Abstract
Exposure to environmental contaminants, including various pesticides and trace metals, can disrupt critical olfactory-driven behaviors of fish such as homing to natal streams, mate selection, and an ability to detect predators and prey. These neurobehavioral injuries have been linked to reduced survival, and population declines. Despite the importance of maintaining proper olfactory signaling processes in the presence of chemical exposures, little is known regarding chemical detoxification in the salmon olfactory system, and in particular, the antioxidant defenses that maintain olfactory function. An understudied, yet critical component of cellular antioxidant defense is phospholipid hydroperoxide glutathione peroxidase (PHGPx/GPx4), an isoform within the family of selenium-dependent glutathione peroxidase (GPx) enzymes that can directly reduce lipid peroxides and other membrane-bound complex hydroperoxides. In this study, we cloned two gpx4 isoforms (gpx4a and gpx4b) from Coho salmon olfactory tissues and compared their modulation in olfactory and liver tissues by cadmium, an environmental pollutant and olfactory toxicant that cause oxidative damage as a mechanism of toxicity. Amino acid sequence comparisons of the two gpx4 isoforms shared 71% identity, and also relatively high sequence identities when compared with other fish GPx4 isoforms. Sequence comparisons with human GPx4 indicated conservation of three important active-sites at selenocysteine (U46), glutamine (Q81), and tryptophan (W136), suggesting similar catalytic activity between fish and mammalian GPx4 isoforms. Tissue profiling confirmed the expression of gpx4a and gpx4b in all ten Coho tissues examined. The expression of gpx4 mRNAs in the Coho olfactory system was accompanied by comparably high initial rates of GPx4 enzymatic activity in mitochondrial and cytosolic fractions. Exposure to low (3.7 ppb) and high (347 ppb) environmental Cd concentrations for 24–48 hrs significantly decreased gpx4a expression in Coho olfactory rosettes, whereas olfactory gpx4b mRNA expression was not modulated by exposures at these concentrations. In summary, Coho salmon express two paralogs of gpx4, a key enzyme in the maintenance of signal transduction processes that protect against cellular oxidative damage. The Cd-associated downregulation of salmon olfactory gpx4a expression in particular, may be associated with the loss of olfactory signal transduction that accompanies metal-associated loss of olfaction in salmonids.
Keywords: GPx4, cadmium, olfactory injury, Coho salmon
1. Introduction
The decline of Pacific salmon populations in the Western United States has been associated with the loss of habitat quality and contamination of surface waters and sediments (reviewed in (Lackey, 2003)). Environmental pollutants, including trace metals and pesticides are a particular concern due to their neurotoxic effects on the salmon olfactory system (Baatrup, 1991; Baldwin et al., 2003; Jarrard et al., 2004; Scott et al., 2003; Tierney et al., 2006). The olfactory rosettes constitute the major component of the salmon peripheral olfactory system and are in direct contact with the surrounding water, which renders this system vulnerable to the toxicity of dissolved pollutants. Damage to olfactory sensory neurons (OSNs) located within the olfactory epithelium (OE) (Laberge and Hara, 2001) leads to impaired olfactory function, resulting in the loss of critical behaviors such as normal swimming, detection of predators and prey, recognition of reproductive chemical cues, and homing to natal streams, which ultimately affects species survival (Laetz et al., 2009; Palm and Powell, 2010; Sandahl et al., 2007).
Despite their critical function in mediating behavioral processes that can affect salmon survival, little is known regarding the chemical biotransformation capacity of the salmon olfactory system. We have shown that the peripheral olfactory system of Coho salmon expresses a number of glutathione S-transferases (GSTs), including several isoforms that detoxify secondary products of oxidative damage (Espinoza et al., 2011). In addition to GSTs, the glutathione peroxidases (GPx) are another important family of glutathione-associated enzymes that participate in cellular defense via enzymatic reduction of reactive cellular hydroperoxides in the presence of glutathione (GSH) (Conrad et al., 2005; Imai and Nakagawa, 2003; Kuhn and Borchert, 2002). Of these enzymes, the phospholipid hydroperoxide metabolizing glutathione peroxidase (PHGPx/GPx4) represents a unique member of the tetrameric selenium-contained GPx family (Savaskan et al., 2007b; Ufer and Wang, 2011). GPx4 possesses a distinctive monomeric structure and hydrophobic surface (Ursini et al., 1985), which underlies a unique catalytic activity capable of reducing complex membrane-bound hydroperoxides and lipid peroxides, such as phospholipid and cholesterol hydroperoxides (Roveri et al., 1994; Schnurr et al., 1996; Thomas et al., 1990). Since its initial purification from pig liver (Ursini et al., 1982), GPx4 has been extensively studied in mammalian models and its function addressed through several genetic approaches. For example, GPx4+/− knockout mice have an increased sensitivity to oxidative stress (Imai et al., 2003; Yant et al., 2003), whereas mice that overexpress GPx4 are protected from liver oxidative damage (Ran et al., 2004), and this effect is also observed in cortical neurons (Ran et al., 2006).
GPx4 isoforms have been identified in several fish species, including common carp (Cyprinus carpio, gpx4a and gpx4b) (Hermesz and Ferencz, 2009), zebrafish (Danio rerio, gpx4a and gpx4b) (Kryukov and Gladyshev, 2000), Atlantic salmon (Salmo salar, gpx4b) and southern bluefin tuna (Thunnus maccoyii, gpx4) (Thompson et al., 2010). However, little is known about how these unique GSH-dependent enzymes respond to environmental stressors or if they are functional in the fish olfactory system. The goal of the present study was to characterize gpx4 expression and distribution in Coho salmon (Oncorhynchus kisutch), with a particular emphasis on function in the salmon olfactory system. A secondary goal of the study was to examine gpx4 modulation in salmon olfactory tissues on exposure to a prototypical toxicant that causes oxidative stress. In this regard, cadmium (Cd) is a ubiquitously distributed trace metal that is toxic to liver and kidney tissues and also disrupts normal behavioral and alarm responses at environmental exposure levels (Scott et al., 2003; Stromberg et al., 1983). These neurobehavioral injuries are associated with the accumulation of Cd in the olfactory system and disruption of normal olfactory function (Gottofrey and Tjälve, 1991; Tjälve et al., 1986; Tjälve and Henriksson, 1999). Studies in numerous species, including fish, have demonstrated that a major mechanism of Cd toxicity involves enhancing oxidative stress via increased production of cellular reactive oxygen species (ROS) (Hart et al., 1999; Risso-de Faverney et al., 2004; Thevenod et al., 2000; Wang et al., 2004; Watanabe and Suzuki, 2002). After characterizing the presence and functionality of the gpx4 paralogs, we challenged Coho with environmentally-relevant levels of Cd associated with modulation of cell signaling and also oxidative stress to determine such effects on gpx4 mRNA expression.
