Abstract
A typical eukaryotic genome harbors a rich variety of repetitive elements. The most abundant are retrotransposons, mobile retroelements that utilize reverse transcriptase and an RNA intermediate to relocate to a new location within the cellular genomes. A vast majority of the repetitive mammalian genome content has originated from the retrotransposition of SINE (100–300 bp short interspersed nuclear elements that are derived from the structural 7SL RNA or tRNA), LINE (7kb long interspersed nuclear element), and LTR (2–3 kb long terminal repeats) transposable element superfamilies. Broadly labeled as “evolutionary junkyard” or “fossils”, this enigmatic “dark matter” of the genome possesses many yet to be discovered properties.
Keywords: retrotransposons, SINE, genomic boundaries, Pol III transcription, TFIIIC, chromatin architecture, intrachromosomal interactions, tRNA genes
Introduction
Historically, retrotransposons were first characterized as “controlling elements” of neighboring genes (McClintock 1956). Our understanding of the biology of retrotransposal elements and the mechanisms of retrotransposition, as well as assessment of their abundance within eukaryotic genomes, has increased over the last several years. We have learned that these elements serve as a driving force of cell diversification and specification during development and in response to environmental stimuli (Muotri et al. 2005, 2007; Coufal et al. 2009), as well as that they are responsible for genomic rearrangements contributing to genomic diversity (Kidd et al. 2008; Damert et al. 2009; Xing et al. 2009). Indeed, it is becoming increasingly apparent that the retrotransposal portion of the genome has an impact on many biological processes, from gene transcription to genome stability, and evidence is mounting that it is more then insertional mutagenesis (Morrish et al. 2002). Although we are still far from a complete understanding of genome “dark matter”, cumulative evidence suggests that the retrotransposons can serve as regulatory sequences (both positive and negative) by providing mobile RNA polymerase (Pol) II and Pol III promoters (Muotri et al. 2005; Walters et al. 2009; Ponicsan et al. 2010) and DNA methylation centers (Englander et al. 1993; Hasse and Schulz 1994; Arnaud et al. 2000; Suzuki et al. 2007). Retrotransposal elements can also mediate diverse cellular responses through a variety of embedded transcriptional factor (TF) binding sites (Tomilin 2008; Bourque 2009), participate in the establishment and maintenance of the pericentric heterochromatin and imprinted X chromosome inactivation (Wheeler et al. 2009; Chow et al. 2010), act as a chromatin boundary and insulator elements (Lunyak et al. 2007; Roman et al. 2008), and drive carcinogenic transformation and human aging (Zhao and Bourque 2009; Han et al. 2008; Lin et al. 2009). The full spectrum of the biological consequences of the transcriptional activation and repression of retrotransposons and the mechanistic aspects of their action within genic versus intergenic portions of genomes are still poorly understood.
In this review we intend to convey some of the recent excitement in the field that points to the functional significance of these elements in a variety of cellular processes and discuss the remaining mysteries. Taking into account the non-random distribution patterns of retrotransposons found in many eukaryotic genomes, we suggest that the biological utility of these elements is similar to linguistic “punctuation marks”, which are aimed at separating words into sentences, clauses, and phrases and are vital to clarifying their meaning. It is plausible that retrotransposal elements participate in the packaging of the genome and, owing to their higher degree of sequence conservation, are capable of mediating long-range interactions by clustering of coordinately regulated loci. We believe that, as with linguistic punctuation, these elements guide and direct genomic information to formulate a variety of nuclear processes.
Abundance of retrotransposal transcription in the mammalian genome
It should be stressed that retrotransposons, together with other so-called junk DNA such as DNA transposons and pseudogenes, have long been considered as lacking function (Feng et al. 1996) and even having a deleterious effect on the host genome through their transcriptional activity. Starting from the work of Chandler and Walbot (1986), who described the role of DNA methylation in controlling the transposon’s activity in plants, it was generally accepted that the potentially deleterious transcriptional activity of retrotransposons in eukaryotic cells must be suppressed by DNA methylation-dependent silencing, which has evolved as a cell defense mechanism against retrotransposal proliferation within the host genome. The dominant view that retrotransposal RNA expression is largely restricted to developing germ cells has been challenged in recent years.
Large-scale approaches to exploring the transcriptional status of the retrotransposal portion of genomes leave no doubts that the initial rate and scale of their transcriptional activity were highly underestimated. Pervasive, tissue-specific retrotransposal transcription was recorded by using CAGE sequencing, where most of the retrotransposons initiate transcription from a single, well-defined initiation site (Faulkner et al. 2009). Overall, approximately 35% of all retrotransposon-associated transcriptional start sites have shown spatially or temporally restricted expression. Such nonrandom expression could be related to some functional implications of the retrotransposons in the genomes, despite the traditional assumption that their transcription is “simply a noise”.
Interestingly, the vast majority of retrotransposal transcription was initiated in previously unidentified sense and anti-sense promoters, although canonical 5′ promoters of short interspersed nuclear elements SINE B2 and L1 were found to be active as well (Faulkner et al. 2009). A recent study of genome-wide occupancy of the 5 components of the Pol III machinery in human K562 cells revealed that the striking majority (90%) of the otherwise nonannotated Pol III-associated loci are near SINE retrotransposal elements (Moqtaderi et al. 2010). There are hundreds of thousands of SINEs in many eukaryotic genomes (Kramerov and Vassetzky 2005), with extensive variability between individual elements (Umylny et al. 2007), and therefore a transcription of just a small proportion of them would generate thousands of different transcripts. The SINE-associated genomic loci show much lower levels of Pol III factors compared with tRNA genes, but these levels are still clearly above the background. Importantly, it has been reported that retrotransposal transcripts in K562 cells are less abundant than protein-coding mRNA as shown by RNA-seq experiments, but this could be due to stability of these transcripts, their processing, or their turnover. There is no assessment of the function of retrotransposal transcriptional activity in the K562 system just yet, although there has been much debate about whether transcripts, the act of transcription, or both are relevant.
