Abstract
Macromolecular interactions are central to the regulation and execution of many key biological processes, and therefore, they are attractive targets for drug discovery. Previously, we identified an RNA aptamer for the heat shock factor (HSF1), which is capable of interfering with the binding of HSF1 to its cognate DNA elements. Here we report the significant enhancement of avidity through dimerization of this aptamer. In particular, we describe the effect of 2 factors in designing a multivalent aptamer: the distance between active subunits and the flexibility of the linkage.
Introduction
A key issue in both drug discovery and basic biological research is to develop approaches and reagents capable of modulating macromolecular interactions (Juliano et al., 2001). Due to their physical limitations, very few small molecular drugs are able to interfere with interactions between proteins or nucleic acids (Juliano et al., 2001; Egner et al., 2005). As a result, macromolecular interactions have been dismissed as “undruggable” in many cases (Juliano et al., 2001). In contrast, monoclonal antibodies have been efficacious in targeting cell surface protein targets, but their intracellular applications are restricted by currently available delivery systems (Juliano et al., 2001; Egner et al., 2005). Compared with both small molecules and protein-based reagents, RNA aptamers have some special features (Ellington and Szostak, 1990; Tuerk and Gold, 1990). Generally, they possess high affinity and specificity for a targeted protein, evoke little immune response, and can be overproduced in specific cell types (Shi et al., 1999; Brody and Gold, 2000). These advantages led to the successful utilization of RNA aptamers to inhibit interactions involving protein and/or nucleic acids in a number of cases (Shi et al., 1999; Santulli-Marotto et al., 2003; Fan et al., 2004).
The interactions between transcription activators and their target DNA elements are good examples of macromolecular interactions and are essential for the induction of most genes. Among such interactions, those involving binding of multimeric complexes are usually very strong and particularly challenging to disrupt with drugs (Egner et al., 2005). The HSF1 trimer–HSE3 interaction is known as one of the strongest binding events and critical for the transcription activation of heat shock genes (WU, 1995). The apparent dissociation constant (Kd) of a typical HSF1 trimer interacting with the DNA heat shock elements, HSE3, is about 10–12 M (Xiao et al., 1991). HSF1 is highly conserved from yeast to humans and functions in a variety of important biological processes (Xiao et al., 1991; Pirkkala et al., 2001; Hahn et al., 2004). In mammals, it plays a unique role in supporting highly malignant cancers, including the most aggressive forms of breast, lung, and colon cancer (Mendillo et al., 2012). Elevated levels of HSF1 have been shown to be associated with poorer prognosis in some forms of breast cancer (Santagata et al., 2012); and down-regulation of HSF1 can sensitize tumor cells to anti-cancer drugs (Zaarur et al., 2006). Moreover, Hsf1–/– mice are resistant to tumor inducing agents (Dai et al., 2007). Therefore, modulating HSF1 activity in vivo by interfering with the HSF1/HSE3 interaction has important potential clinical significance (Mendillo et al., 2012).
For this purpose, we previously isolated an RNA aptamer for HSF1 named AptHSF-RA1 (Zhao et al., 2006). This aptamer has a Kd around 30 nM and interferes with the DNA binding of HSF1 in multiple species, including humans. To enhance its potency to disrupt the HSF-HSE interaction, we had sought to increase the avidity of the aptamer to HSF1 by homodimerization with various degrees of success (Wang et al., 2010; Salamanca et al., 2011). Here we report a series of constructs we designed and constructed in searching for the optimal arrangement of the 2 monomeric aptamers in the dimeric composite. Our data indicates that the spacing and linker flexibility are critical to the improved avidity, and our results demonstrate an effective strategy in designing high affinity multivalent aptamers to multimeric target proteins.
Materials and Methods
Aptamer and aptamer-derived constructs
The monomeric AptHSF-RA1 was described previously (Zhao et al., 2006). The full-length isolate and the “Core” used in this study have the following sequences:
RA1: 5′-GGGAGAAUUCAACUGCCAUCUAGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGU UCUCCAUCUAGUACUACAAGCUUCUGGACUCGAU-3′.