2. Material and methods
2.1 Animals and tissue processing
Juvenile Coho salmon (~1 year of age) and adult Coho (~2 year old) were provided by the National Oceanic and Atmospheric Administration (NOAA), Seattle, Washington. Fish were raised in cylindrical tanks with recirculated freshwater under natural photoperiod in dechlorinated municipal water. Fish were fed commercial dry food pellets (BioOregon, Warrenton, OR, USA) once a day. Water quality conditions were typically ~120 mg/L as CaCO3, pH 6.6, 10–12 °C, and 8.1 mg/L dissolved oxygen content. Twenty-four hr prior to initiation of exposures, the juvenile Coho were transferred to 120 L aquaria receiving filtered city water (pH=7.1 ± 0.1, 12 ± 1 °C, alkalinity 80 mg/L, hardness 50 mg/L, dissolved oxygen 8.6 ± 0.58 mg/L). Following the exposures, fish were anesthetized with tricaine methanesulfonate (MS-222, Argent Chemical Laboratories, Redmond, WA) prior to cervical dislocation. For tissue expression analysis, gills, liver, olfactory rosettes, kidney, spleen, ovary or testis, heart, brain and pyloric caeca were collected from eight adult Coho salmon, rinsed in 1X PBS (pH=7.0), placed in 1 ml Trizol (Invitrogen, Carlsbad, CA) and snap-frozen in liquid nitrogen. For enzyme activity analysis, liver and olfactory rosettes were collected from 20 adult Coho salmon, snap-frozen in liquid nitrogen, and store stored at −80 °C until processing.
2.2 Cloning of gpx4 isoforms by RACE
Total RNA was isolated from Coho salmon tissues using the Trizol method according to manufacturer’s instructions. RNA concentrations and purity were verified on a ND100 nanodrop spectrophotometer (Thermo Fischer Scientific, Waltham, MA). First strand cDNA was synthesized from 1 μg of total RNA using oligo (dT) primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad CA.), and 1 μL of cDNA was used as templates for all PCR amplifications. All amplified products were cloned into a TOPO cloning vector (Invitrogen) and sequenced at the University of Washington, Department of Biochemistry Sequencing Facility. Rapid amplification of cDNA ends (RACE-PCR) using the SMART™ RACE cDNA amplification kit (Clontech, Mountain View, CA) together with MMLV Reverse Transcriptase was used to amplify gpx4, according to the supplier’s protocol. Coho liver cDNA was used to generate all RACE products. Gene-specific RACE primers (primers 1 and 2, Table 1) for gpx4b were designed against Salmo salar gpx4b partial sequence (GenBank accession no. BT044014). Full-length gpx4b was cloned from olfactory and liver cDNA libraries with primer 3 set and sequenced in both directions.
Table 1.
Primers for Coho gpx4 cloning by RACE and for real-time PCR studies.
| Oligo | Function | Primer (5′ to 3′ ) |
|---|---|---|
| 1 | gpx4b 5′Race reverse | CACCACCTGCCCTTCTCTGTTGATC |
| 2 | gpx4b 3′Race forward | GAATGCACGCCTCCTACGCTGACAA |
| 3 | gpx4b full-length | Forward: AGGATGTGGATCGTAACGCGTG |
| Reverse: TCAGAGACATCACAGGTATTTAGG | ||
| 4 | gpx4a 3′Race forward | CACMGMYAGAGGACTGGC |
| 5 | gpx4a 5′Race reverse | ATCCTTCTCCACCACACTGG |
| 6 | gpx4a full-length | Forward: CAATTCAATGTCGTTGTGGC |
| Reverse: GGATCTTCCTAAGTACCTGTAA | ||
| 7 | gpx4b RT primer | Forward: ATCACCAACGTTGCCTCTAAAT |
| Reverse: CCTTGATTTCCACCTCTGTACC | ||
| 8 | gpx4a RT primer | Forward: GTACGCTGAGAAAGGTTTACGC |
| Reverse: TTGATGCCATTTCCCAGG | ||
| 9 | β-actin RT primer | Forward: GACCCACACAGTGCCCATCT |
| Reverse: GTGCCCATCTCCTGCTCAAA | ||
| 10 | GAPDH RT primer | Forward: TCTGTGTTGGAATCAACGGA |
| Reverse: TGAAGAAGACTCCGGTGGAC | ||
| 11 | rpL9 RT primer | Forward: AAAAAGCTGCGTGTGGATAAAT |
| Reverse: GATCGCATCTTATAGCGGAAAC |
A degenerate primer (primer 4, Table 1) for gpx4a 3′RACE was designed by identifying highly conserved regions of gpx4a sequences from three other fish species, including zebrafish (GenBank accession no. NM001007282); carp (GenBank accession no. FJ656211); southern bluefin tuna (GenBank accession no. EF452498), and by omitting conserved regions present in gpx4b. For 5′RACE, a PCR primer was designed (primer 5) from a 3′RACE PCR product. Full-length gpx4a was cloned from olfactory and liver cDNA libraries with primer set 6 (Table 1) and sequenced. Sequence homology searches were carried out using BLAST (Basic Local Alignment Search Tool), whereas sequence alignments were performed using ClustalW (San Diego Biology Workbench 3.2, http://workbench.sdsc.edu/). The final contiguous sequences were compiled using DNASTAR Lasergene8 sequence analysis software (DNASTAR, Madison, WI).
2.3 Gpx4a and gpx4b tissue-specific mRNA expression
Primers for quantitative real-time PCR analysis of gpx4 isoforms in salmon tissues were designed using Primer 3 software program (San Diego Biology Workbench 3.2, http://workbench.sdsc.edu/). Each primer pair produced a single PCR product as evidenced by melt curve analysis and gel electrophoresis (primers 7–11, Table 1). After PCR product validation and assay optimization, qPCR assays were conducted and analyzed using the relative standard curve method (Espinoza et al., 2011). Standards for quantification were created from gel-purified PCR products using QIAX II kit (Qiagen Inc.) and quantified before serial dilutions from 100 to 0.001 pg. All standard curves were generated revealed high coefficients of determination (R2 > 0.99). DNA SYBR Green master mix (Finnzymes), 0.3 μM of each primer, and molecular grade water were used to achieve the final reaction volume of 20 uL for each PCR triplicate. PCR amplifications were performed using a Bio-Rad IQ5 thermocycler (Hercules, CA) for 40 cycles with denaturation at 94 °C for 10 s, annealing at optimum temperature for primers (55–58 °C) for 20 s, and extension at 72 °C for 12 s. For quality control purposes, no template controls, as well as melt curve analyses were completed for all reactions. For gpx4 tissue profiling, gene expression quantities were normalized against β-actin, and ratios were calculated for the comparison between two gpx4 isoforms.
2.4 Analysis of GPx4 catalytic activities
To verify the presence of functional GPx4-associated catalytic activity, liver tissues from three juvenile Coho were thawed and rinsed in ice-cold 0.9% KCl buffer to remove the blood. Olfactory rosettes were pooled from a total of six individual Coho to provide enough tissue for n=3 pools. Tissue samples were homogenized with Potter–Elvehjem Teflon tissue homogenizer in three volumes of homogenization buffer (250 mM sucrose, 10 mM tris, 1 mM EDTA, 0.2 mM DTT, 0.1 mM PMSF, pH=7.4) at 0–4°C. The homogenate was centrifuged at 600 g for 15 min to discard cell debris and the nuclear fraction. The supernatant fluid was centrifuged at 10,000 g for 30 min to isolate the bulk of mitochondria, and the resulting supernatant fluid was centrifuged at 13,000 g for 20 min. The mitochondrial fractions were prepared by washing in homogenization buffer, re-centrifuging, and resuspending in homogenization buffer. The 13,000 g supernatants were centrifuged at 105,000 g for 1 h to obtain the cytosolic fractions. All cellular fractions were stored at −80 °C until use.
