There are several biological sources which could be used for DNA isolation and genetic testing with potential application in the clinical setting [1]. Increasingly, DNA isolated from buccal swabs and, more recently, saliva samples is under investigation for potential use in the clinical setting rather than traditional blood samples due to ease of access, storage, and transport, as well as relative comfort for participants with low cost of acquisition [1, 2]. Reports to date indicate that DNA isolated from saliva may be more reliable for genotyping by PCR and electrophoresis [3–6], whereas buccal DNA from mouth swabs has produced variable genotyping success as low as 23% [4]. However, there is a paucity of information available on the comparison of buccal and/or saliva DNA for chromosomal microarrays, a commonly used tool for genetic testing. In response to Durdiaková et al. [1], we wish to report our experience with DNA isolation using non-traditional sources such as buccal cells, saliva, and even plasma. Plasma is not intended to be a recommended source of DNA isolation other than a possible alternative for DNA isolation if peripheral blood, the more conventional source of DNA for genetic testing, is not available from an individual. Their utility in both genotyping and microarray analysis will be illustrated.
Informed consent approved by the local Institutional Review Board was obtained from all subjects prior to collection of biological specimens. Table 1 summarizes the specimen sources used, methods of DNA isolation, storage and measures. Buccal DNA was extracted from two cotton swabs using either a silica membrane-based DNA extraction kit (QIAamp DNA Investigator Kit from Qiagen, Valencia, CA, USA) or an isopropanol-based precipitation kit (MasterPure DNA Purification Kit from Epicentre, Madison, WI, USA) from control individuals and infants with developmental delay of unknown cause. Blood and lymphoblast DNA were routinely isolated using the traditional phenol-chloroform protocol or more recently silica membrane-based DNA extraction; high quality intact DNA was recovered successfully using either method for genotyping purposes [7]. Plasma was separated from whole blood collected in EDTA tubes from the control individuals and DNA was extracted using the DNeasy Blood & Tissue Kit from Qiagen. Saliva DNA extraction was carried out using the Oragene DNA Collection Kit with collection tubes and DNA preservatives provided by DNA Genotek (Ontario, Canada). All DNA extractions were performed according to the manufacturers' instructions. The DNA was re-suspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). Spectrophotometer Biophotometer Plus from Eppendorf (Hamburg, Germany) was used for determining the DNA OD 260/280 ratio and electrophoresis performed using 1% agarose gel containing 0.5 μg/mL of ethidium bromide for detection of DNA.
Table 1.
Summary of biological specimen used for DNA isolation with results.
| Source | Number | Method | Storage at −80°C | Adequate DNA yielda | Adequate DNA qualityb | DNA degradationc | Successful PCR amplificationd |
|---|---|---|---|---|---|---|---|
| Blood | 100 | Phenol extraction; Qiagen | Mixede (0–10 years) | 95% | 90% | 10% | 90% |
| Lymphoblast | 100 | Phenol extraction; Qiagen | Mixede (0–10 years) | 95% | 90% | 10% | 90% |
| Saliva | 30 | Oragene | Mixed (0–5 years) | 90% | 90% | 20% | 90% |
| Buccal | 84 | Epicenter (MasterPure) | 4–6 months | 90% | 50% | 42% | 70% |
| Buccal | 27 | Qiagen | 4–6 months | 80% | 70% | 74% | 50% |
| Stored plasma | 38 | Qiagen | 4–6 years | 80% | 45% | 90% | 20% |
| Fresh plasma | 4 | Qiagen | Fresh/not stored | 75% | 50% | 50% | 50% |
Estimated at ≥ 1 μg of DNA per extraction.
OD 260/280 ratio between 1.6 and 2.1.
Based on gel electrophoresis with visible intact DNA (i.e., 8000–10,000 bp) with ethidium bromide stain.
PCR amplification success is correlated with the size of the PCR fragment generated (e.g., larger PCR fragments require more intact DNA).
There were no obvious differences in the quality of DNA collected using the two DNA extraction methods in blood or lymphoblast specimens or differences related to short-term (0–5 years) vs. long-term (6–10 years) storage at −80°C.