Given the abundance of the transcribed retrotransposal genomic sequences, it has been suggested that retrotransposal transcription and (or) transcript processing may represent a genome-wide strategy for the control of nuclear architecture and epigenetic memory regulated by an RNA-directed process (Amaral et al. 2008).
Mechanistic aspects of Pol III transcriptional complex assembly
It is useful to examine the mechanisms through which this strategy is carried out. In eukaryotes, Pol III is devoted to the transcription of small RNAs participating in basic cellular functions such as protein synthesis (tRNAs, 5S rRNA), RNA processing (e.g., U6 small nuclear RNA, the RNA subunit of RNase P), protein secretion (7SL RNA) (Paule and White 2000; Geiduschek and Kassavetis 2001), vault RNA (Chugani et al. 1993; van Zon et al. 2003), repression of the Pol II elongation complex (7SK) (Nguyen et al. 2001; Yang et al. 2001), and a family of small, nuclear factor 90 (NF90)-associated RNAs (snaRs) (Parrott et al. 2007). Recently, it has been shown that Pol III can drive the expression of micro-RNAs (Borchert et al. 2006) and brain-specific lncRNAs (BC1 and BC200) (Martignetti and Brosius 1993, 1995). Despite the very simplistic view held 10 years ago regarding what Pol III transcribes, the Pol III transcriptome now appears to be a functionally heterogeneous group of non-protein-coding RNAs with a few well-known, abundant members and an elusive and unexplored realm of nonabundant transcripts such as SINE retrotransposons.
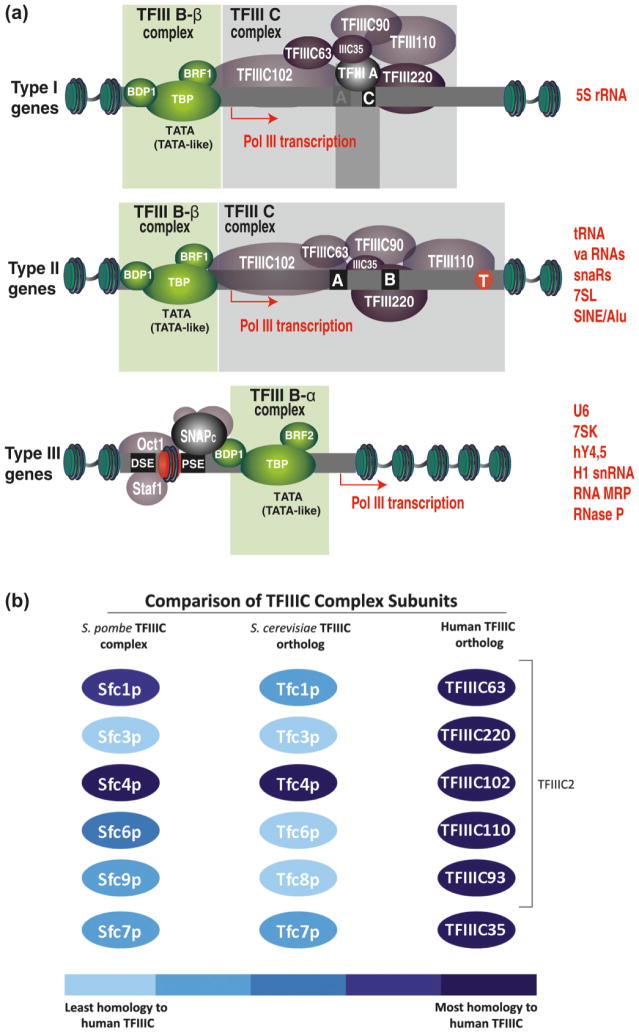
Extensive work on Pol III transcription in yeast and higher eukaryotes has revealed the transcriptional factors required for directing Pol III enzymatic machinery to its targets, which allowed for identification of 3 types of Pol III gene promoters (Fig. 1A). The Pol III machinery makes extensive use of promoter elements, located both within (internal) and outside (external) the transcribed region, which generates remarkable complexity in the delivery of Pol III enzymatic machinery to the gene. This, in turn, reflects how the transcription of the diverse genes can be executed and (or) regulated.
Fig. 1.