Core-2 (sequence starting and ending on stem-loop 2): 5′-GGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCC-3′.
The dimeric constructs have the following sequences:
3-2S: 5′-GGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUGGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCCAUUCAACUGCCC-3′
3-2: 5′-GGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUUCGCCUCUCGCCGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCGGCGAGAGGCGAAUUCAACUGCCC-3′.
3-2H: 5′-GGGCAAGCGCUAUGUUUUAAUUCAACUUGCGGAGUUCGCCUCUCGCCGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCGGCGAGAGGCGAAUUCAACUGCCC-3′.
3-2L: 5′-GGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUUCGCCUCUCGCCGGCAGAGUUCGCCUCUCGCCGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCGGCGAGAGGCGAACUCUGCCGGCGAGAGGCGAAUUCAACUGCCC-3′.
3-2PS: 5′-GGGCGAAUUCAACUGCCUUCGGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUUCGCCUCUCGCCGGCAUCGCGAUACAAAAUUAAGUUGAACGCGAGUCUUCGGAUUCAACUGCCGGC-3′.
All aptamers in this study were produced by in vitro transcription using T7 RNA polymerase from synthetic DNA templates, as described in detail below.
Electrophoretic mobility shift assay and competition assay
The preparation of yeast HSF and Drosophila HSF was described previously (Zhao et al., 2006; Salamanca et al., 2011).
For electrophoretic mobility shift assay (EMSA), RNA probes were internally labeled with [α-32P] UTP using a T7 in vitro transcription kit (MAXIscript Kit, Ambion). The binding solution contained binding buffer (10 mM Tris, 40 mM KOAc, 1mM MgCl2, pH 7.6), 1 μg carrier yeast RNA, 4 μg carrier bovine serum albumin, 5 mM dithiothreitol, 10% glycerol, 6 units of SUPERase-In (Ambion), plus the HSF protein and labeled RNA. The concentration of the labeled RNA probe was below 1 nM in most experiments to ensure an excess protein concentration. Protein and RNA were incubated at room temperature for 30 minutes, and then at 4°C for 10 minutes before loading onto a 6% or 9% native polyacrylamide gel or a 2% agarose gel. The polyacrylamide gels contained 1/4 TBE (Tris/Borate/EDTA) buffer and 1 mM MgCl2, and the agarose gels contained 1× TAE (Tris/acetate/EDTA) buffer. Gels were run at 100–150 V at 4°C for 1–2 hours. They were then dried and the bands were visualized with the aid of a storage phosphor screen and the Typhoon™ phosphoimager system.
Competition assays were performed according to a previously published protocol (Salamanca et al., 2011). DNA probe (HSE3) was end-labeled with [γ-32P] ATP and T4 polynucleotide kinase. An excess of a particular cold RNA was co-incubated with the labeled DNA and the HSF protein at 22°C for 1 hour for the reaction to reach equilibrium, and DNA–protein complex was measured by EMSA.