Whereas all GPx isozymes can catalyze the reduction of CuOOH, only GPx4 isoforms have the ability to reduce phosphatidylcholine hydroperoxide (PCOOH) with GSH, and thus this substrate can reflect GPx4-specific activity. PCOOH was prepared by oxidation of 1,2-dilinoleoyl-3-phosphatidylcholine (PC) with soybean lipoxidase as described previously (Garry et al., 2008). The concentration of PCOOH was determined spectrophotometrically using an extinction coefficient of 25,000 M−1 cm−1 at 234 nm. GPx4 catalytic activity was determined spectrophotometrically by a coupled enzyme assay (Maiorino et al., 1990; Weitzel et al., 1990). Briefly, 10 uL of GPx4-containing sample were added to a total volume of 1.0 mL buffer mixture containing 0.1 M Tris-HCl (pH=7.6), 5 mM EDTA, 1 mM sodium azide, 3 mM GSH, 0.2 mM NADPH, 0.1% peroxide free Triton X-100 (AMRESCO, Solon, OH), 1.2 U glutathione reductase (Sigma, specific activity 230 U/mg protein). The reaction mixture was pre-incubated for 3 min at room temperature and the initial slope was recorded as non-specific NADPH and glutathione oxidation. The reaction was then initiated by addition of substrate (4.2–5.9 nmol PCOOH, or 100 nmol CuOOH), and the oxidation of NADPH was monitored at ΔA340nm for 3 min. Negative controls were conducted in parallel in the absence of enzyme, and the resulting GPx4 activity was calculated from the difference between the two slopes using an extinction coefficient of 6220 M−1 cm−1. Protein concentrations were determined using the Bradford method (Bradford, 1976).
2.5 Effect of in vivo Cd exposures on gpx4 mRNA expression in Coho liver and olfactory rosettes
To analyze the effects of Cd on Coho liver and olfactory gpx4 mRNA expression, 8 juvenile Coho salmon (15.0 g ± 5.7) per treatment group were exposed to the intended concentrations of 0, 3.1 and 310 ppb cadmium (CdCl2, Mallinckrodt Baker, Phillipsburg, NJ) in 120 L aquaria contained within a large chilled re-circulating water bath and aerated by individual air stones for 24 and 48 hrs. The exposures were accomplished using a 90% renewal with water containing test agent at 24 hours. Water samples were taken pre- and post-exposures to assess the nominal concentrations (Frontier GeoSciences Inc. Seattle WA) and the Cd concentrations reported herein reflect measured waterborne levels (Espinoza et al., 2011). Specifically, the actual measured waterborne cadmium concentrations in the tanks at the initiation of exposures were 3.7 and 347 ppb in the low and high dose exposure tanks, respectively. Hereafter, measured cadmium concentrations are used in all text, tables and figures. Following exposures, the olfactory rosettes and liver tissues were collected from n=8 anesthetized fish per treatment as described above. Analysis of olfactory β-actin mRNA expression showed unacceptable variation among the treatments, thus 3 internal reference genes (β-actin, Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein L9 (rpL9)) were used to create geometric mean values for normalization of the gpx4 mRNA expression. Normalization factors were determined using geNorm (version 3.5) software (http://medgen.ugent.be/~jvdesomp/genorm/).
2.6 Statistical Analysis
Prior to statistical analyses, all data were inspected for homogeneity of variances using the Bartlett’s test. In some cases, such as tissue specific profile data, the distributions did not follow parametric distributions. Accordingly, tissue differences among gpx4a and gpx4b were assessed by using the (Wilcoxon) Mann-Whitney U test. To analyze the effects of Cd exposures on gpx4 mRNA expression, data sets conforming to normal distributions were assessed for significance using a one-way ANOVA followed by a Dunnett’s test. For non-parametric distributions, data were assessed for significance using Kruskal-Wallis nonparametric one-way analysis of variance test followed by a Dunn’s test. All data were considered statistically significant at p ≤ 0.05. All statistical analyses were conducted using GraphPad Prism Ver 5.0 (Graph Pad Software Inc, San Diego, CA, USA).
3. Results
3.1 Cloning of gpx4a and gpx4b isoforms
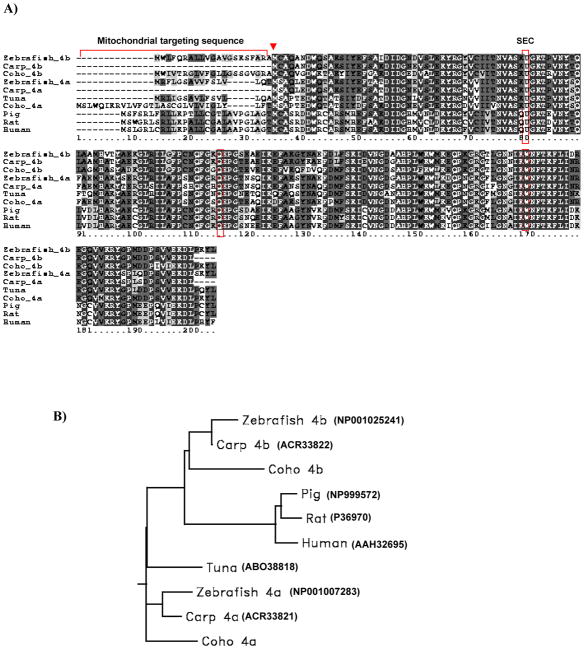
Full-length gpx4a and gpx4b cDNAs were successfully cloned from Coho salmon olfactory and liver total RNA by RACE-PCR. The full length gpx4a cDNA contained a 5′-UTR of 31 bp, an open reading frame (ORF) of 603 bp and also a 3′-UTR of 333 bp (GenBank accession no. JN967674). Inspection of the gpx4a ORF revealed two potential protein translation start sites (5′-ATG and 3′-ATG) encoding deduced proteins of 170 and 200 amino acids in length. The gpx4b cDNA contained a 5′-UTR of 62 bp, an open reading frame (ORF) of 576 bp, and a 3′-UTR of 436 bp (GenBank accession no. JN967675). The ORF has two possible protein translation start sites encoding deduced proteins of 170 and 191 amino acids in length (Figure 1A). Amino acid sequence comparisons of the two isoforms revealed 71% identities. The phylogenetic analysis of the two isoforms revealed high sequence identities when compared with other fish and higher vertebrate GPx4 isoforms (Figure 1B). For example, Coho GPx4a shared 83% identity with carp GPx4a, and Coho GPx4b shared 81% identity with carp GPx4b. Both Coho GPx4 isoforms shared ~60% identity with human GPx4. Similar to mammalian GPx4, the predicted Coho GPx4 protein showed a potential mitochondrial targeting sequence (30 amino acids in length for GPx4a and 21 amino acids in length for GPx4b, respectively, Figure 1A).
Figure 1.
(A) Predicted amino acid sequence alignment of the two Coho GPx4 isoform with other fish and mammalian GPx4s. The selenocysteine residue is indicated as U in sequence. The three boxes highlight conserved residues at selenocysteine (U), glutamine (Q) and tryptophan (W). Black shading indicates completely conserved residues and grey shading indicates similar residues (similarity threshhold fraction ≥0.7). The arrow indicates the second translational initiation site. Mitochondrial targeting sequences are underlined. (B) Phylogenetic rooted-tree showing the relationship of Coho GPx4 with other fish and mammalian GPx4s. GenBank accession numbers are shown in parentheses. Graph generated from San Diego Biology Workbench 3.2.