(A) Highly schematic representation of different classes of Pol III transcribed genes. The important function of the various promoter elements and their associated transcriptional factors is to bring TFIIIB to the start site of the transcription and to stabilize it there for the recruitment of Pol III enzymatic machinery. Type I genes are composed of a major internal element, the C box, as well as additional elements that vary among species. The single-factor TFIIIA mediates the recruitment of TFIIIC, which binds along the length of the 5S gene in the presence of TFIIIA and is required for transcription. The exact mechanism of TFIIIC recruitment to the type 1 promoter remains unclear. Type II genes consist of 2 highly conserved sequence elements, a proximal A box and a more distal B box, within the transcribed region. The distance between the A and B boxes is variable, and these elements are responsible for the recruitment of the TFIIIC complex. TFIIIC binds along the entire length of a tRNA gene, beginning just upstream of the start site of transcription and extending through the terminator (shown in red circle (dark grey in print version)), a separate control element that resides 20–25 bp downstream of the B box. For type I and type II gene promoters the proximal subunits of TFIIIC direct TFIIIB to bind upstream of the transcription start site. TFIIIB then recruits and positions Pol III over the initiation site and remains stably bound to the DNA through multiple rounds of reinitiation by Pol III. Type III genes utilize an upstream TATA box, a proximal sequence element (PSE), and a distal sequence element (DSE). The PSE functions with the TATA element to recruit the SNAPc complex (Mittal et al. 1999). The transcriptional activator Oct1 is recruited to DSE and functions in part by promoting SNAPc complex recruitment. TFIIIB, SNAPc, and Oct-1 cooperative interactions promote a stable initiation complex mediated, in part, by a positioned nucleosome (red nucleosome (dark grey in print version)) between DSE and PSE (Zhao et al. 2001). Oct1, Staf1, and SNAPc can also participate in the activation of Pol II transcription (Schaub et al. 1997). In some Pol III transcribed genes the combination of promoter element differs from those described above. Green nucleosomes represent the histone marks associated with active transcription recently observed in genome-wide studies (Barski et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010). (B) Schematic representation of the homology of the TFIIIC complex subunits in Schizosaccharomyces pombe, Saccharomyces cerevisiae, and human cells. The comparison analysis for homolog identifications was reported by Huang and Maraia (2001) and completed by Dumay-Odelot et al. (2010). Intensity of color corresponds to strength of homology (Dumay-Odelot et al. 2007). The brackets enclose the 5 subunits of human TFIIIC that correspond most strongly to the yeast TFIIIC (Dumay-Odelot et al. 2007).
Pol III is brought to the transcriptional start sites of the genes through interactions with its central 3-subunit initiation factor, TFIIIB (subunits TBP, Bdp1, and Brf). Only a single isoform of TFIIIB has been described in yeast (Huang and Maraia 2001). In contrast, human cells contain 2 isoforms of TFIIIB (TFIIIB-α and TFIIIB-β) (Teichmann and Seifart 1995) that are specifically recruited either to gene type 1 and 2 (TFIIIB-β) promoters or to type 3 promoters (TFIIIB-α) shown in Fig. 1A. Human TFIIIB-α and TFIIIB-β both contain TATA-binding proteins TBP and Bdp1, but differ with respect to the presence of TFIIIB-related factor Brf1 (in TFIIIB-β complex) and Brf2 (In TFIIIB-α complex). In most Pol III genes, these are the so-called box A and box B (or A-block and B-block) regions, which were initially identified in yeast tRNA genes and later shown to be evolutionarily conserved in multiple species. Box B is always located at a variable distance (usually 30–60 bp) downstream of box A, which itself is located downstream of the transcriptional start site. These 2 sites are responsible for the oriented positioning of the 6-subunit assembly factor transcription factor TFIIIC. TFIIIC in turn directs the association of the Pol III-recruiting transcription factor TFIIIB to a ~50 bp DNA region immediately upstream of the transcription start site in a largely sequence-independent process. In addition to box A, transcription of the 5S rRNA gene does not require a box B, but rather a box C and an intermediate element, recognized by the gene-specific transcription factor TFIIIA. This additional initiation factor, TFIIIA, serves solely as the adaptor for assembling TFIIIC on 5S rRNA genes. Some genes (such as the 7SL RNA or the vault RNA genes) are not strictly related to tRNA genes, yet they possess internal, tRNA-like box A – box B combinations that are essential for transcription and sometimes even an external, downstream box B (Saccharomyces cerevisiae U6 snRNA gene). In other cases (e.g., S. cerevisiae RPR1), the A and B boxes are situated within a transcribed leader sequence that is processed from the mature transcript. Sometimes, a double box B is present in combination with box A (Geiduschek and Kassavetis 2001; Schramm and Hernandez 2002; Dieci et al. 2007) (for review, see Dieci et al. 2007).
A key component of TFIIIB is the TATA box-binding protein (TBP) or a TBP-related factor (such as Drosophila melanogaster TRF1) that interacts with upstream DNA around position −30. Accordingly, a TATA box or a TATA-like sequence element is found at this position upstream of many class III genes. Not only is an essential TATA element present upstream of all genes that lack internal promoter elements (such as the 7SK RNA gene), but a functional TATA element can also be found upstream of box A- and box B-containing genes, as in many tRNA genes or in the vault RNA gene. Composite, lineage-specific upstream sequence motifs centered around the TBP-interacting region have been noted upstream of tRNA genes in many eukaryotes (Giuliodori et al. 2003).
A well-characterized upstream promoter element of type III Pol III genes is the proximal sequence element (PSE) that interacts with a multisubunit factor variously called SNAPc, PBP, or PTF (Schramm and Hernandez 2002). The PSE is generally located ~20 bp upstream of the TATA box. Vertebrates contain an additional form of TFIIIB in which its paralogue Brf2 replaces Brf1 for transcription of a class of genes that use the SNAPc complex in place of TFIIIC as their TFIIIB-assembly factor (Geiduschek and Kassavetis 2001; Schramm and Hernandez 2002). In human cells, SNAPc binding to PSE facilitates the TATA box-mediated association to DNA of a specific TFIIIB variant. The expression of type III genes with a completely external TATA/PSE-based promoter is enhanced by the so-called distal sequence element (SDSE). The distal sequence element can contain several protein binding sites, most frequently an SPH element and an octamer sequence, recruiting the transcription factors STAF and OCT1, respectively (Schramm and Hernandez 2002). Upstream binding sites for other transcription factors (such as Sp1 and ATF) have been found to stimulate transcription of certain type III genes (Fig. 1A).