Results and Discussion
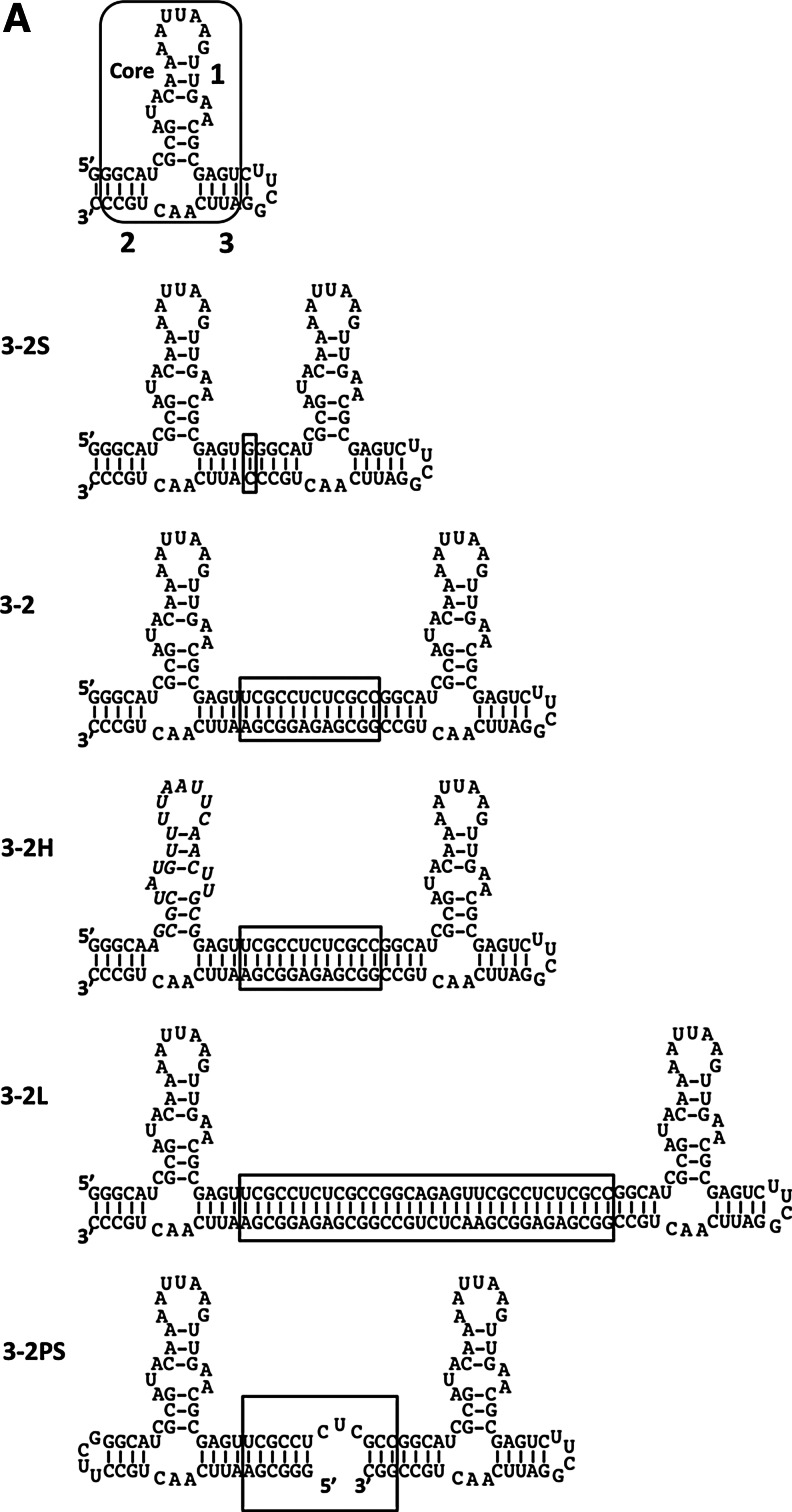
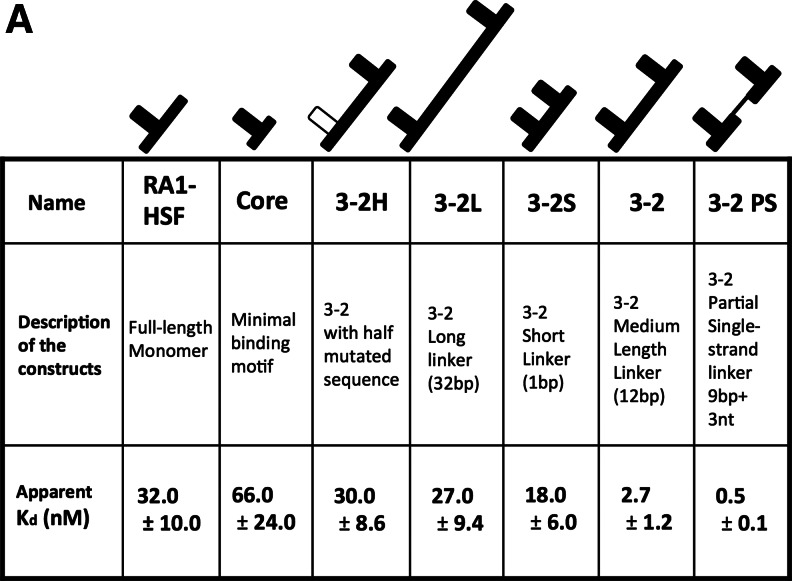
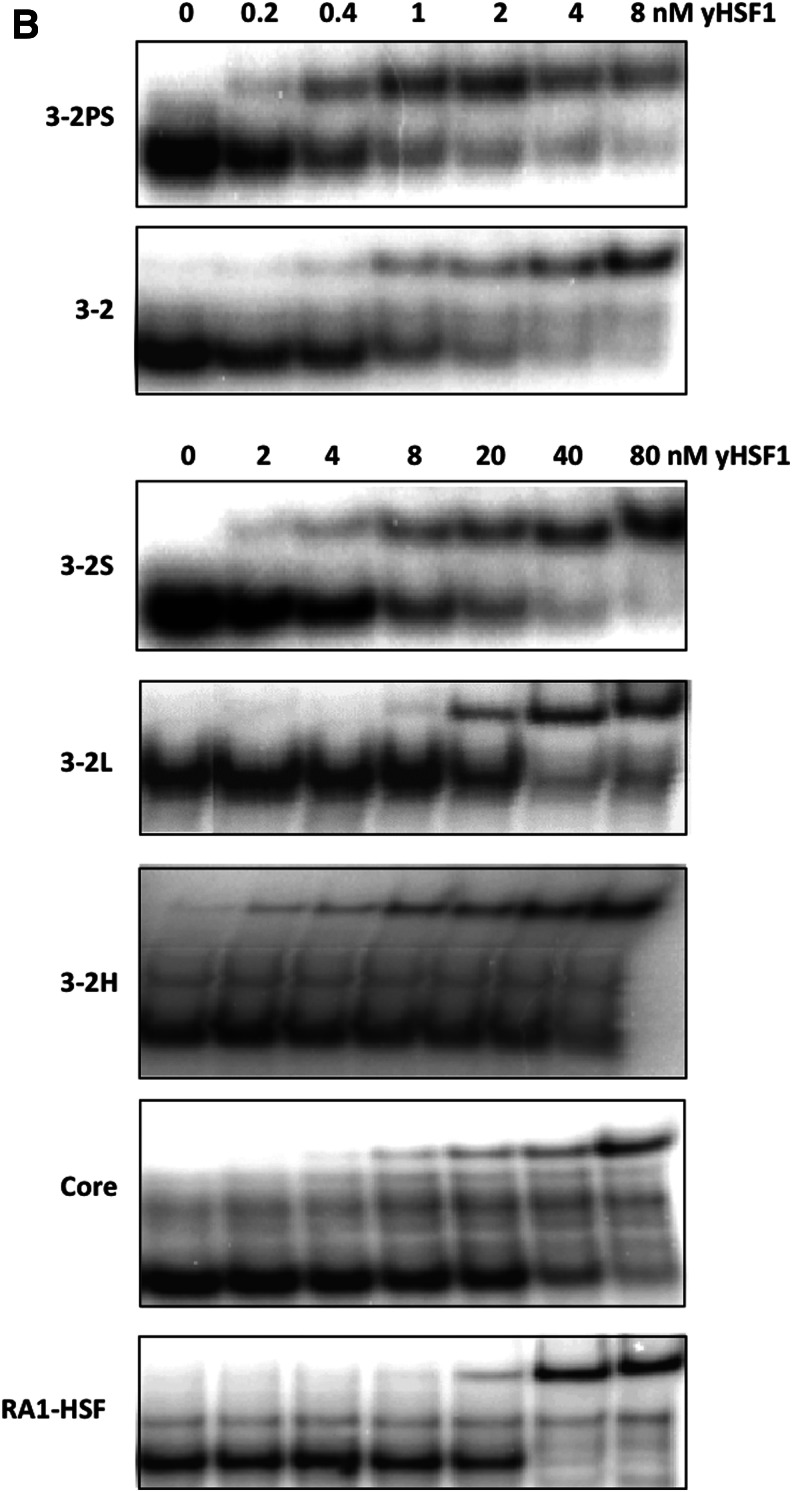
The AptHSF-RA1 aptamer binds to the DNA binding domain and a flanking peptide sequence of HSF1 protein with modest affinity (Kd 32.0±10.0 nM for full-length aptamer; 66.0±24.0 nM for the minimal “Core” aptamer), and it competes with the DNA element HSE for binding to HSF1 in vitro (Zhao et al., 2006). Because the HSF1 protein is itself a trimer, we tested the binding of a series of dimeric constructs of the aptamer core in an attempt to optimize the linker spacing between subunits. As shown in Fig. 1A, two AptHSF-RA1 aptamers were joined by connecting stem 3 of one to stem 2 of the other with either a 1-bp linker between the two “Core” subunits for construct 3-2S, or a 12-bp linker in construct 3-2, or a 32-bp linker in construct 3-2L. Construct 3-2H is a control construct derived from 3-2, in which the sequence of one “Core” subunit was partially mutated through A↔U and G↔C transversions to abolish its activity. In theory, if the 2 binding sites of a homo-bivalent construct are identical and independent, the first occupation of the free dimer would have an apparent dissociation constant that is half of the intrinsic dissociation constant of the monomeric aptamer, (i.e., the first binding would appear to be twice as tight as with a monomer) (Mack et al., 2008). The behavior of 3-2S agreed with this prediction. Although only one shifted band was visible in the EMSA, the apparent Kd (18.0±6.0 nM) was roughly half of the original full-length aptamer (Fig. 2). The