCR for the absolute quantification of long noncoding RNA
Two percent of the human genome is encoded by mature mRNA and 30 % by pre-mRNA (Lander et al., 2001; Venter et al., 2001). Analysis has revealed, however, that >80% of the genome is transcribed (ENCODE Project Consortium, 2012). Most of these transcripts have no obvious potential to code for proteins and those with lengths greater than 200 nucleotides are termed long noncoding RNAs (lncRNAs). Some of these lncRNAs are intergenic while others overlap the coding regions of known mRNA.
While the function of some lncRNAs, such as HOTAIR or XIST, has been described (Brown et al. 2001; Rinn et al. 2007) function has not been assigned to the vast majority. Accurate quantitation of lncRNA abundance might suggest function and discriminate between lncRNAs that have a biological role and those that may be biological noise due to trace action of RNA polymerase. Accurate quantitation would also help separate lncRNAs that affect gene expression in cis relative to chromosomal DNA (likely requiring one or a few RNAs) relative to those that act in trans (likely to require hundreds or thousands).Accurate quantitation of a transcript relies on a proper distribution of positive (cDNA from the target RNA present) to negative (no cDNA) droplets. If a cDNA is present, TaqMan PCR will yield a fluorescent signal. Should the concentration of target cDNA be too low or too high, statistical analysis becomes less reliable. For example, concentrations that are too low will yield few positive droplets, while concentrations that are too high will yield many droplets with 2 or more templates, obscuring the potential for digital identification. In testing the dynamic range of ddPCR, we found that the instrument would give useful information from between ∼10 and ∼10,000 copies per μL of reaction mixture. This corresponds to CT values in the range of approximately 18–28. Droplet digital PCR, therefore, is effective over a concentration range similar to that characterizing qPCR.
For determining transcript numbers by qPCR, developing a standard curve requires serial dilutions that require multiple pipetting steps that can lead to introduction of experimental error. Droplet digital PCR does not require serial dilution steps, reducing one source of error.
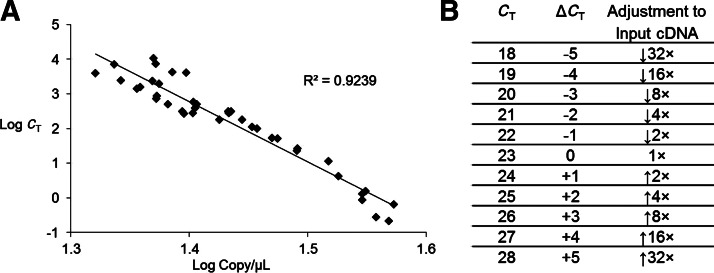
Using a log plot of CT values against absolute quantification by ddPCR involving 8 separate genes and primer sets, a general estimate of copy number can be obtained from a CT value determined by traditional qPCR (Fig. 2A). Optimal precision is found when the sample lies at the center of this plotted range (CT ∼23). We used these data to develop a guide for how to adjust the concentration of samples that were previously analyzed by qPCR to obtain the best possible results from ddPCR (Fig. 2B).
FIG. 2.
Comparison of droplet digital PCR (ddPCR) and quantitive PCR (qPCR). (A) Correlation of log base 10 copy number obtained from ddPCR with CT value obtained from qPCR. Greater variation at the lower and upper end of concentrations tested can be seen. (B) A guide for using qPCR data to choose an appropriate volume for ddPCR. For example, a sample with a CT value of 18 should be diluted 32-fold for best results. A sample with a CT value of 28 should be increased in concentration 32-fold.
Detection of cyclooxygenase-2 lncRNA
We have observed that a lncRNA overlapping the cyclooxygenase 2 (COX-2) transcription start site and COX-2 promoter is a target for an endogenous micro RNA and multiple synthetic RNAs that cause activation of COX-2 gene transcription (Matsui et al., submitted). This COX-2 lncRNA is expressed 50-fold less than COX-2 mRNA and has been determined to be distinct from the COX-2 mRNA by 5′-rapid amplification of cDNA ends experiments and can be detected by traditional PCR using a forward primer complementary to a site 81 base pairs upstream of the mRNA transcription start site (Matsui et al., submitted). Because of the clear role for this lncRNA in controlling COX-2 expression, and the therapeutic relevance of the gene with regards to inflammatory response, we used ddPCR to characterize its expression levels.