Once it has been assembled onto DNA, the core initiation complex recruits Pol III enzymatic machinery (RNA PIII), the largest and most complex among RNA polymerases. Pol III is highly conserved from yeast to humans. The yeast enzyme is composed of 17 subunits with an overall mass of 700 kDa (Fernández-Tornero et al. 2007). Of the 17 Pol III subunits, 5 (ABC27(hRPC25), ABC23(hRPC15), ABC14.5 (hRPC14), ABC10α(hRPC10), and ABC10β(hRPC8)) are shared among polymerases I, II, and III, another 2 are shared with Pol I (AC19 (hRPC19) and AC40(hRPC40)), 4 are homologous to subunits found in Pol I and (or) Pol II (C160 (hRPC155), C128(hRPC128), C25(homologs in human are not identified), and C11(hRPC25)), and 6 are unique to Pol III (C82(hRPC62), C53(BN51), C37(not identified in humans), C34(hRPC39), C31(hRPC32), and C17(hRPC17)) with no apparent homology with the other polymerases (Huang and Maraia 2001; Dumay-Odelot et al. 2010). Most of the latter group, the Pol III-specific subunits, appear to function in recognizing the TFIIIC–TFIIIB–DNA initiation complex. In S. cerevisiae yeast, the 2 largest polypeptides in the complex, C160 and C128, form the binding cleft for DNA and harbor the active site of the enzyme. Humans have the homolog of yeast subunit C160- hRPC155. Three of these Pol III subunits (C82(hRPC62), C34(hRPC39), and C31(hRPC32)) form a subassembly that interacts with the TFIIIB–DNA complex and is required specifically for initiating transcription (for review, see Fernández-Tornero et al. 2007). Studies with recombinant proteins further showed that hRPC62 interacts in vitro with TFIIIC63 and TFIII90 (Hsieh et al. 1999a, 1999b). In addition, hRPC39 interacts in vitro with TFIIIC90, thus further reinforcing the hypothesis that the ternary subcomplex of Pol III participates in establishing protein–protein contact with transcriptional factors. Two subunits, C53(BN51) and C37, also form a subassembly that limits the processivity of Pol III during elongation and contributes to the ability of Pol III to terminate transcription. The association of C53(BN51) and C37 with Pol III is mediated at least in part through interaction with the Rpb9- and TFIIS-related subunit, C11(hRPC11) (Chédin et al. 1998; Landrieux et al. 2006).
Our understanding of the diversity of the Pol III complex assembly on its genomic targets in higher eukaryotes has benefited tremendously from recent genome-wide analysis performed by several groups (Barski et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010) in a number of human transformed cells and cells immortalized with TERT. These studies not only confirmed the existence of the complexity (Fig. 1A) in the recruitment of basic transcription machinery for Pol III but also largely agreed on (i) the existence of cell-type specificity of the Pol III expression (Han et al. 2008; Dieci et al. 2007), (ii) the condition that not all genomic units with assembled TFIIIC complexes might be actively transcribed (Barski et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010), and demonstrated (iii) the astonishing correlation between Pol III-associated loci and the presence of the Pol II transcription machinery. Intriguingly, this association also extends to the presence of genomic histone marks associated with Pol II gene regulation (such as histone acetylation and histone H3 lysine 4 methylation, among many). The most exciting observation in the context of this review that comes from these studies is the notion that a vast majority (about 90%) of the previously unreported Pol III-associated loci are located near SINE retrotransposons (Han et al. 2008).
However, there are still a number of open questions related to these recently published data. One of them is whether Pol III and Pol II association has a functional relevance for genome organization and whether the presence of a Pol II or Pol III genomic unit is functionally relevant to transcription by Pol III holoenzyme.
Pol III-controlled elements as genome organizers
Cytological studies pinpoint distinct functional territories within the nucleus of the cells dedicated to Pol III transcriptional activities (Pombo et al. 1999). The initial finding that 274 Pol III-transcribed tRNA genes in S. cerevisiae that are dispersed throughout the genome are localized to a single nuclear substructure, the nucleolus, was an astonishing observation (Thompson et al. 2003). Such localization is associated with more than tRNA maturation, raising the possibility that this clustering has a major impact on the spatial organization of the genome. Recently obtained data in the same system demonstrate that (i) general transcriptional factors for Pol III, such as TFIIIC and TFIIIB, can mediate genomic association with condensin complexes (Haeusler et al. 2008; D’Ambrosio et al. 2008a); (ii) the introduction of an ectopic TFIIIC binding site (B-box element GTTCXAXXC) into the budding yeast genome generates a new condensin binding site (D’Ambrosio et al. 2008a); and (iii) chemical inhibition of Pol III transcription has little effect on the ability of the condensin to bind tDNA (D’Ambrosio et al. 2008a), thus suggesting that active transcription and tRNA production is not required for the clustering event. On the other hand, the fluorescent in situ hybridization (FISH) microscopy analysis shows dispersed and mislocalized distribution of tDNA of the temperature-sensitive mutants of all 5 subunits of condensin (Haeusler et al. 2008). Similar to budding yeast, TFIIIC has been shown to be involved in tDNA clustering in fission yeast, thus implying a role for this factor in the maintenance of genome structure through interchromosomal interactions among Pol III-transcribed genes (Tsang et al. 2007). The involvement of highly conserved protein complexes such as cohesin and condensin at tRNA gene clusters brings into question whether a similar function of Pol III-controlled elements can be found in higher eukaryotes. If TFIIIC can mediate its action through the recruitment of highly evolutionarily conserved condensin and (or) cohesin complexes (for review, see Wood et al. 2010), then the role of Pol III-dependent genomic units in organizing chromatin in the nucleus is more general then previously anticipated.