binding of one bulky HSF to the construct might have made the other aptameric binding site no longer available, as we did not observe the second occupancy of the dimer. Interestingly, the insertion of 12-bp double-stranded RNA between the 2 HSF aptamers produced a dimeric aptamer with an apparent Kd of 2.7±1.2 nM, which is about 10-fold tighter in binding than the full-length monomer (Fig. 2). Since the A-form helix has 11–12 bp per turn, the relative orientation of the two “Core” domains in constructs 3-2S and 3-2 should be similar (i.e., an 12-bp insert should have roughly preserved the relative orientation of the two individual aptamers with regard to the axis formed by the rigid double helical linker. Therefore, the increased distance between the 2 units seems to be responsible for the dramatic improvement in avidity beyond theoretical prediction. As a control, the mutant construct 3-2H showed the same affinity as a full-length monomer (Fig. 2), indicating that the improvement of the construct 3-2 was not simply through a more stabilized structure or the extra sequence carried by the dimers. The construct with a long, 32-bp linker, 3-2L, did not show much improvement in affinity over the monomer. Thus, positioning the aptamers too far apart appears not to be beneficial to the effective interaction with target in the HSF trimer.
FIG. 1.
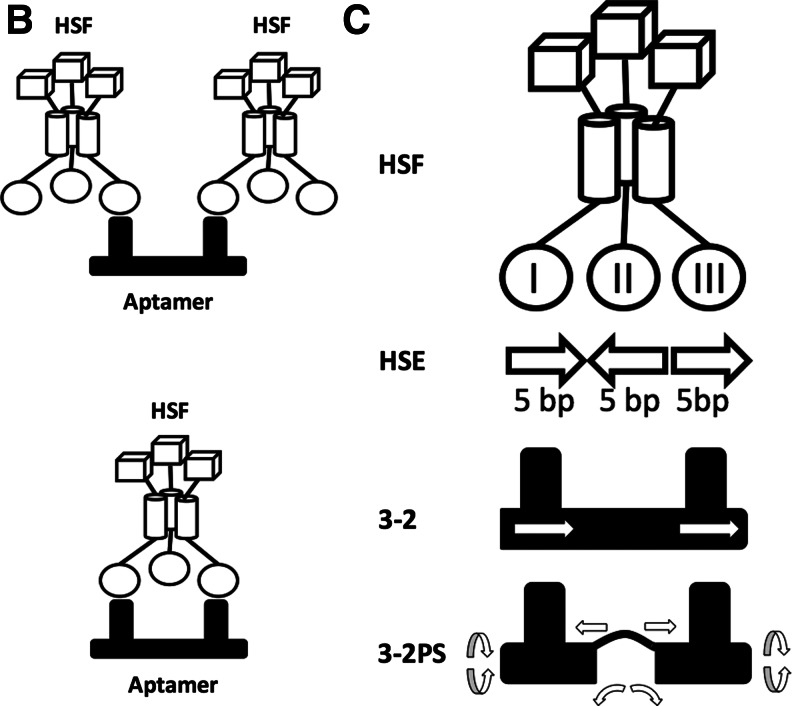
