Abstract
Bacterial cells, like their eukaryotic counterparts, are capable of constructing lipid-based organelles that carry out essential biochemical functions. The magnetosomes of magnetotactic bacteria are one such compartment that is quickly becoming a model for exploring the process of organelle biogenesis in bacteria. Magnetosomes consist of a lipid-bilayer compartment that houses a magnetic crystal. By arranging magnetosomes into chains within the cell, magnetotactic bacteria create an internal compass that is used for navigation along magnetic fields. Over the past decade, a number of studies have elucidated the possible factors involved in the formation of the magnetosome membrane and biomineralization of magnetic minerals. Here, we highlight some of these recent advances with a particular focus on the cell biology of magnetosome formation.
Introduction
In 1884, Karl August Mobius first coined the term ‘organula’ to describe the reproductive structures of protists [1]. In the subsequent centuries, the distinction of organelle has expanded to include subcellular structures ranging from membrane-bounded compartments to molecular machines comprised of harmonious assemblages of proteins and other macromolecules. Such intra-cellular organization is primarily thought to be unique to the eukaryotic lineage, but research over the past two decades has identified complex subcellular compartments and cytoskeletal elements in bacteria [2,3]. The presence of these features in bacteria raises the question of whether principles governing eukaryotic organelle formation, including membrane shaping and protein targeting, also hold for bacterial organelles. If not, an understanding of bacterial compartments promises to reveal a number of unique cell biological mechanisms.
Magnetotactic bacteria (MTB), a phylogenetically diverse cohort of microorganisms characterized by their ability to orient in magnetic fields, provide one of the clearest examples of cytoplasmic compartmentalization in bacteria in the form of organelles called magnetosomes. Electron cryotomography (ECT) and other electron microscopy studies have shown that magnetosomes are lipid-bounded and derived from the inner cell membrane [4,5••] (Figure 1). In addition, magnetosomes have a specific protein content that allows for biomineralization of the crystalline magnetic minerals magnetite (Fe3O4) and/or greigite (Fe3S4). Finally, individual magnetosomes are aligned into chains by dynamic cytoskeletal filaments, whose ancestry and activities are reminiscent of eukaryotic actin systems [6]. These characteristics have made magnetosomes a prime target for understanding the cell biology of bacterial organelles.
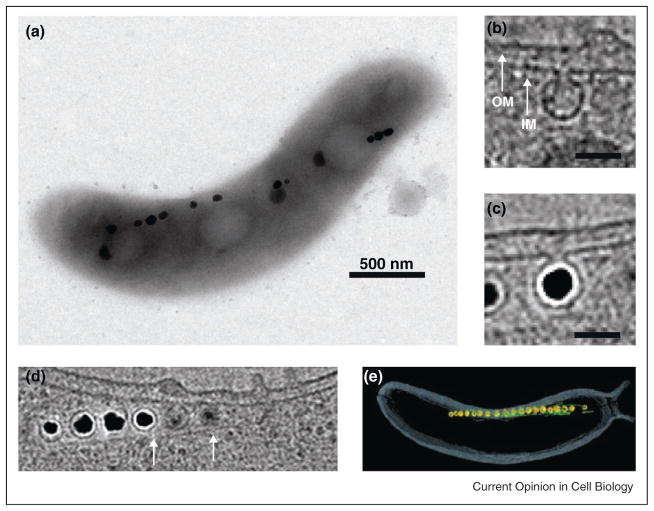
Figure 1.
Cell biological features of magnetotactic bacteria. (A) Transmission electron micrograph (TEM) of Magnetospirillum magneticum AMB-1 reveals a linear chain of electron-dense magnetite crystals [44]. (B) Single section of an electron cryotomographic (ECT) image of AMB-1 shows that magnetosome membranes invaginate from the inner membrane before biomineralization [5••]. (C) Inner membrane invaginations remain even when filled with a mature magnetite crystal [5••]. (D) ECT images also reveal cytoskeletal filaments flanking the magnetosome chain [5••]. (E) Magnetosome membranes (yellow), magnetite crystals (orange) and filaments (green) are highlighted in a 3D reconstruction of AMB-1 from an ECT image [5••].
Biochemical analyses of magnetosome membrane protein content, genetic screens and comparative genomic studies of various MTB led to the identification of a conserved suite of magnetosome-associated genes organized in a large genomic island called the Magnetosome Island (MAI) [7–11]. A subset of these genes, encoded by the mamAB gene cluster, has been identified as not only essential for magnetosome formation but also sufficient for this process [12••,13••]. Individual deletions of genes within this cluster produce strains arrested at various stages of magnetosome biogenesis and hint at a stepwise assembly of the organelle where membrane formation, protein sorting, biomineralization, and chain formation are distinct processes [12••] (Figure 2). In this review we highlight some of the recent developments in understanding of the molecular pathways that govern the formation, organization and intracellular dynamics of magnetosomes.