Human TFIIIC complex was separated by B-box-based affinity purification or by Mono-Q chromotography into 2 functional modules, TFIIIC1 and TFIIIC2 (Dean and Berk 1987; Yoshinaga et al. 1987). Although the exact composition of TFIIIC1 remains unknown (Dumay-Odelot et al. 2007), the experiments performed upon F9 embryonic stem cell differentiation suggest that the TFIIIB subunit BDP1 is essential for TFIIIC1 activity (Weser et al. 2004). TFIIIC1 is generally required for transcription of all types of Pol III genes (Dumay-Odelot et al. 2007; Oettel et al. 1997) (Fig. 1A) and has shown to stimulate binding of the TFIIIC2 complex. Human TFIIIC2 was resolved into a 5-subunit complex, encompassing TFIII220, TFIIC110, TFIIIC102, TFIIIC90, and TFIIIC63 (Yoshinaga et al. 1987; Kovelman and Roeder 1992; see recent review by Dumay-Odelot et al. 2010) (Fig. 1B). Three subunits of human TFIIIC2 (hTFIIIC220, hTFIIIC110, and hTFIIIC90) each harbor intrinsic histone acetylation activity that enables TFIIIC to combat the repressive effects of chromatin (Hsieh et al. 1999b; Kundu et al. 1999). These hTFIIIC subunits exhibit the least homology with the scTFIIIC and spTFIIIC subunits (Giuliodori et al. 2003) (Fig. 1B). hTFIIIC90 exhibits a low level of homology with the putative Sfc9p but not with any S.cerevisiae protein, and as noted previously, hTFIIIC110 and TFC6p appear only distantly related. In many higher eukaryotes, particularly vertebrates, the most abundant Pol III elements are SINE retrotransposons (Okada 1991). Some of the better-studied SINEs are derived from pre-tRNA or 7SL RNA and make up about 7% of the murine and about 10% of the human genome (Lander et al. 2001; Waterston et al. 2002). These data raise speculation of the possible role of Pol III-regulated retrotransposons in orchestrating the coordination chromatin dynamic within the cells via the recruitment of TFIIIC and (or) by generating the transcripts. Several observations support this hypothesis. Transcripts from a tRNA promoter-driven small RNA construct were found throughout the nucleoplasm and resembled a punctate foci (Good et al. 1997). Preliminary attempts to broadly localize the positions of SINE elements in human HeLa cells and SINE B2 elements in mouse embryonic fibroblasts by FISH also showed that these genomic elements form a granular pattern throughout the nucleoplasm, with no particular association with the nucleolus or nuclear periphery (Kaplan et al. 1993; Haeusler and Engelke 2006; Pai and Engelke 2010). In addition, in human HeLa cells, primary fibroblasts, and myogenic cells, the transcription factor TFIIIB has been localized to concentrated foci throughout nuclei (Kelter et al. 2000).
No evidence has yet been reported to confirm that the multiple linearly dispersed Pol III transcription units or the highly repetitive SINE elements influence subnuclear organization of DNA in higher eukaryotes, but their potential influence on local and long-distance chromatin organization has not yet been systematically explored. The next several examples give a hint as to what these influences might be.
SINE repeats as chromosomal boundary elements
In mice, the strong developmental regulation of B1 and B2 SINE transcription has received considerable interest. SINE B2 retrotransposons derived from pre-tRNA and SINE B1 (human Alu) derived from 7SL RNA can function as chromatin “boundary elements” (Lunyak et al. 2007; Román et al. 2011), effectively acting as a road block for the propagation of heterochromatic spreading similar to the function of tRNA genes in yeast (Donze et al. 1999).
The B2 SINE family constitutes approximately 0.7% of total mouse genomic DNA (Bennett et al. 1984). An interesting feature of SINE B2 repeats is that, in addition to serving as a Pol III promoter, SINE B2 contains an active Pol II promoter located outside the tRNA region (Ferrigno et al. 2001). The 70 bp minimal Pol II promoter was initially delineated within SINE B2 allocated in the Lama3 gene. This sequence has substantial nucleotide similarity within the B2 SINE family. Moreover, Pol II activity of SINE B2 does not preclude the Pol III transcription that originated in the tRNA portion of the repeat (Ferrigno et al. 2001).
The data obtained from the functional analysis of the SINE B2 repeat in the murine GH locus demonstrate that the repeat element is able to generate short overlapping Pol II- and Pol III-driven transcripts. The striking difference between the transcriptional activity of this mouse SINE B2 repeat and the data on tRNA gene transcription reported in the yeast system is that Pol II transcription from SINE B2 is activated in a developmental and tissue-specific fashion and correlates with restructuring of the chromosomal domain of the GH gene. Physical repositioning of the GH gene locus from the heterochromatic to the euchromatic compartment was observed by FISH analysis within specific cell type. This repositioning coincides with Pol II transcriptional activity from the SINE B2 repeat and is accompanied by changes in histone modification within the locus (Lunyak et al. 2007).