Rational design of dimeric aptamers to enhance the binding avidity. (A) Secondary structures for the “Core” and the dimeric aptamers constructed by linking 2 “Core” subunits (stem 3 of one to stem 2 of the other). The “Core-2” construct used in this study is depicted with structural annotations. The linker of each aptamer dimer is enclosed in a box. For the construct 3-2H, the mutated sequence is presented in italics. All structures were predicted by the mfold program (Zuker, 2003). (B) Two possible binding modes. The top panel shows dimerization of 2 heat shock factor (HSF) trimers. The bottom panel shows bidentate binding of one HSF trimer. (C) The bidentate binding model of the dimeric aptamer with supporting structural information. The white arrows in the construct 3-2 signify the topological relationship between the two “Core” units. The arrows around the construct 3-2PS indicate the flexibility of the linker. HSE, heat shock elements.
FIG. 2.
A comparison of apparent dissociation constant for the constructs designed. (A) A sketch of the constructs is shown above each construct. Thicker lines represent the stem-loops or stems of the construct; the thin line in 3-2PS signifies the single stranded linker; the hollow part signifies the mutated “Core” unit in 3-2H. The dissociation constant (Kd) (average±standard deviation) for each construct was measured by electrophoretic mobility shift assay (EMSA) in at least 3 independent experiments. (B) Representative examples of EMSA results used to generate Kds in panel A. (Note: the top 2 panels used much lower yeast HSF1 concentrations than the lower panels.)