To test the suitability of isolated DNA from several sources for PCR amplification and genotyping, we used the growth hormone receptor (GHR) gene from chromosome 5p, which carries a polymorphic 2.7 kb genomic deletion (d3) spanning exon 3 in approximately two-thirds of Caucasian controls [8, 9]. Selected PCR primers for the GHR gene will generate two fragment sizes (full length=935 bp; d3=532 bp) following electrophoresis [8]. For statistical analysis, one-way, single factor ANOVA, Student t- and χ2-tests were performed using Microsoft Excel 2007 (Redmond, WA, USA).
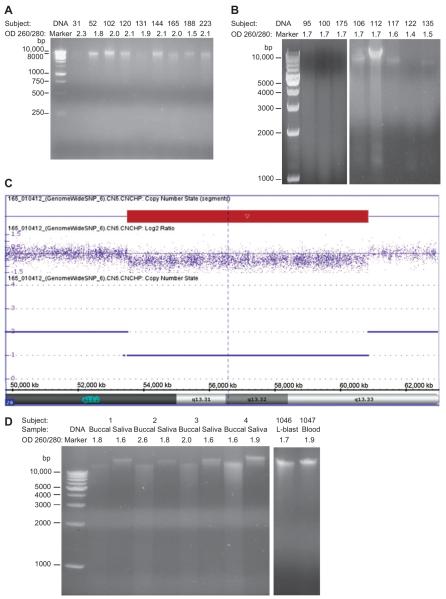
Nine of 15 representative samples of buccal DNA were isolated from a total of 27 infants with developmental delay of unknown cause using the QIAamp kit from Qiagen and discernible DNA signals were produced by agarose gel electrophoresis meeting standardized laboratory criteria for chromosomal microarray analysis (Figure 1A). Six DNA samples were degraded and not suitable for microarray hybridization (three representative samples shown in Figure 1B). Buccal DNA was successfully hybridized using the Affymetrix Genome-Wide Human SNP Array 6.0 (Santa Clara, CA, USA) to identify genomic deletions or duplications and one infant (Subject 165) showed a 7 Mb deletion of chromosome 20q. This region includes several genes, such as the complex GNAS imprinted loci which can cause developmental delay [10] (Figure 1C).
Figure 1.
Electrophoretic analysis of genomic DNA from buccal cell and saliva samples showing mixed results with DNA (100–200 ng) loaded onto 1% agarose gel and visualized using 0.5 μg/mL of ethidium bromide. (A) Stored buccal cell DNA isolated with the QIAamp (Qiagen) kit from nine infants with developmental delay in which seven DNA samples (Subjects 52, 102, 120, 144, 165, 188, 223) met standardized laboratory criteria for chromosomal microarray analysis based on DNA quantity or yield (e.g., 0.75 μg), purity or qualtiy (i.e., spectrophotometer OD 260/280 ratios; e.g., 1.6–2.1) and sufficient intact high molecular weight DNA using gel electrophoresis. (B) Stored buccal cell DNA isolated with the QIAamp (Qiagen) kit from three representative infants (Subjects 95, 100, 175) with developmental delay showing the degree of degradation from 10,000 bp to 1000 bp range as designated by known DNA markers and not meeting laboratory criteria whereas a greater yield of high quality intact DNA was found with MasterPure DNA kit in five representative infants (Subjects 106, 112, 117, 122, 135). No intact DNA fragments were visualized below 1000 bp. (C) Chromosomal microarray analysis of buccal DNA from Subject 165 using the Affymetrix Genome-Wide Human SNP Array 6.0 (Santa Clara, CA, USA) to identify genomic deletions or duplications showed a 7 Mb deletion (copy number of 1) of the 20q13.2–20q13.33 region occurring at 53,512,484–60,850,110 bp from the p-terminus of the chromosome. (D) DNA isolated from freshly-collected buccal and saliva using the QIAamp (Qiagen) kit from four control subjects meeting laboratory criteria except for one buccal sample (Subject 2) with a high OD ratio of 2.6 in comparison with DNA isolated from blood and lymphoblasts (L-blast), more conventional sources for DNA, obtained from two different representative control subjects (Subjects 1046, 1047) showing the typical DNA pattern with gel electrophoresis ranging from 10,000 bp to 1000 bp designated by known DNA markers. No intact DNA fragments were visualized below 1000 bp.