The SINE B2 repeat region within the GH locus provides context-independent insulator activity (based on enhancer-blocking analysis) and can also buffer the spread of heterochromatic modifications from facultative heterochromatin regions flanking the murine GH locus at the 5’ end. Therefore, SINE B2 repeats can be viewed as true genomic boundaries. Promoter deletion and substitution analysis demonstrates that both Pol II and Pol III transcription are required to mediate the insulator function (Lunyak et al. 2007). How does Pol II gain access to sequences that are packaged as heterochromatin? Several models have been proposed (Yasuhara and Wakimoto 2006) to explain the mechanisms of heterochromatic transcription (Yasuhara and Wakimoto 2006; Lunyak 2008). One can argue that the promoters driving the transcription of SINE repeats, unlike the promoters of protein-coding euchromatic genes, have evolved to become somewhat impervious to heterochromatic repression and might be marked by an epigenetic signature specifically dedicated to this occasion. Indeed, one of the strands of SINE B2 repeat in the GH locus is always transcribed at a low level by Pol III transcriptional machinery even when the locus is heterochromatic (Lunyak et al. 2007). Can the formation of Pol III-mediated transcripts from SINE B2 create a clamp in the Pol II promoter portion of the repeat by engaging RNAi molecular complexes, thus denying the recruitment of Pol II- or Pol II-recruiting factors to the site? Could this restriction be lifted by development-specific changes in histone modifications or by the recruitment of development-specific DNA binding factor(s) and their coregulators?
To date there is no experimental evidence that can support the molecular basis for development-specific Pol II transcriptional activity of SINE B2 in the context of the GH locus, although a recent study of the boundary activity of B1_X35S that has shown the recruitment of zing-finger transcription factor SLUG is important for the insulation function (Roman et al. 2008). The insulation mechanisms of B1_X35S are complex and involve Pol III and Pol II transcription, with 2 major differences that set this genomic element apart from SINE B2 in the GH locus. First, both Pol III and Pol II transcription are working in the same orientation (e.g., transcribing the same strand of DNA). Second, the transcription factor AHR increases B1_X35S transcription by an exchange mechanism that recruits a Pol II and releases a Pol III complex (Román et al. 2011), suggesting the role for the transcriptional factors involved in cell growth, proliferation, apoptosis, and migration in the regulation of transcriptional activities of retrotransposons. Since dynamic restructuring of the nucleus by arming or disarming genomic boundary and insulator elements appears to be both a cause and consequence of alterations in gene expression, DNA replication, and DNA damage repair, these observations might imply a role for SINE retrotransposons in all these processes.
Pol III-regulated network and repertoire of transcription factor binding in retrotransposons
Multiple reports have demonstrated that chromosomal sites of Pol III transcription have “extratranscriptional” functions. For instance, the assembled Pol III complexes can act as pause sites for replication forks (Deshpande and Newlon 1996), alter nucleosome positioning (Oki and Kamakaka 2005; Dhillon et al. 2009), affect the transcription of neighboring genes (Raab and Kamakaka 2010), act as genomic boundaries or insulators (Lunyak et al. 2007; Donze et al. 1999), and partition the genome into distinct chromatin domains by bringing the elements into proximity at the nuclear periphery (Noma et al. 2006). In addition, Pol III transcription units can play a role in sister chromatid cohesion and chromatin condensation (D’Ambrosio et al. 2008a, 2008b).
Recent studies have indicated that a large fraction of bona fide binding sites for a number of transcriptional factors (TFs) that are development or cell-type specific, such as ER, TP 53, myc, Oct4, Sox2, and CTCF, are embedded in distinct families of retrotransposons. By leveraging the ability of the ChiP sequencing platforms to detect TF binding sites, Bourgue et al. (2008) have demonstrated that a significant portion of binding sites of the transcriptional regulator are embedded within the different classes of retrotransposal elements. In cross-linking experiments, Dumay-Odelot et al. (2010) have found that c-myc is often present at Pol III promoters, indicating that it might have a role in initiating Pol III transcription. In addition, Pol III-transcribed genes (along with c-myc and Pol III polymerases) are often found nearby Pol II transcriptional activators (Dumay-Odelot et al. 2010). These observations suggest that there is much more to the regulation of the Pol III and Pol II transcriptional activity of the retrotransposons than meets the eye and that many of these regulations can directly impact the course of the developmental event.
In mice, the strong developmental regulation of B1 and B2 SINE transcription has received considerable interest. Regulation of B2 RNA expression is especially intriguing in relation to its demonstrated role in Pol II transcriptional regulation. B2 RNA binds directly to Pol II and represses transcription from specific genes after heat shock (Goodrich and Kugel 2006). Another interesting example of Pol III developmental regulation is provided by the BC1 RNA, whose gene has been suggested to act as a master gene for the SINE repetitive DNA family whose members (ID elements) are interspersed throughout rodent genomes. The BC1 RNA is expressed in a subset of male germ cells (a prerequisite for the generation of repetitive elements through retrotransposition), and its expression pattern was found to change during spermatogenic development (Muslimov et al. 2002). Interestingly, several novel Pol III-dependent ncRNAs recently identified in Caenorhabditis elegans display developmentally variable expression. In particular, snoRNA-like transcripts, whose expression is driven by upstream tRNA-like promoters, showed the highest expression during the middle stages of development, whereas the genes of the other class of noncoding RNA, stem-bulge RNA (sbRNA), tended to be expressed more in the later stages of worm development (Deng et al. 2006). Therefore, an assumption that developmental and tissue-specific regulation of Pol III transcription must rely on transcriptional regulatory activities is probably correct, although the mechanistic aspects of this events are largely unknown and deserve further exploration.