When the binding mixture contains a trimeric protein and a divalent aptamer, there are two possible modes of interaction: bidentate binding of one aptamer to a single protein trimer or dimerization of 2 trimers by one homo-bifunctional aptamer (Fig. 1B). (At high concentration, the second mode may lead to further RNA–protein conglomeration.) Our data suggests bidentate binding for the construct 3-2, because there was only 1 shifted band observed in EMSA with similar mobility whether we used an aptamer monomer or an aptamer dimer as the probe (Fig. 2B). With additional structural considerations that follow, we proposed the model as shown in Fig. 1C. Theoretically, the HSF trimer should have cyclic symmetry with a 3-fold rotational axis. However, the C3 symmetry must be broken to form 3 non-identically poised DNA binding domains (I, II, and III in Fig. 1C) when the trimer binds HSE3, which is comprised of 3 contiguous inverted repeats of 5 bp (Xiao et al., 1991). In the DNA-bound form of a HSF, each DNA-binding domain recognizes the HSE in the major groove of the double helix. This fact further suggests that protein–DNA interactions at positions I and III are similar to each other, in contrast with that at position II, as indicated in Fig. 1C. Based on this information, we postulate that the dimeric aptamer may interact with 1 HSF trimer in a manner spatially analogous to DNA binding at positions I and III. Supporting this model, it is noticeable that the 2 aptamer units in our series of dimeric constructs are topologically identical, and the distance between the centers of the first and the third pentameric sequence elements in HSE3 is approximately recapitulated by a linker of 12 bp in the construct 3-2 (considering that A-form RNA double helix has more base pairs per turn and lower helical rise per base pair than B-form DNA).