3.2 Tissue expression profiles of gpx4 genes
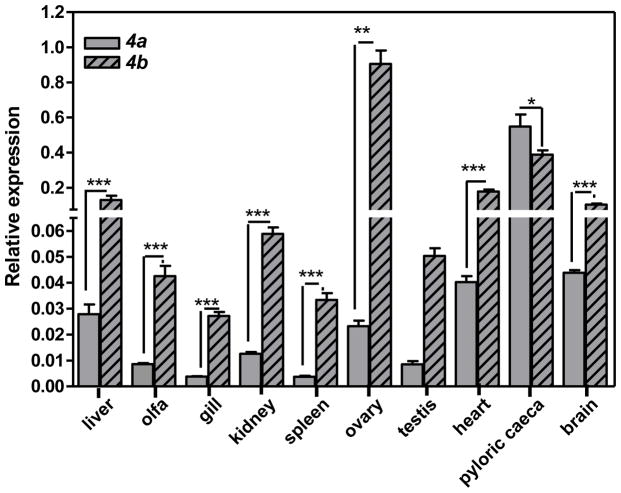
Expression of the gpx4a and gpx4b were analyzed in 10 tissues using qRT-PCR. In these experiments, β-actin mRNA expression levels did not differ among the tissues and was consequently used as the internal reference gene for normalization purposes. The highest gpx4a mRNA expression was found in pyloric caeca, followed by brain, heart and liver (Figure 2). By contrast, gpx4b showed highest mRNA expression in ovary, followed by pyloric caeca, heart, liver and brain (Figure 2). In general, gpx4b mRNA expression exceeded that of gpx4a in all tissues with the exception of the pyloric caeca. The ratio of gpx4b/gpx4a was highest (−39) in ovary, and lowest in pyloric caeca (−0.7). With the exception of the gonads, no sex differences in constitutive gene expression were observed.
Figure 2.
Tissue-specific gpx4 isoform expression. Gene expression normalized to β-actin. All data represent the mean ± SEM of measurement of 3 to 8 individuals. Asterisks indicate statistically significant difference compared to control samples (* p <0.05; **p<0.01 and *** p<0.001).
3.4 GPx catalytic enzyme activities
The results of the total GPx and GPx4-specific catalytic activity analyses in adult Coho liver and olfactory rosette tissues are shown in Table 2. As observed, the mitochondrial and cytosolic GPx4 isoforms were distinguished from total tissue GPx by using the substrate PCOOH, a specific substrate of GPx4. As observed, the total tissue initial rate GPx activities were approximately 2- fold higher in olfactory mitochondrial and cytosolic fractions compared to those in the corresponding liver subcellular fractions (Table 2). Further, higher GPx activities were observed in cytosolic fractions than mitochondrial fractions in both the olfactory and liver tissues (Table 2). The presence of GPx4-specific catalytic activity accompanied gpx4 mRNA expressions in olfactory and liver subcellular fractions, and did not markedly differ among the two tissues.
Table 2.
Total GPx and GPx4-specific activity in Coho liver and olfactory rosettes using different hydroperoxide substrates*
| Tissue | Substrate | Mitochondrial GPx or GPx4 (μmol/min per mg protein) | Cytosolic GPx or GPx4 (μmol/min per mg protein) |
|---|---|---|---|
| Liver1 | CuOOH | 28.4 ± 3.90 | 53.2 ± 3.51 |
| PCOOH | 10.6 ± 0.53 | 12.3 ± 1.01 | |
| Olfactory rosettes2 | CuOOH | 62.6 ± 6.38 | 96 ± 7.72 |
| PCOOH | 7.8 ± 1.70 | 9.1 ± 1.04 |
Activities were measured as described under methods, section 2.4. The GSH concentrations in all assays were fixed as 3 mM.
Enzymatic values in the liver tissue reflect the mean ± S.E. of 3 individual fish.
Enzymatic values in the olfactory rosettes reflect the mean ± S.E. of n=3 pools.
3.5 Effect of Cd on olfactory and liver gpx4 mRNA expression
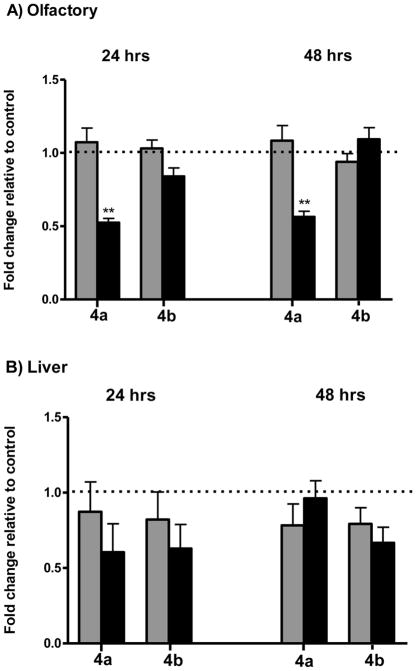
As observed in Figure 3, gpx4 mRNA expressions in olfactory and liver tissues were modulated by exposure to both doses of Cd. In olfactory rosettes, exposure to 347 ppb Cd significantly decreased gpx4a expression (48% loss by 24hr; 44% loss by 48 hr exposure, Figure 3A). By contrast, olfactory gpx4b mRNA expression was not extensively modulated by Cd. A transient effect of Cd in the expression of liver gpx4 was also observed (Figure 3B). By 24 hr, exposure to the low dose of Cd caused mild changes of gpx4a and gpx4b expression (13% and 18% loss, respectively). As similarly observed in olfactory tissues, the higher Cd dose elicited a more extensive effect on liver gpx4a and gpx4b expression (40% and 37% loss by 24 hr, respectively). Longer exposure periods caused minor (20–22%) losses of liver gpx4a and gpx4b expression by the low Cd dose, whereas at higher Cd exposures, a 33% loss in gpx4b expression was observed (Figure 3).
Figure 3.
Modulation of gpx4 expression by Cd in Coho olfactory rosettes (A) and liver (B). Expression of gpx4a and gpx4b were measured in the olfactory rosettes and liver of Coho salmon exposed to 0, 3.7, and 347 ppb Cd for 24 and 48 hr periods. All data represent the mean ± SEM of n=8 individuals. Graphs are presented as fold-change relative to the corresponding controls, with the 3.7 ppb and 347 ppb datasets represented by light and dark bars, respectively. In the olfactory rosettes, data was normalized to a normalization factor comprised of multiple housekeeping genes as described in the methods. Liver mRNA data was normalized to the expression of β-actin. Asterisks indicate statistically significant difference compared to control samples ( **p<0.01).