Conclusion
This is an exciting time in genome biology. Aspects of genomic form and function that were largely inconceivable only a few decades ago are now being revealed on a daily basis. It should come as no surprise (and indeed, it probably does not) that new roles are being discovered for noncoding DNA and that some of yesterday’s buzzwords — including “junk DNA” — are destined for the junkyard. Long-standing assumptions and dogmas about how transcriptional machineries operate are destined to be revised in light of current knowledge about genome complexity. New experimental evidence makes room for new paradigms regarding how these machineries might function on portions of the genome far greater than the ~2% that codes for protein. In our assumptions, we have artificially constrained ourselves to a limited and incomplete definition of the genes that are parsed throughout the genome in a linear fashion. With every new step in genome research we are learning that genomes are far more complex than we imagined, and there is now little use in approaching them from a simplistic point of view.
Acknowledgments
We apologize to our colleagues for the omission of so many important research contributions owing to the space constraints of this review. We thank Warren Schonfeld for the critical reading of the manuscript and valuable comments. We also thank the members of the Lunyak lab for their constructive and helpful comments. V.V.L. and M.A. are supported by Buck Institute startup funding to V.V.L. and by UL1 DE019608 Pilot.
References
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science (Washington, D C) 2008;319(5871):1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Arnaud P, Goubely C, Pelissier T, Deragon JM. SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol. 2000;20(10):3434–3441. doi: 10.1128/MCB.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17(5):629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Schmid CW, Deininger PL. Evolutionary analyses of repetitive DNA sequences. Methods Enzymol. 1993;224:213–232. doi: 10.1016/0076-6879(93)24017-O. [DOI] [PubMed] [Google Scholar]
- Bennett KL, Hill RE, Pietras DF, Woodworth-Gutai M, Kane-Haas C, Houston JM, et al. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus — the polyA connection. Nat Genet. 1997;16(1):6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19(6):607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Walbot V. DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci USA. 1986;83(6):1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinwalla AT, Delehaunty KD, Fewell GA, Fulton LA, Fulton RS, Graves TA, et al. Initial sequencing and comparative analysis of the mouse genome. Nature (Lond) 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Chédin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12(24):3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141(6):956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Chugani DC, Rome LH, Kedersha NL. Evidence that vault ribonucleoprotein particles localize to the nuclear pore complex. J Cell Sci. 1993;106(Pt. 1):23–29. doi: 10.1242/jcs.106.1.23. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature (Lond) 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008a;22(16):2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Kelly G, Shirahige K, Uhlmann F. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol. 2008b;18(14):1084–1089. doi: 10.1016/j.cub.2008.06.058. [DOI] [PubMed] [Google Scholar]
- Damert A, Raiz J, Horn AV, Löwer J, Wang H, Xing J, et al. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19(11):1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Berk AJ. Separation of TFIIIC into two functional components by sequence specific DNA affinity chromatography. Nucleic Acids Res. 1987;15(23):9895–9907. doi: 10.1093/nar/15.23.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Zhu X, Skogerbø G, Zhao Y, Fu Z, Wang Y, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16(1):20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science (Washington, D C) 1996;272(5264):1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- Dhillon N, Raab J, Guzzo J, Szyjka SJ, Gangadharan S, Aparicio OM, et al. DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 2009;28(17):2583–2600. doi: 10.1038/emboj.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13(6):698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumay-Odelot H, Marck C, Durrieu-Gaillard S, Lefebvre O, Jourdain S, Prochazkova M, et al. Identification, molecular cloning, and characterization of the sixth subunit of human transcription factor TFIIIC. J Biol Chem. 2007;282(23):17179–17189. doi: 10.1074/jbc.M611542200. [DOI] [PubMed] [Google Scholar]
- Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9(18):3687–3699. doi: 10.4161/cc.9.18.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander EW, Wolffe AP, Howard BH. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J Biol Chem. 1993;268(26):19565–19573. [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41(5):563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fernández-Tornero C, Böttcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, et al. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol Cell. 2007;25(6):813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Virolle T, Djabari Z, Ortonne JP, White RJ, Aberdam D. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet. 2001;28(1):77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310(1):1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- Giuliodori S, Percudani R, Braglia P, Ferrari R, Guffanti E, Ottonello S, Dieci G. A composite upstream sequence motif potentiates tRNA gene transcription in yeast. J Mol Biol. 2003;333(1):1–20. doi: 10.1016/j.jmb.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Good PD, Krikos AJ, Li SX, Bertrand E, Lee NS, Giver L, et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4(1):45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7(8):612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Engelke DR. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34(17):4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22(16):2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci USA. 2008;105(49):19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse A, Schulz WA. Enhancement of reporter gene de novo methylation by DNA fragments from the α-fetoprotein control region. J Biol Chem. 1994;269(3):1821–1826. [PubMed] [Google Scholar]
- Hsieh YJ, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999a;19(7):4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YJ, Kundu TK, Wang Z, Kovelman R, Roeder RG. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol Cell Biol. 1999b;19(11):7697–7704. doi: 10.1128/mcb.19.11.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 2001;29(13):2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan FS, Murray J, Sylvester JE, Gonzalez IL, O’Connor JP, Doering JL, et al. The topographic organization of repetitive DNA in the human nucleolus. Genomics. 1993;15(1):123–132. doi: 10.1006/geno.1993.1020. [DOI] [PubMed] [Google Scholar]