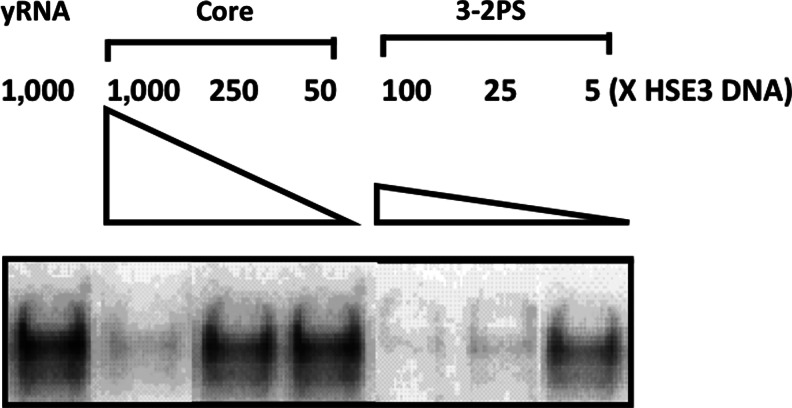
The model as depicted in Fig. 1C suggested a strategy to further improve the avidity of the aptamer dimer. It seems intuitive that the optimal distance and orientation between aptamer subunits would depend on the size and configuration of the HSF1 trimer. While the construct 3-2 might have had roughly matched the distance and orientation of 2 DNA binding domains to allow bidentate binding to occur, the steric constraint imposed by the double-stranded linker may not have been presenting the 2 aptamers with both optimal distance and optimal orientation at the same time. To add more flexibility to the linker, we introduced a partial single-stranded RNA sequence (Fig. 1A, C) as a hinge between the 2 monomers to provide rotational, bending, and stretching capability. As predicted, the resulting construct, 3-2PS, attained 5-fold further increase in avidity over its rigid counterpart 3-2, and about 100-fold increase over the “Core” (Fig. 2). To confirm these results, we employed a different assay and a different system. Whereas our initial analysis of the AptHSF-RA1 (Zhao et al., 2006) and the testing of dimeric constructs herein were carried out largely using the yeast protein, the HSF–HSE interaction had been characterized in more detail with the Drosophila system (Xiao et al., 1991), and competitive inhibition of this interaction was used before to study this aptamer in Drosophila (Salamanca et al., 2011). Therefore, to gauge the utility of the improved dimer in future functional investigations, we examined the performance of the construct 3-2PS in the previously established competition assay in comparison with the “Core.” As shown in Fig. 3, unlabeled 3-2PS competed with labeled HSE3 double-stranded DNA for binding to HSF1 trimer much more effectively than did unlabeled “Core.”
FIG. 3.
Enhanced ability of construct 3-2PS to compete with HSE3 DNA. A competition assay using excessive unlabeled yeast RNA (yRNA), “Core” or the construct 3-2PS to compete with labeled HSE3 double-stranded (ds)DNA binding to the dHSF1 protein. The concentration of dHSF1 is 50nM. Labeled HSE3 dsDNA is 1 nM in concentration, and the relative concentrations of cold competitors to HSE3 dsDNA are as indicated. Yeast RNA was used as negative control in competition.
Competitive inhibition of strong interactions, such as that between HSF1 and HSE3, requires that the inhibitor have a high avidity for target surfaces. Physically linking small molecule inhibitors into multimers can create a reagent that binds cooperatively to a target, if the target is itself multimeric (Mammen et al., 1998). In the field of RNA aptamers, however, the successes of multivalent design have been modest, where improvements have been 10-fold or less (Shi et al., 1999; Santulli-Marotto et al., 2003; Di Giusto and King, 2004). Shi et al. first designed a pentavalent B52 aptamer that increased the avidity by 10-fold (Shi et al., 1999). In the case of designing a tetravalent aptamer to cytotoxic T cell antigen-4 (CTLA-4), which exists on cell surface as a dimer, 4 monomeric modified RNA aptamers were linked with a double-stranded oligonucleotide scaffold (Santulli-Marotto et al., 2003). The distance between 2 CTLA-4 was given consideration; however, the improvement in avidity was only about 10-fold, which is most likely the result of the rigid double-stranded DNA scaffold that lacks sufficient rotational or stretching flexibility (Santulli-Marotto et al., 2003). Other examples included designing a multivalent circular DNA aptamer, which was successful in increasing the half-life of the aptamer in serum, but improved the avidity only several-fold (Di Giusto and King, 2004). Again, perhaps insufficient consideration was given to the distance and flexibility of the aptamer linkage. In addition, a dimeric construct of RNA aptamers for the prostate-specific membrane antigen was generated using a double stranded RNA linker (Wullner et al., 2008). However, the purpose was to deliver small interfering RNA rather than to improve avidity. Because the features of these multivalent RNAs that are critical for cooperative binding have not been systematically investigated previously, here we tested 2 parameters in designing a multivalent aptamer: the distance between active subunits and the flexibility of the linkage. Our study demonstrated that a proper design considering these features helped to improve RNA aptamer avidity by 2 orders of magnitude, which made the aptamer more useful both as a reagent to study the function of HSF and as a lead for anti-HSF therapeutics.