4. Discussion
Our recent study showing the expression of eight GST isoforms in Coho olfactory tissues, including isoforms that detoxify secondary byproducts of oxidative stress, suggests that the cellular antioxidant pathways that maintain redox status and protect against cellular oxidative damage may be important in maintaining olfactory processes (Espinoza et al., 2011). Among GSH-dependent antioxidant enzymes, the unique substrate profile of cellular GPx4 is of particular importance in maintaining olfactory signal transduction profiles by nature of their functional ability to modulate signal transduction, as well as reduce potentially toxic intracellular lipid peroxides (Ursini et al., 1985; Ursini et al., 1982). Our demonstration of two gpx4 paralogs in Coho olfactory tissues, accompanied by comparably high GPx4 catalytic activities, is consistent with an active GSH-associated olfactory antioxidant defense system, with a function in maintaining normal olfactory signal transduction in the face of environmental stress.
Studies in other species indicate a substantial role for GPx4 in maintaining neuronal cell function, especially under conditions of oxidative stress (Ran et al., 2006; Savaskan et al., 2007a; Savaskan et al., 2007b; Seiler et al., 2008). Furthermore, GPx4 is highly expressed in several regions of the rodent brain (i.e. cerebral cortex, hippocampus) and olfactory tissues, including olfactory epithelium and olfactory bulb (Schneider et al., 2006; Zhang et al., 2008), supporting a potential role in neuroprotective function (Savaskan et al., 2007b). In mammals, the GPx4 protein maintains high stability and catalytic function in brain, even under conditions when selenium levels are compromised (Brigelius-Flohe, 1999). Knockout of GPx4 in neural cells using CamKIIα-Cre transgenic mice resulted in increased lipid peroxidation leading to cell death (Seiler et al., 2008), whereas brain tissues from a mouse model of Alzheimer’s disease showed reduced GPx4 associated with increased oxidized lipid by-products (Yoo et al., 2010). In addition to its role as an antioxidant enzyme, GPx4 modulates cell signaling and regulatory events (Imai and Nakagawa, 2003; Ufer and Wang, 2011; Ursini et al., 1997) including regulation of lipoxygenase and cyclooxygenase activity and apoptotic pathways (Chen et al., 2003; Huang et al., 1999; Imai et al., 1998; Schnurr et al., 1996; Seiler et al., 2008; Sutherland et al., 2001). Moreover, GPx4 has been shown to modulate the redox–regulated transcription factors nuclear factor-κB (NF-κB) and nuclear factor erythroid 2-related factor 2 (Nrf2) (Brigelius-Flohe, 2006; Ufer and Wang, 2011). In vitro studies have suggested that GPx4 inhibits NF-κB activation and up-regulates heme oxygenase-1 via the Nrf2 pathway in vascular smooth muscle cells (Banning and Brigelius-Flohe, 2005). Interestingly, both NF-κB and Nrf2 are involved in heavy metal-induced stress responses (Chen et al., 2001; Liu et al., 2009; Lushchak, 2011). Collectively, the regulatory mechanisms of gpx4 are consistent with a role in the maintenance of signal transduction processes as well as protecting against cellular oxidative stress during normal olfactory function.
Our hypothesis of a protective role of GPx4 in maintaining Coho olfactory function under oxidative stress was supported by the results of our chemical challenges in which we observed that exposure to Cd generally decreased gpx4 mRNA expression in olfactory tissues. In this regard, Cd is a well-established olfactory toxicant in aquatic species (Blechinger et al., 2007; Matz and Krone, 2007) as well as in rodents (Bondier et al., 2008; Kumar et al., 1996; Sun et al., 1996). Exposure to Cd in mice leads to extensive lipid peroxidation in the mouse olfactory bulb which is accompanied by decreased levels of GSH (Kumar et al., 1996). Although the linkage between Cd-mediated oxidative damage and gpx4 expression has not been well documented in fish, several studies have indicated a decrease in overall tissue GPx expression and/or catalytic activity following Cd exposure (Banni et al., 2011; Choi et al., 2007; Karaytug et al., 2011). Of relevance to the current project was a study showing decreased gpx4a mRNA expression in the olfactory lobes of common carp over a 96 hr period of Cd exposure (10 mg/L), accompanied by a complete elimination of gpx4b expression (Hermesz and Ferencz, 2009). The fact that we observed a loss of gpx4 expression at low environmentally- relevant levels of Cd suggests that this phenomenon is of relevance to field scenarios and exposures encountered in polluted waterways.
Although there is little information concerning the translational control of gpx4 in aquatic species, we have recently observed a general loss of several GST forms expression in Coho exposed to Cd (Espinoza et al., 2011). In our GST study, the loss of olfactory GST expression was accompanied by a strong and concomitant increase in the expression of metallonthionein, a sensitive marker of heavy metal exposures in fish (Bigot et al., 2011; Falfushynska et al., 2011; Gagne et al., 2007; Kim et al., 2010). In addition to observed effects at the mRNA level, Cd can inhibit GPx catalytic activities in several species, with the loss of catalytic capacity associated with GSH depletion or the formation of a chemical complex between Cd and selenium at the active site of the enzyme (Cuypers et al., 2010; Gambhir and Nath, 1992; Quig, 1998).
In addition to the comparably high levels of gpx4 mRNA expression observed in liver and olfactory tissues, we also observed the presence of the gpx4 paralogs in other tissues, including pyloric caeca, brain, heart, gonad, and kidney. While the functional significance of these differences are unknown, the broad tissue distribution of Coho gpx4 is consistent with conservation of important physiological functions (Brigelius-Flohe, 1999; Schneider et al., 2006; Thompson et al., 2010; Zhang et al., 1989), such as protecting against free radical attack on lipid-rich membranes in these tissues. Others have reported that the comparably high GPx4 expression in gonads is associated with a role in maintaining favorable redox status within the reproductive and endocrine systems, especially during sexual maturation or during spermatozoa development (Beckett and Arthur, 2005; Schneider et al., 2009).
In mammalian cells, the GPx4 gene gives rise to three different isoforms that associate with different subcellular compartments, including mitochondrial GPx4 (m- GPx4), cytosolic GPx4 (c-GPx4), and nuclear GPx4 (n-GPx4). The alternative usage of the translational initiation site of GPx4 genes controls the ultimate subcellular localizations of GPx4 proteins (Arai et al., 1999; Knopp et al., 1999; Maiorino et al., 2003; Nam et al., 1997; Pfeifer et al., 2001). The sequence between the first two initiation sites is thought to encode for a mitochondrial targeting sequence responsible for directing m-GPx4 import of the mitochondria (Thomas et al., 1990). Thus, m-GPx4 and c-GPx4 can not be distinguished at a protein level once this targeting sequence is cleaved. The fact that we observed total GPx activity and GPx4-specific catalytic activities in both the mitochondrial and cytosolic fractions of salmon tissues indicates subcellular targeting of GPx4 isoforms in salmon. However, in the present study we were unable to differentiate between the two GPx4 isoforms based upon substrate specificities. An existing structural model of the human GPx4 protein indicates that certain residues, including selenocysteine (U46), glutamine (Q81) and tryptophan (W136) are functionally critical to its catalytic activity (Scheerer et al., 2007). These three amino acids are considered a catalytic triad and are well conserved among fish and mammalian species, suggesting similar GPx4 catalytic activities or substrate specificities across species. Although GPx4-associated catalytic activities have not been extensively investigated in fish species, our studies in Coho are consistent with a similar catalytic profile with GPx4 in mammalian cells.