The first step was to obtain an accurate count of the number of cells. We compared 2 methods, automated cell counting, and manual measurement using a hemocytometer. We found that the manual measurement was superior because automated cell counting, while faster and more convenient, does not distinguish well between single cells and clumps. Clumping may not be an issue for some cell lines, but it was a feature of the A549 lung cancer cells used in our study.
After counting cells, we obtained cell extracts. Because we were interested in the amount of lncRNA throughout the cell, we obtained extracts from cytoplasm and nuclei. We further purified nuclei to obtain nucleoplasmic (soluble nuclear) RNA and RNA associated with chromatin. These 3 samples were analyzed by both ddPCR and qPCR. For qPCR, we calculated a standard curve to enable calculation of transcript number. While the generation of a single standard curve is not prohibitively laborious, if one were to study several transcripts the process would rapidly become so. One of the advantages of ddPCR is that up to 96 genes can be analyzed per plate for absolute quantification.
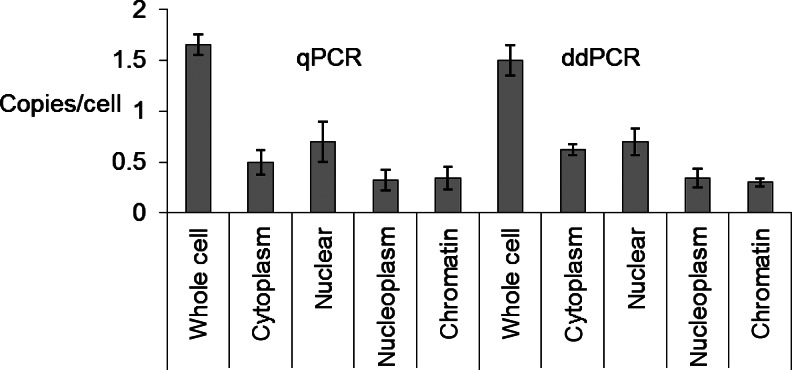
Droplet digital PCR and qPCR produced similar results (Fig. 3). The lncRNA is detected at between 1 and 2 copies per cell, can be associated with chromatin, and is evenly distributed between cytoplasm and nuclei. Relevant to COX-2 regulation, the low copy number of the promoter lncRNA favors action in cis immediately after synthesis rather than a mechanism in which the lncRNA would leave the chromosome and then diffuse back or affect some other target. For digital determination of RNA levels, these data confirm that (1) ddPCR can detect rare transcripts, and (2) ddPCR performs as well as qPCR but does not require calculation of a standard curve.
FIG. 3.
Quantification of cyclooxygenase 2 long noncoding RNA (COX-2 lncRNA) by qPCR and ddPCR showing good agreement in total copy number per cell and distribution across compartments. Experiments were performed in triplicate. Error is given as±standard deviation from the mean.
Detection of MALAT1, HOTAIR, and NEAT1 lncRNAs and U14 snoRNA
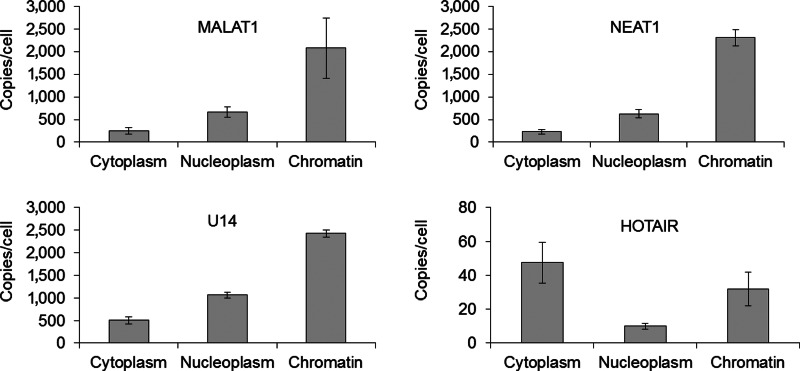
After validating the ddPCR platform in comparison to qPCR for COX-2 lncRNA, we used ddPCR to evaluate the quantities of abundant lncRNAs MALAT1, HOTAIR, and NEAT1, and the snoRNA U14 (SNORD14A) in different compartments within HeLa cells (Fig. 4). The lncRNAs were chosen because of their potential for therapeutic significance. MALAT1 has been implicated in lung and colon cancers while HOTAIR has been shown to promote the metastasis of breast and hepatocellular tumors (for review see Prensner and Chinnaiyan, 2011). All 4 noncoding RNAs were much more abundant than the COX-2 promoter lncRNA, suggesting a different mechanism of action. MALAT1, NEAT1, and U14 were present at ∼3000 copies per cell, while HOTAIR was present at ∼100 copies per cell. Compared with COX-2 lncRNA, action in trans seems much more feasible for these lncRNA transcripts.