We measured the concentration, quality and degradation of buccal DNA isolated from two cotton swabs each from 111 infants and stored at room temperature in plastic bags for approximately 4 months without a preservative added then frozen at −80°C until used. DNA from 27 samples were isolated with the QIA amp kit and 84 samples with the MasterPure kit. Spectrophotometer OD measurements ranged from 1.5 to 3.2 A260/280 ratio with a mean of 2.1 for the QIAamp kit and from 1.3 to 4.1 A260/280 ratio with a mean of 1.7 for the MasterPure kit. Total DNA collected per sample based on OD readings ranged from 0.1 to 3.5 μg with a mean of 1.4 μg for the QIAamp kit and from 0.4 to 17.2 μg with a mean of 3.7 μg for the MasterPure kit.
The DNA quality based on degradation by gel electrophoresis of 111 buccal DNA samples using both the QIAamp kit and MasterPure kit were analyzed statistically with the χ2-test. Significant differences were found with fewer samples showing DNA degradation with the MasterPure kit (i.e., 20 or 74% of 27 samples were degraded using QIAamp and 35 or 42% of 84 samples were degraded using MasterPure; χ2=8.53, p<0.01; see Figure 1B). The purity of representative buccal DNA samples using the QIAamp kit was compared with representative lymphoblast and blood DNA samples and no significant differences were detected in the three DNA categories (F=3.13, p>0.05). However, the purity of buccal DNA using the two DNA isolation kits (QIAamp and MasterPure) was compared and significant differences were detected with higher OD 260/280 ratios with greater variance found in the QIAamp kit (t=2.00, p<0.05).
Our second study compared the quality of DNA isolated from buccal cells from freshly collected swabs (n=4) and saliva (n=4) from collection kits without previous storage or preservative added. Electrophoretic analysis indicated greater degradation in buccal DNA than in saliva DNA (Figure 1D) with the quality of the representative buccal DNA samples ranging from 1.6 to 2.6 A260/280 ratio with a mean of 2.0 and saliva DNA ranging from 1.6 to 1.9 with a mean of 1.7.
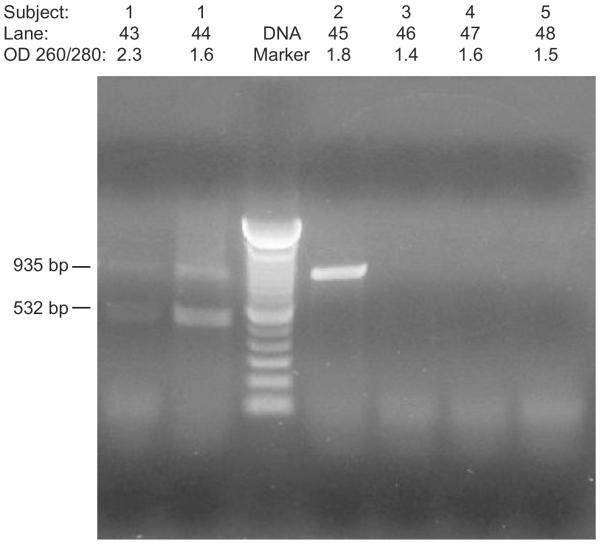
Our third study tested the subjects' genotype using GHR as a representative gene with PCR amplification of DNA isolated from buccal, saliva, blood, and plasma specimens. Electrophoretic analysis indicated less DNA degradation from buccal cells than from plasma. Both buccal and plasma DNA produced the subject's genotype for GHR with PCR primers generating up to two DNA fragments representing the heterozygous state. However, plasma DNA yielded poorer DNA quality with OD 260/280 ratios from 1.4 to 2.3 when compared with saliva or blood DNA. Plasma DNA was also impacted by storage and handling. For example, DNA isolated from plasma stored for at least 4 years was generally unsuccessfully amplified by PCR using GHR primers while DNA isolated from fresh plasma were more likely to produced genotyping results (Figure 2).
Figure 2.
PCR amplification of the growth hormone receptor (GHR) gene with DNA isolated from both plasma and buccal cells. Lanes 43 and 44 show two PCR fragments representing heterozygosity of the GHR gene from Subject 1 using fresh plasma DNA and fresh buccal DNA, respectively; Lane 45 shows a single PCR fragment representing homozygosity using fresh buccal DNA from Subject 2, and Lanes 46–48 represent unsuccessful PCR amplification using stored frozen plasma DNA from Subjects 3, 4, and 5.