Acknowledgments
This work was supported by grants from the National Institutes of Health [GM40918 and CA140730].
Author Disclosure Statement
No competing financial interests exist.
References
- BRODY E.N. GOLD L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- DAI C. WHITESELL L. ROGERS A.B. LINDQUIST S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1008. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI GIUSTO D.A. KING G.C. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004;279:46483–46489. doi: 10.1074/jbc.M408037200. [DOI] [PubMed] [Google Scholar]
- EGNER U. KRATZSCHMAR J. KREFT B. POHLENZ H.D. SCHNEIDER M. The target discovery process. Chembiochem. 2005;6:468–479. doi: 10.1002/cbic.200400158. [DOI] [PubMed] [Google Scholar]
- ELLINGTON A.D. SZOSTAK J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- FAN X. SHI H. ADELMAN K. LIS J.T. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6934–6939. doi: 10.1073/pnas.0401523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAHN J.S. HU Z. THIELE D.J. IYER V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JULIANO R.L. ASTRIAB-FISHER A. FALKE D. Macromolecular therapeutics: emerging strategies for drug discovery in the postgenome era. Mol. Interven. 2001;1:40–53. [PubMed] [Google Scholar]
- MACK E.T. PEREZ-CASTILLEJOS R. SUO Z. WHITESIDES G.M. Exact analysis of ligand-induced dimerization of monomeric receptors. Anal. Chem. 2008;80:55505555. doi: 10.1021/ac800578w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAMMEN M. CHOI S.-K. WHITESIDES G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- MENDILLO M.L. SANTAGATA S. KOEVA M. BELL G.W. HU R. TAMIMI R.M. FRAENKEL E. INCE T.A. WHITESELL L. LINDQUIST S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIRKKALA L. NYKANEN P. SISTONEN L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- SALAMANCA H.H. FUDA N. SHI H. LIS J.T. An RNA aptamer perturbs heat shock transcription factor activity in Drosophila melanogaster. Nucleic Acids Res. 2011;39:6729–6740. doi: 10.1093/nar/gkr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTAGATA S. HU R. LIN N.U. MENDILLO M.L. COLLINS L.C. HANKINSON S.E. SCHNITT S.J. WHITESELL L. TAMIMI R.M. LINDQUIST S. INCE T.A. High levels of nuclear heat-shock factor 1(HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANTULLI-MAROTTO S. NAIR S.K. RUSCONI C. SULLENGER B. GILBOA E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003;63:7483–7489. [PubMed] [Google Scholar]
- SHI H. HOFFMAN B.E. LIS J.T. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUERK C. GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- WANG S. ZHAO X. SURAN R. VOGT V.M. LIS J.T. SHI H. Knocking down gene function with an RNA aptamer expressed as part of an intron. Nucleic Acids Res. 2010;38:e154. doi: 10.1093/nar/gkq529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU C. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- WULLNER U. NEEF I. ELLER A. KLEINES M. TUR M.K. BARTH S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets. 2008;8:554–565. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- XIAO H. PERISIC O. LIS J.T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- ZAARUR N. GABAI V.L. PORCO J.A., JR. CALDERWOOD S. SHERMAN M.Y. Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer Res. 2006;66:1783–1791. doi: 10.1158/0008-5472.CAN-05-3692. [DOI] [PubMed] [Google Scholar]
- ZHAO X. SHI H. SEVILIMEDU A. LIACHKO N. NELSON H.C. LIS J.T. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZUKER M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]