- Kelter AR, Herchenbach J, Wirth B. The transcription factor-like nuclear regulator (TFNR) contains a novel 55-aminoacid motif repeated nine times and maps closely to SMN1. Genomics. 2000;70(3):315–326. doi: 10.1006/geno.2000.6396. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature (Lond) 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovelman R, Roeder RG. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267(34):24446–24456. [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19(2):1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature (Lond) 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25(1):118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139(6):1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV. Boundaries. Boundaries…Boundaries??? Curr Opin Cell Biol. 2008;20(3):281–287. doi: 10.1016/j.ceb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Núñez E, Cramer T, Ju BG, Ohgi KA, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317(5835):248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci USA. 1993;90(24):11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol. 1995;15(3):1642–1650. doi: 10.1128/mcb.15.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13(14):1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17(5):635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31(2):159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature (Lond) 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum Mol Genet. 2007;16(R2):R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- Muslimov IA, Lin Y, Heller M, Brosius J, Zakeri Z, Tiedge H. A small RNA in testis and brain: implications for male germ cell development. J Cell Sci. 2002;115(Pt. 6):1243–1250. doi: 10.1242/jcs.115.6.1243. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/ cyclin T complexes. Nature (Lond) 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125(5):859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Oettel S, Hartel F, Kober I, Iben S, Seifart KH. Human transcription factors IIIC2, IIIC1 and a novel component IIIC0 fulfil different aspects of DNA binding to various pol III genes. Nucleic Acids Res. 1997;25(12):2440–2447. doi: 10.1093/nar/25.12.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991;1(4):498–504. doi: 10.1016/S0959-437X(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Oki M, Kamakaka RT. Barrier function at HMR. Mol Cell. 2005;19(5):707–716. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17(5):620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai DA, Engelke DR. Spatial organization of genes as a component of regulated expression. Chromosoma. 2010;119(1):13–25. doi: 10.1007/s00412-009-0236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Walsh MR, Mathews MB. Analysis of RNA:protein interactions in vivo: identification of RNA-binding partners of nuclear factor 90. Methods Enzymol. 2007;429:243–260. doi: 10.1016/S0076-6879(07)29012-3. [DOI] [PubMed] [Google Scholar]
- Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28(6):1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KE, Deka N, Schmid CW, Misra R, Schindler CW, Rush MG, et al. A transposon-like element in human DNA. Nature (Lond) 1985;316(6026):359–361. doi: 10.1038/316359a0. [DOI] [PubMed] [Google Scholar]
- Perlman PS, Boeke JD. Molecular biology. Ring around the retroelement. Science (Washington, DC) 2004;303(5655):182–184. doi: 10.1126/science.1093514. [DOI] [PubMed] [Google Scholar]
- Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18(8):2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20(2):149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Kamakaka RT. Insulators and promoters: closer than we think. Nat Rev Genet. 2010;11(6):439–446. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman AC, Benitez DA, Carvajal-Gonzalez JM, Fernandez-Salguero PM. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc Natl Acad Sci USA. 2008;105(5):1632–1637. doi: 10.1073/pnas.0708366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román AC, González-Rico FJ, Moltó E, Hernando H, Neto A, Vicente-Garcia C, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21(3):422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16(1):173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16(20):2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246(3):401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3(4):e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14(23):5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science (Washington, DC) 2003;302(5649):1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilin NV. Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. Bioessays. 2008;30(4):338–348. doi: 10.1002/bies.20741. [DOI] [PubMed] [Google Scholar]
- Tsang CK, Wei Y, Zheng XF. Compacting DNA during the interphase: condensin maintains rDNA integrity. Cell Cycle. 2007;6(18):2213–2218. doi: 10.4161/cc.6.18.4733. [DOI] [PubMed] [Google Scholar]
- Umylny B, Presting G, Efird JT, Klimovitsky BI, Ward WS. Most human Alu and murine B1 repeats are unique. J Cell Biochem. 2007;102(1):110–121. doi: 10.1002/jcb.21278. [DOI] [PubMed] [Google Scholar]
- van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EA. The vault complex. Cell Mol Life Sci. 2003;60(9):1828–1837. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life. 2009;61(8):831–837. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weser S, Gruber C, Hafner HM, Teichmann M, Roeder RG, Seifart KH, Meissner W. Transcription factor (TF)-like nuclear regulator, the 250-kDa form of Homo sapiens TFIIIB”, is an essential component of human TFIIIC1 activity. J Biol Chem. 2004;279(26):27022–27029. doi: 10.1074/jbc.M312790200. [DOI] [PubMed] [Google Scholar]
- Wheeler BS, Blau JA, Willard HF, Scott KC. The impact of local genome sequence on defining heterochromatin domains. PLoS Genet. 2009;5(4):e1000453. doi: 10.1371/journal.pgen.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11(6):391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19(9):1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature (Lond) 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22(6):330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Boulanger PA, Berk AJ. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Bourque G. Recovering genome rearrangements in the mammalian phylogeny. Genome Res. 2009;19(5):934–942. doi: 10.1101/gr.086009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Pendergrast PS, Hernandez N. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol Cell. 2001;7(3):539–549. doi: 10.1016/S1097-2765(01)00201-5. [DOI] [PubMed] [Google Scholar]