In conclusion, we have shown that buccal cell DNA can be used for chromosomal microarray analysis requiring high quality intact DNA by evidence of a 20q deletion detected in one of seven infants studied with developmental delay of unknown cause but buccal DNA may also be too degraded to produce useful microarray or PCR results. Thus, buccal DNA was also not as dependable as blood, lymphoblast or saliva DNA for such purposes. However, in our experience, the MasterPure DNA isolation kit based on isopropanol precipitation generally produced DNA with less degradation from stored buccal cell samples than the DNA isolated with the QIAamp kit using silica membrane-based extraction procedures. DNA isolated from stored plasma samples yielded low quality DNA but fresh samples were more suitable for PCR amplification. Further improvement and standardization of collection methods will aid in the use of buccal DNA and more specifically saliva DNA as practical alternatives to blood DNA in the clinical setting for genetic testing, particularly in young patients where access to blood may be difficult to obtain or is limited.
Acknowledgments
This study was partially supported by the Fogarty International Research Grant no. HD060500, NICHD HD02528, and NICHD U54 HD61222.
Research funding played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Research funding: No other funding declared.
Footnotes
Conflict of interest statement
Authors' conflict of interest disclosure: The authors have no conflicts of interest regarding the publication of this article.
Employment or leadership: None declared.
Honorarium: None declared.
Contributors: All authors contributed to experimental design, data analysis, and participated equally in manuscript preparation. All authors reviewed and approved the study.
References
- 1.Durdiaková J, Kamodyová N, Ostatníková D, Vlková B, Celec P. Comparison of different collection procedures and two methods for DNA isolation from saliva. Clin Chem Lab Med. 2012;50:643–7. doi: 10.1515/CCLM.2011.814. [DOI] [PubMed] [Google Scholar]
- 2.King IB, Satia-Abouta J, Thornquist MD, Bigler J, Patterson RE, Kristal AR, et al. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev. 2002;11:1130–3. [PubMed] [Google Scholar]
- 3.Nemoda Z, Horvat-Gordon M, Fortunato CK, Beltzer EK, Scholl JL, Granger DA. Assessing genetic polymorphisms using DNA extracted from cells present in saliva samples. BMC Med Res Methodol. 2011;11:170. doi: 10.1186/1471-2288-11-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen TV, Simonsen MK, Nielsen FC, Hundrup YA. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol Biomarkers Prev. 2007;16:2072–6. doi: 10.1158/1055-9965.EPI-07-0611. [DOI] [PubMed] [Google Scholar]
- 5.Halfon P, Ouzan D, Khiri H, Pénaranda G, Castellani P, Oulés V, et al. Detection of IL28B SNP DNA from buccal epithelial cells, small amounts of serum, and dried blood spots. PLoS One. 2012;7:e33000. doi: 10.1371/journal.pone.0033000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maloney B, Ray B, Hayden EP, Nurnberger JI, Jr., Lahiri DK. Development and validation of the high-quality `rapid method for swab' to genotype the HTTLPR serotonin transporter (SLC6A4) promoter polymorphism. Psychiatr Genet. 2009;19:72–82. doi: 10.1097/YPG.0b013e3283208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler MG, Fischer W, Kibiryeva N, Bittel DC. Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. Am J Med Genet. 2008;146:854–60. doi: 10.1002/ajmg.a.32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantel J, Machinis K, Sobrier M, Duquesnoy P, Goossens M, Amselem S. Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J Biol Chem. 2000;275:18664–9. doi: 10.1074/jbc.M001615200. [DOI] [PubMed] [Google Scholar]
- 9.Padidela R, Bryan SM, Abu-Amero S, Hudson-Davies RE, Achermann JC, Gudrun EM, et al. The growth hormone receptor gene deleted for exon 3 (GHRd3) polymorphism is associated with birth and placental weight. Clin Endocrinol. 2012;76:236–40. doi: 10.1111/j.1365-2265.2011.04207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genevieve D, Sanlaville D, Faivre L, Kottler ML, Jamou M, Gosset P, et al. Paternal deletion of the GNAS imprinted locus (includomg Gnasxl) in two girls presenting with severe pre- and post-natal growth retardation and intractable feeding difficulties. Eur J Hum Genet. 2005;13:1033–9. doi: 10.1038/sj.ejhg.5201448. [DOI] [PubMed] [Google Scholar]