Summary and conclusions
In summary, we have cloned two paralogs of gpx4 (gpx4a and gpx4b) from Coho salmon olfactory and liver tissues. The amino acid sequence of the two GPx4 isoforms shared 71% identity and exhibited high homology with other fish and mammalian GPx4 enzymes. Both isoforms showed a ubiquitous expression pattern among the tissues investigated, and catalytic function analysis substantiated relatively rapid initial rates of GPx4 enzymatic activity in olfactory and liver subcellular fractions. Accordingly, our results demonstrate a functional role of GPx4 in Coho salmon. Furthermore, the transient modulation of gpx4 isoforms in liver and olfactory tissues on exposure to the prototypical environmental contaminant Cd, while not showing an inductive or adaptive response, was suggestive of an overwhelming of cellular antioxidant responses by Cd. Our present study also extends observations in mammalian species indicating an important role of GPx4 in protecting against cellular oxidative damage in olfactory system of vertebrates. Of importance in future studies is the need to address the regulatory mechanisms of salmon gpx4 in the context of chemical exposures, as well as functional studies addressing the role of gpx4 knockdown on metal injury to the fish olfactory system.
Highlights of the current research.
Cloned two gpx4 isoforms (gpx4a and gpx4b) from the Coho salmon peripheral olfactory system
Developed qPCR assays for a comprehensive analysis of gpx4 expression in 10 tissues
High initial rates of GPx4 enzymatic activity in Coho olfactory and liver tissues
Examined the effect of cadmium on gpx4 expression in olfactory and liver tissues
Acknowledgments
This work was supported in part by the University of Washington NIEHS Superfund Basic Sciences Grant NIEHS P42-004696. The authors appreciate the assistance of Dr. Brian Beckman and Abby Tillotson at NOAA fisheries, Seattle, WA, who provided the juvenile Coho salmon for these experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai M, Imai H, Koumura T, Yoshida M, Emoto K, Umeda M, Chiba N, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J Biol Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- Baatrup E. Structural and functional effects of heavy metals on the nervous system, including sense organs, of fish. Comp Biochem Physiol C. 1991;100:253–257. doi: 10.1016/0742-8413(91)90163-n. [DOI] [PubMed] [Google Scholar]
- Baldwin DH, Sandahl JF, Labenia JS, Scholz NL. Sublethal effects of copper on coho salmon: impacts on nonoverlapping receptor pathways in the peripheral olfactory nervous system. Environ Toxicol Chem. 2003;22:2266–2274. doi: 10.1897/02-428. [DOI] [PubMed] [Google Scholar]
- Banni M, Chouchene L, Said K, Kerkeni A, Messaoudi I. Mechanisms underlying the protective effect of zinc and selenium against cadmium-induced oxidative stress in zebrafish Danio rerio. Biometals. 2011;24:981–992. doi: 10.1007/s10534-011-9456-z. [DOI] [PubMed] [Google Scholar]
- Banning A, Brigelius-Flohe R. NF-kappaB, Nrf2, and HO-1 interplay in redox-regulated VCAM-1 expression. Antioxid Redox Signal. 2005;7:889–899. doi: 10.1089/ars.2005.7.889. [DOI] [PubMed] [Google Scholar]
- Beckett GJ, Arthur JR. Selenium and endocrine systems. J Endocrinol. 2005;184:455–465. doi: 10.1677/joe.1.05971. [DOI] [PubMed] [Google Scholar]
- Bigot A, Minguez L, Giamberini L, Rodius F. Early defense responses in the freshwater bivalve Corbicula fluminea exposed to copper and cadmium: Transcriptional and histochemical studies. Environ Toxicol. 2011;26:623–632. doi: 10.1002/tox.20599. [DOI] [PubMed] [Google Scholar]
- Blechinger SR, Kusch RC, Haugo K, Matz C, Chivers DP, Krone PH. Brief embryonic cadmium exposure induces a stress response and cell death in the developing olfactory system followed by long-term olfactory deficits in juvenile zebrafish. Toxicol Appl Pharmacol. 2007;224:72–80. doi: 10.1016/j.taap.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Bondier JR, Michel G, Propper A, Badot PM. Harmful effects of cadmium on olfactory system in mice. Inhal Toxicol. 2008;20:1169–1177. doi: 10.1080/08958370802207292. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R. Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Huang HS, Chang WC. Depletion of phospholipid hydroperoxide glutathione peroxidase up-regulates arachidonate metabolism by 12(S)-lipoxygenase and cyclooxygenase 1 in human epidermoid carcinoma A431 cells. FASEB J. 2003;17:1694–1696. doi: 10.1096/fj.02-0847fje. [DOI] [PubMed] [Google Scholar]
- Chen F, Ding M, Castranova V, Shi X. Carcinogenic metals and NF-kappaB activation. Mol Cell Biochem. 2001;222:159–171. [PubMed] [Google Scholar]
- Choi CY, An KW, Nelson ER, Habibi HR. Cadmium affects the expression of metallothionein (MT) and glutathione peroxidase (GPX) mRNA in goldfish, Carassius auratus. Comp Biochem Physiol C Toxicol Pharmacol. 2007;145:595–600. doi: 10.1016/j.cbpc.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Conrad M, Moreno SG, Sinowatz F, Ursini F, Kolle S, Roveri A, Brielmeier M, Wurst W, Maiorino M, Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol Cell Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers A, Plusquin M, Remans T, Jozefczak M, Keunen E, Gielen H, Opdenakker K, Nair AR, Munters E, Artois TJ, Nawrot T, Vangronsveld J, Smeets K. Cadmium stress: an oxidative challenge. Biometals. 2010;23:927–940. doi: 10.1007/s10534-010-9329-x. [DOI] [PubMed] [Google Scholar]
- Espinoza HM, Williams CR, Gallagher EP. Effect of cadmium on glutathione S-transferase and metallothionein gene expression in coho salmon liver, gill and olfactory tissues. Aquat Toxicol. 2011 doi: 10.1016/j.aquatox.2011.12.012. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falfushynska HI, Gnatyshyna LL, Stoliar OB, Nam YK. Various responses to copper and manganese exposure of Carassius auratus gibelio from two populations. Comp Biochem Physiol C Toxicol Pharmacol. 2011;154:242–253. doi: 10.1016/j.cbpc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Gagne F, Gagnon C, Turcotte P, Blaise C. Changes in metallothionein levels in freshwater mussels exposed to urban wastewaters: effects from exposure to heavy metals? Biomark Insights. 2007;2:107–116. [PMC free article] [PubMed] [Google Scholar]
- Gambhir J, Nath R. Effect of cadmium on tissue glutathione and glutathione peroxidase in rats: influence of selenium supplementation. Indian J Exp Biol. 1992;30:597–601. [PubMed] [Google Scholar]
- Garry MR, Kavanagh TJ, Faustman EM, Sidhu JS, Liao R, Ware C, Vliet PA, Deeb SS. Sensitivity of mouse lung fibroblasts heterozygous for GPx4 to oxidative stress. Free Radic Biol Med. 2008;44:1075–1087. doi: 10.1016/j.freeradbiomed.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Gottofrey J, Tjälve H. Axonal transport of cadmium in the olfactory nerve of the pike. Pharmacol Toxicol. 1991;69:242–252. doi: 10.1111/bcpt.1991.69.4.242. [DOI] [PubMed] [Google Scholar]