FIG. 4.
Quantification and subcellular distribution of metastasis associated lung adenocarcinoma transcript 1 (MALAT-1), nuclear enriched abundant transcript 1 (NEAT-1), HOX antisense intergenic RNA (HOTAIR), and small nucleolar RNA U14 (snoRNA U14). Experiments were performed in triplicate. Error is given as±standard deviation from the mean.
We expected these transcripts to reside primarily in the nucleus, as the 3 lncRNAs have known nuclear functions and snoRNAs, such as U14, are involved in the biogenesis of ribosomes in the nucleolus (Terns and Dahlberg, 1994). We found that all four RNAs were present in both the nucleus and the cytoplasm and could be found associated with chromatin. MALAT1, NEAT1, and U14 were primarily in the chromatin fraction, while most HOTAIR transcripts were cytoplasmic (Fig 4.).
By contrast, the protein-encoding genes GAPDH and RPL30 were present at 3,500 and 10,000 copies per cell respectively, and were primarily distributed to the cytoplasm (Fig. 5). These data suggest that care should be taken when choosing a gene for normalization. Our qPCR-derived CT values show no single gene was evenly distributed across all cellular compartments. Unless a gene of this nature can be found, normalization of CT values across cellular compartments is not possible without the use of a spike-in control. Droplet digital PCR avoids this complication by directly giving a count of the number of RNA copies.
FIG. 5.
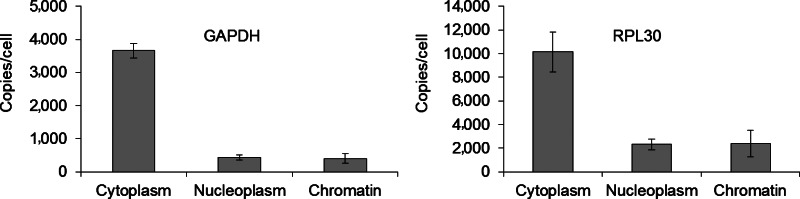
Quantification and subcellular distribution of housekeeping genes commonly used as controls for qPCR experiments. Experiments were performed in triplicate. Error is given as±standard deviation from the mean.
Quantitation of HTT mRNA
We also examined applications of ddPCR for quantifying specific mutant alleles of huntingtin (HTT) mRNA. Huntington's Disease (HD) is an incurable neurological disorder caused by an expanded CAG trinucleotide repeat in one allele of the HTT gene (MacDonald et al., 1993). Many neurological disorders, including Alzheimer's disease, Parkinson's disease, and depression are usually caused by a combination of multiple genetic influences and environmental causes. Because HD is caused by a single genetic defect, reducing mutant HTT expression has become a major target for gene-specific therapeutics (Matsui and Corey, 2012).
One strategy for treating HD is to develop agents that reduce the expression of mutant HTT mRNA that contains the expanded CAG repeat tract. Evaluating the copy number of HTT mRNA transcripts per cell would directly benefit drug development by providing a basis for estimating how many molecules will be needed to achieve inhibition. Indirectly, knowledge of HTT mRNA copy number may also facilitate the design of studies aimed at probing mRNA structure inside of cells or cell extracts. HTT is a large protein and detailed insights into the structure of its mRNA may require full-length message rather than small model RNAs. We applied ddPCR to evaluate HTT mRNA in homozygous wild-type human fibroblast cells and heterozygous patient derived cells. We detected ∼400 copies per cell for wild-type fibroblasts and ∼250 copies of HTT mRNA/cell in heterozygous fibroblasts containing an expanded repeat region.
We reasoned that it might be possible to determine the ratio of mutant to normal mRNA transcripts by exploiting the difference in repeat length of the alleles. Droplet digital PCR uses a TaqMan probe containing a 5′-FAM fluorophore and a 3′ quencher. As the polymerase reads the cDNA, when it reaches the hybridized probe, the 5′-FAM fluorophore is released. With both the wild-type and mutant alleles, there is just 1 probe binding event and the probe is complementary to the region immediately adjacent to the 3′ end of the repeat. An excess of primer and probe in the reaction ensures that an end point would be reached when all dNTPs are consumed.
Under such conditions, the wild-type transcript (shorter repeat—consuming fewer nucleotides per cycle) could potentially go through more rounds of replication and hydrolyze more FAM probe than the mutant transcript (longer repeat—consuming more nucleotides per cycle). Greater hydrolysis would result in increased fluorescence for reactions containing the wild-type HTT allele, as more rounds of replication are possible with the limited pool of dNTPs.
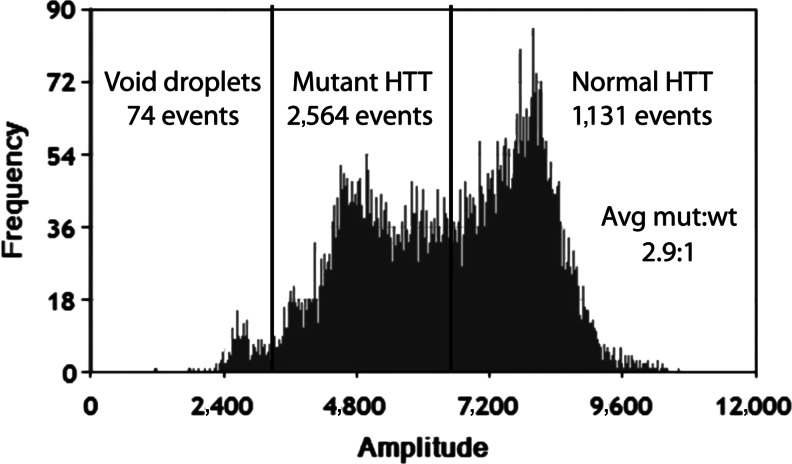
With primer sets at three times the typical concentration to ensure that primer concentration was not rate limiting we were able to see some separation of positive droplets into two populations (Fig. 6). Negative droplets (containing no template) can be seen at the far left of the histogram while positive droplets (containing mutant, wild-type, or both alleles) to the right of the histogram appear to display a bimodal distribution. We consistently observed a droplet ratio of ∼3:1 for what we posit to be mutant allele (lower fluorescence) to normal allele (higher fluorescence) HTT mRNA. Although these data are preliminary, this finding is consistent with reports that mutant Huntington transcript may accumulate in cells and thereby assert a toxic effect through the sequestration of RNA binding proteins (de Mezer et al., 2011).
FIG. 6.
Distribution of fluorescence events after ddPCR analysis of complementary DNA from heterozygous Huntington's Disease patient-derived cells. Frequency is the number of droplets. Amplitude is the signal in a given droplet generated by the release of fluorophore from hydrolyzed probe and is proportional to the number of amplicons within a droplet. Events due to both mutant and wild-type huntingtin (HTT) protein mRNA are shown.
Discussion
A full appreciation for how RNAs function inside of cells and how they can be most efficiently targeted by therapeutics requires an accurate determination of how many RNA molecules are present and where they are functioning in the cell. Such accurate numerical determinations, however, are rare. Quantitative PCR methods are indirect, only providing an estimate of transcript number, and require standard curves for each sequence of interest. In addition, differences in primer efficiency can still arise in the presence of a complex pool of nucleic acids, such as is found in a biologically relevant sample. Cognate or partially cognate sequences likely compete for binding, and native secondary structures may be present that are not in the template used for standard curve generation. As CT values are a rate-based measurement, these differences may lead to false conclusions.
When considering whether to use ddPCR, the major question facing any experimentalist is whether the benefits outweigh the cost of spending resources becoming familiar with a new technology and obtaining an instrument capable of quantifying single molecules. Droplet digital PCR has several benefits that suggest it as a valuable option for some experiments. Droplet digital PCR does not require a standard curve. While the development of standard curves is not particularly laborious, generation of multiple standard curves can be time consuming, and the need for serial dilutions requires unusual care when pipetting samples. A key advantage of ddPCR is its convenience for multiple samples. We were able to quantify multiple RNA species without the time consuming generation of multiple standard curves.
Also, when calculating a standard curve for qPCR, a synthetic template is used. Because it is not the natural template, alternate secondary structures might affect amplification. Even if qPCR is selected as a routine method, use of ddPCR to confirm results may be well advised for critical applications, and ddPCR may be helpful even if throughput does not need to be high. Taken together, ddPCR is a good option for the robust quantitation of multiple RNA samples. This may be especially true in a clinical setting where an absolute determination of RNA using a robust protocol is desired.
There are many applications for precise and accurate quantitation of RNA. An RNA species present in 1 copy per 100 cells might be more likely to be considered transcriptional noise or an artifact. A transcript present in 2 copies per cell and associated with chromatin might act in cis, while one present in a thousand copies per cell might suggest action in trans. An appreciation of numbers directly generates functional hypotheses. For important disease targets like HTT mRNA, knowing the number of transcripts aids studies of disease mechanism, mRNA structural analyses, and design and testing of therapeutics.