- Hart BA, Lee CH, Shukla GS, Shukla A, Osier M, Eneman JD, Chiu JF. Characterization of cadmium-induced apoptosis in rat lung epithelial cells: evidence for the participation of oxidant stress. Toxicology. 1999;133:43–58. doi: 10.1016/s0300-483x(99)00013-x. [DOI] [PubMed] [Google Scholar]
- Hermesz E, Ferencz A. Identification of two phospholipid hydroperoxide glutathione peroxidase (gpx4) genes in common carp. Comp Biochem Physiol C Toxicol Pharmacol. 2009;150:101–106. doi: 10.1016/j.cbpc.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Huang HS, Chen CJ, Suzuki H, Yamamoto S, Chang WC. Inhibitory effect of phospholipid hydroperoxide glutathione peroxidase on the activity of lipoxygenases and cyclooxygenases. Prostaglandins Other Lipid Mediat. 1999;58:65–75. doi: 10.1016/s0090-6980(99)00017-9. [DOI] [PubMed] [Google Scholar]
- Imai H, Hirao F, Sakamoto T, Sekine K, Mizukura Y, Saito M, Kitamoto T, Hayasaka M, Hanaoka K, Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305:278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- Imai H, Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- Imai H, Narashima K, Arai M, Sakamoto H, Chiba N, Nakagawa Y. Suppression of leukotriene formation in RBL-2H3 cells that overexpressed phospholipid hydroperoxide glutathione peroxidase. J Biol Chem. 1998;273:1990–1997. doi: 10.1074/jbc.273.4.1990. [DOI] [PubMed] [Google Scholar]
- Jarrard HE, Delaney KR, Kennedy CJ. Impacts of carbamate pesticides on olfactory neurophysiology and cholinesterase activity in coho salmon (Oncorhynchus kisutch) Aquat Toxicol. 2004;69:133–148. doi: 10.1016/j.aquatox.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Karaytug S, Sevgiler Y, Karayakar F. Comparison of the protective effects of antioxidant compounds in the liver and kidney of Cd- and Cr-exposed common carp. Environ Toxicol. 2011 doi: 10.1002/tox.20779. [DOI] [PubMed] [Google Scholar]
- Kim JH, Dahms HU, Rhee JS, Lee YM, Lee J, Han KN, Lee JS. Expression profiles of seven glutathione S-transferase (GST) genes in cadmium- exposed river pufferfish (Takifugu obscurus) Comp Biochem Physiol C Toxicol Pharmacol. 2010;151:99–106. doi: 10.1016/j.cbpc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Knopp EA, Arndt TL, Eng KL, Caldwell M, LeBoeuf RC, Deeb SS, O’Brien KD. Murine phospholipid hydroperoxide glutathione peroxidase: cDNA sequence, tissue expression, and mapping. Mamm Genome. 1999;10:601–605. doi: 10.1007/s003359901053. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Gladyshev VN. Selenium metabolism in zebrafish: multiplicity of selenoprotein genes and expression of a protein containing 17 selenocysteine residues. Genes Cells. 2000;5:1049–1060. doi: 10.1046/j.1365-2443.2000.00392.x. [DOI] [PubMed] [Google Scholar]
- Kühn H, Borchert A. Regulation of enzymatic lipid peroxidation: the interplay of peroxidizing and peroxide reducing enzymes. Free Radic Biol Med. 2002;33:154–172. doi: 10.1016/s0891-5849(02)00855-9. [DOI] [PubMed] [Google Scholar]
- Kumar R, Agarwal AK, Seth PK. Oxidative stress-mediated neurotoxicity of cadmium. Toxicol Lett. 1996;89:65–69. doi: 10.1016/s0378-4274(96)03780-0. [DOI] [PubMed] [Google Scholar]
- Laberge F, Hara TJ. Neurobiology of fish olfaction: a review. Brain Res Brain Res Rev. 2001;36:46–59. doi: 10.1016/s0165-0173(01)00064-9. [DOI] [PubMed] [Google Scholar]
- Lackey RT. Pacific Northwest Salmon: Forecasting Their Status in 2100. Reviews in Fisheries Science. 2003;11:35–88. [Google Scholar]
- Laetz CA, Baldwin DH, Collier TK, Hebert V, Stark JD, Scholz NL. The synergistic toxicity of pesticide mixtures: implications for risk assessment and the conservation of endangered Pacific salmon. Environ Health Perspect. 2009;117:348–353. doi: 10.1289/ehp.0800096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qu W, Kadiiska MB. Role of oxidative stress in cadmium toxicity and carcinogenesis. Toxicol Appl Pharmacol. 2009;238:209–214. doi: 10.1016/j.taap.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak VI. Environmentally induced oxidative stress in aquatic animals. Aquat Toxicol. 2011;101:13–30. doi: 10.1016/j.aquatox.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Maiorino M, Gregolin C, Ursini F. Phospholipid hydroperoxide glutathione peroxidase. Methods Enzymol. 1990;186:448–457. doi: 10.1016/0076-6879(90)86139-m. [DOI] [PubMed] [Google Scholar]
- Maiorino M, Scapin M, Ursini F, Biasolo M, Bosello V, Flohe L. Distinct promoters determine alternative transcription of gpx-4 into phospholipid-hydroperoxide glutathione peroxidase variants. J Biol Chem. 2003;278:34286–34290. doi: 10.1074/jbc.M305327200. [DOI] [PubMed] [Google Scholar]
- Matz CJ, Krone PH. Cell death, stress-responsive transgene activation, and deficits in the olfactory system of larval zebrafish following cadmium exposure. Environ Sci Technol. 2007;41:5143–5148. doi: 10.1021/es070452c. [DOI] [PubMed] [Google Scholar]
- Nam S, Nakamuta N, Kurohmaru M, Hayashi Y. Cloning and sequencing of the mouse cDNA encoding a phospholipid hydroperoxide glutathione peroxidase. Gene. 1997;198:245–249. doi: 10.1016/s0378-1119(97)00321-1. [DOI] [PubMed] [Google Scholar]
- Palm RC, Jr, Powell DB. Alarm substance recognition and predator avoidance by chinook salmon (Oncorhynchus tschawytscha) following exposure to an organophosphate pesticide. Environ Toxicol Chem. 2010;29:1113–1122. doi: 10.1002/etc.142. [DOI] [PubMed] [Google Scholar]
- Pfeifer H, Conrad M, Roethlein D, Kyriakopoulos A, Brielmeier M, Bornkamm GW, Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- Quig D. Cysteine metabolism and metal toxicity. Altern Med Rev. 1998;3:262–270. [PubMed] [Google Scholar]
- Ran Q, Gu M, Van Remmen H, Strong R, Roberts JL, Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J Neurosci Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, 2nd, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- Risso-de Faverney C, Orsini N, de Sousa G, Rahmani R. Cadmium-induced apoptosis through the mitochondrial pathway in rainbow trout hepatocytes: involvement of oxidative stress. Aquat Toxicol. 2004;69:247–258. doi: 10.1016/j.aquatox.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Roveri A, Maiorino M, Nisii C, Ursini F. Purification and characterization of phospholipid hydroperoxide glutathione peroxidase from rat testis mitochondrial membranes. Biochim Biophys Acta. 1994;1208:211–221. doi: 10.1016/0167-4838(94)90106-6. [DOI] [PubMed] [Google Scholar]
- Sandahl JF, Baldwin DH, Jenkins JJ, Scholz NL. A sensory system at the interface between urban stormwater runoff and salmon survival. Environ Sci Technol. 2007;41:2998–3004. doi: 10.1021/es062287r. [DOI] [PubMed] [Google Scholar]