Obtaining reliable quantitative estimates will likely never be as easy as existing qualitative or semiquantitative methods for RNA measurement but should nevertheless be adopted as a more widely used tool for understanding and studying RNA.
Acknowledgments
This work was supported by grants from the Robert A. Welch Foundation (I-1244), the Cure Huntington's Disease Initiative (CHDI), and the National Institutes of Health (NIGMS 73042). DWD acknowledges an Isis Pharmaceuticals sponsored fellowship from the Life Sciences Research Foundation. We thank Rachelle Alderson and BioRad for loaning our laboratory a BioRad QX Droplet Digital PCR to complete these studies.
Author Disclosure Statement
No competing financial interests exist.
References
- BROWN C.J. BALLABIO A. RUPERT J.L. LAFRENIERE R.G. GROMPE M. TONLORENZI R. WILLARD H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- CITRI A. PANG Z.P. SÜDHOF T.C. WERNIG M. MALENKA R.C. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat. Protoc. 2012;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENOCDE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSSMANN V. KOHLMANN A. EDER C. HAFERLACH C. KERN W. CROSS N.C. HAFERLACH T. SCHNITTGER S. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- HAYDEN R.T. GU Z. INGERSOLL J. ABDUL-ALI D. SHI L. POUNDS S. CALIENDO A.M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of Cytomegalovirus. J. Clin. Microbiol. 2013;51:540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENRICH T.J. GALLIEN S. LI J.Z. PEREYRA F. KURITZKES D.R. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods. 2012;186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEREDIA N.J. BELGRADER P. WANG S. KOEHLER R. REGAN J. COSMAN A. SAXONOV S. HINDSON B. TANNER S.C. BROWN A.S. KARLIN-NEWMAN G. Droplet digital PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59:S20–S23. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- HINDSON B.J. NESS K.D. MASQUELIER D.A. BELGRADER P. HEREDIA N.J. MAKAREWICZ A.J. BRIGHT I.J. LUCERO M.Y. HIDDESSEN A.L. LEGLER T.C., et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALININA O. LEBEDEVA I. BROWN J. SILVER J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 1997;25:1999–2004. doi: 10.1093/nar/25.10.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISS M.M. ORTOLEVA-DONNELY L. BEER N.R. WARNER J. BAILEY C.G. COLSTON B.W. LINK D.R. LEAMON J.H. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal. Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDER E.S. LINTON L.M. BIRREN B. NUSBAUM C. ZODY M.C. BALDWIN J. DEVON K. DEWAR K. DOYLE M. FITZHUGH W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- MACDONALD M.E. AMBROSE C.M. DUYAO M.P. MYERS R.H. LIN C. SRINIDHI L. BARNES G. TAYLOR S.E. JAMES M. GROOT N., et al. THE HUNTINGTON'S DISEASE COLLABORATIVE RESEARCH GROUP. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- MATSUI M. COREY D.R. Allele-selective inhibition of trinucleotide repeat genes. Drug Discov. Today. 2012;10:443–450. doi: 10.1016/j.drudis.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE MEZER M. WOJCIECHOWSKA M. NAPIERALA M. SOBCZAK K. KRZYZOSIAK A.J. Mutant CAG repeats of huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETRIV O.I. KUCHENBAUER F. DELANEY A.D. LECAULT V. WHITE A. KENT D. MARMOLEJO L. HEUSER M. BERG T. COPLEY M., et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15443–15448. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PINHEIRO L.B. COLEMAN V.A. HINDSON C.M. HERRMANN J. HINDSON B.J. BHAT S. EMSLIE K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRENSNER J.R. CHINNAIYAN A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RINN J.L. KERTESZ M. WANG J.K. SQUAZZO S.L. XU X. BRUGMANN S.A. GOODNOUGH H.L. HELMS J.A. FARNHAM P.J. SEGAL E. CHANG H.L. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 1. 2007;29:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERNS M.P. DAHLBERG J.E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- VENTER J.C. Adams M.D. Myers E.W. Li P.W. Mural R.J. Sutton G.G. Smith H.O. Yandell M. Evans C.A. Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- VOGELSTEIN B. KINZLER K.W. Digital PCR. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARREN L. BRYDER D. WEISSMAN I.L. QUAKE S.R. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]