- Savaskan NE, Borchert A, Brauer AU, Kuhn H. Role for glutathione peroxidase-4 in brain development and neuronal apoptosis: specific induction of enzyme expression in reactive astrocytes following brain injury. Free Radic Biol Med. 2007a;43:191–201. doi: 10.1016/j.freeradbiomed.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Savaskan NE, Ufer C, Kuhn H, Borchert A. Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol Chem. 2007b;388:1007–1017. doi: 10.1515/BC.2007.126. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Borchert A, Krauss N, Wessner H, Gerth C, Hohne W, Kuhn H. Structural basis for catalytic activity and enzyme polymerization of phospholipid hydroperoxide glutathione peroxidase-4 (GPx4) Biochemistry. 2007;46:9041–9049. doi: 10.1021/bi700840d. [DOI] [PubMed] [Google Scholar]
- Schneider M, Forster H, Boersma A, Seiler A, Wehnes H, Sinowatz F, Neumuller C, Deutsch MJ, Walch A, Hrabe de Angelis M, Wurst W, Ursini F, Roveri A, Maleszewski M, Maiorino M, Conrad M. Mitochondrial glutathione peroxidase 4 disruption causes male infertility. FASEB J. 2009;23:3233–3242. doi: 10.1096/fj.09-132795. [DOI] [PubMed] [Google Scholar]
- Schneider M, Vogt Weisenhorn DM, Seiler A, Bornkamm GW, Brielmeier M, Conrad M. Embryonic expression profile of phospholipid hydroperoxide glutathione peroxidase. Gene Expr Patterns. 2006;6:489–494. doi: 10.1016/j.modgep.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Schnurr K, Belkner J, Ursini F, Schewe T, Kuhn H. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase controls the activity of the 15-lipoxygenase with complex substrates and preserves the specificity of the oxygenation products. J Biol Chem. 1996;271:4653–4658. doi: 10.1074/jbc.271.9.4653. [DOI] [PubMed] [Google Scholar]
- Scott GR, Sloman KA, Rouleau C, Wood CM. Cadmium disrupts behavioural and physiological responses to alarm substance in juvenile rainbow trout (Oncorhynchus mykiss) J Exp Biol. 2003;206:1779–1790. doi: 10.1242/jeb.00353. [DOI] [PubMed] [Google Scholar]
- Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Stromberg PC, Ferrante JG, Carter S. Pathology of lethal and sublethal exposure of fathead minnows, Pimephales promelas, to cadmium: a model for aquatic toxicity assessment. J Toxciol Environ Health. 1983;11:247–259. [Google Scholar]
- Sun TJ, Miller ML, Hastings L. Effects of inhalation of cadmium on the rat olfactory system: behavior and morphology. Neurotoxicol Teratol. 1996;18:89–98. doi: 10.1016/0892-0362(95)02013-6. [DOI] [PubMed] [Google Scholar]
- Sutherland M, Shankaranarayanan P, Schewe T, Nigam S. Evidence for the presence of phospholipid hydroperoxide glutathione peroxidase in human platelets: implications for its involvement in the regulatory network of the 12-lipoxygenase pathway of arachidonic acid metabolism. Biochem J. 2001;353:91–100. [PMC free article] [PubMed] [Google Scholar]
- Thevenod F, Friedmann JM, Katsen AD, Hauser IA. Up-regulation of multidrug resistance P-glycoprotein via nuclear factor-kappaB activation protects kidney proximal tubule cells from cadmium- and reactive oxygen species-induced apoptosis. J Biol Chem. 2000;275:1887–1896. doi: 10.1074/jbc.275.3.1887. [DOI] [PubMed] [Google Scholar]
- Thomas JP, Maiorino M, Ursini F, Girotti AW. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation. In situ reduction of phospholipid and cholesterol hydroperoxides. J Biol Chem. 1990;265:454–461. [PubMed] [Google Scholar]
- Thompson JL, See VH, Thomas PM, Schuller KA. Cloning and characterization of two glutathione peroxidase cDNAs from southern bluefin tuna (Thunnus maccoyii) Comp Biochem Physiol B Biochem Mol Biol. 2010;156:287–297. doi: 10.1016/j.cbpb.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Tierney KB, Ross PS, Jarrard HE, Delaney KR, Kennedy CJ. Changes in juvenile coho salmon electro-olfactogram during and after short-term exposure to current-use pesticides. Environ Toxicol Chem. 2006;25:2809–2817. doi: 10.1897/05-629r1.1. [DOI] [PubMed] [Google Scholar]
- Tjälve H, Gottofrey J, Bjorklund I. Tissue disposition of 109Cd2+ in the brown trout (Salmo trutta) studied by autoradiography and impulse counting. Toxicological & Environmental Chemistry. 1986;12:31–45. [Google Scholar]
- Tjälve H, Henriksson J. Uptake of metals in the brain via olfactory pathways. Neurotoxicology. 1999;20:181–195. [PubMed] [Google Scholar]
- Ufer C, Wang CC. The Roles of Glutathione Peroxidases during Embryo Development. Front Mol Neurosci. 2011;4:12. doi: 10.3389/fnmol.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Gregolin C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim Biophys Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Roveri A. Phospholipid hydroperoxide glutathione peroxidase (PHGPx): more than an antioxidant enzyme? Biomed Environ Sci. 1997;10:327–332. [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Valente M, Ferri L, Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim Biophys Acta. 1982;710:197–211. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang J, Leonard SS, Rao KM. Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic Biol Med. 2004;36:1434–1443. doi: 10.1016/j.freeradbiomed.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Suzuki T. Involvement of reactive oxygen stress in cadmium-induced cellular damage in Euglena gracilis. Comp Biochem Physiol C Toxicol Pharmacol. 2002;131:491–500. doi: 10.1016/s1532-0456(02)00036-4. [DOI] [PubMed] [Google Scholar]
- Weitzel F, Ursini F, Wendel A. Phospholipid hydroperoxide glutathione peroxidase in various mouse organs during selenium deficiency and repletion. Biochim Biophys Acta. 1990;1036:88–94. doi: 10.1016/0304-4165(90)90018-r. [DOI] [PubMed] [Google Scholar]
- Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- Yoo MH, Gu X, Xu XM, Kim JY, Carlson BA, Patterson AD, Cai H, Gladyshev VN, Hatfield DL. Delineating the role of glutathione peroxidase 4 in protecting cells against lipid hydroperoxide damage and in Alzheimer’s disease. Antioxid Redox Signal. 2010;12:819–827. doi: 10.1089/ars.2009.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LP, Maiorino M, Roveri A, Ursini F. Phospholipid hydroperoxide glutathione peroxidase: specific activity in tissues of rats of different age and comparison with other glutathione peroxidases. Biochim Biophys Acta. 1989;1006:140–143. doi: 10.1016/0005-2760(89)90336-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou Y, Schweizer U, Savaskan NE, Hua D, Kipnis J, Hatfield DL, Gladyshev VN. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J Biol Chem. 2008;283:2427–2438. doi: 10.1074/jbc.M707951200. [DOI] [PubMed] [Google Scholar]