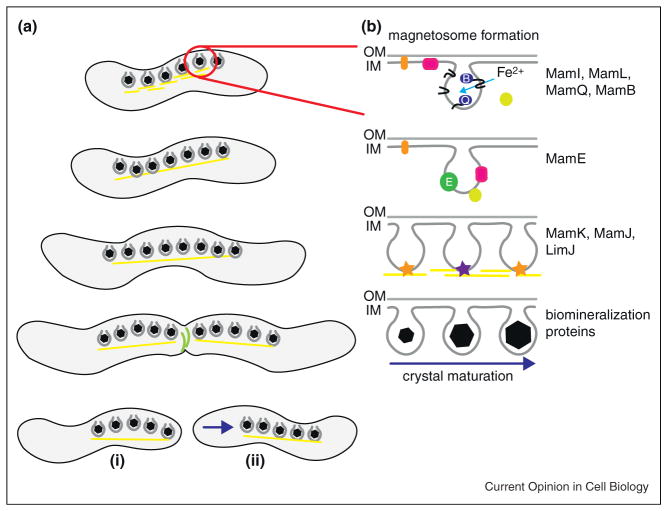
Figure 2.
Magnetosome formation during cell cycle progression. (A) Magnetotactic bacteria increase the number of magnetosomes per cell throughout growth, with the chains centrally located. As the septum forms at the midcell, facilitated by constricting FtsZ rings (green), cytoskeletal filaments flanking the magnetosome chains (yellow) must be separated or stimulated to disassemble. Following cell division, polarly localized magnetosome chains (i) quickly relocalize to the new midcell (ii). (B) The formation of an individual magnetosome is a step-wise process. Magnetosome membrane invagination from the inner cell membrane occurs via the combined actions of MamI, MamL, MamQ, and MamB and other factors. The serine protease MamE is required to properly localize other magnetosome proteins to the compartment. MamK, comprising the cytoskeletal filaments, functions with MamJ and LimJ to coordinate chain organization of the magnetosomes. Protease and putative heme-binding activities of MamE are required for magnetite crystal maturation, with other factors participating in the regulation of crystal number, size, and shape.
Magnetosome membrane formation
A key step in the formation of a functional magnetosome chain is the invagination and shaping of the inner cell membrane. Among eukaryotes, key protein domains have been implicated in generating extensive membrane curvature [14,15]. Within MTB, however, homologs of these factors are not apparent. Thus far, four genes have been identified for their essential roles in magnetosome membrane formation; deletion of any four of these genes individuals results in AMB-1 cells completely devoid of inner membrane decoration [12••]. Two genes, mamI and mamL, encode small (~7–8 kDa), inner membrane proteins of no homology to any proteins beyond the magnetotactic bacteria and their role in magnetosome formation is unclear [12••]. It has been hypothesized that the C-terminal tail of MamL, which is rich in positively charged amino acids, could potentially interact with or insert into the phosphate backbone of the inner membrane to induce local curvature [6]. The magnetosome membrane protein MamQ is homologous to the LemA family of proteins, although no function is known for this group. Interestingly, it bears a potential resemblance to BAR domain proteins, known to be involved in bending membranes in eukaryotic cells [14]. However, this similarity may be due to the presence of coiled-coil domains in MamQ and is not indicative of a specific function in altering membrane architecture.
Only MamB has homology to a family of proteins with a known function. MamB belongs to the cation diffusion facilitator (CDF) superfamily, which is known to transport divalent cation metals and includes a ferrous iron transport system [16]. It has recently been shown that MamB interacts with a number of other magnetosome proteins [17••]. For example, MamB was shown to self-interact via its C-terminal domain. Additionally, it also appears to interact with, and in turn be stabilized by, a second CDF protein MamM [17••]. Intriguingly, MamB potentially interacts with the PDZ1 domain of MamE, a protein involved in the localization of various proteins to the magnetosome. Such an interaction could link the putative transporter to the network of magnetosome proteins managed by MamE [17••]. Thus, MamB may not be directly involved in magnetosome membrane biogenesis and may instead stabilize the membrane by acting as a hub for organization of other magnetosome proteins. Once a magnetosome compartment has been formed MamB might then act as a transporter of iron or other cations into the magnetosome.
Experiments thus far have been unable to establish sufficiency, however, for the four magnetosome membrane proteins MamI, MamL, MamQ, and MamB in establishing structures reminiscent of magnetosome membranes in vivo [12••]. Because no other individual gene deletions yield an absence of magnetosome membranes, additional functionally redundant factors are required. An additional player in shaping the magnetosome membrane may be MamY, an MAI-encoded protein exhibiting weak homology to BAR domain proteins. MamY is capable of inducing liposome tabulation in vitro and has been implicated in maintaining the size of cell membrane invaginations in vivo [18].
Protein localization to the magnetosome
Once the magnetosome membrane invagination has been established, biomineralization can occur if environmental conditions are met. A number of proteins have been implicated in this process and several have been identified as integral or peripheral to the magnetosome membrane itself [8,10,12,19,20••,21,22]. How these proteins localize to the membrane is not well understood, and no evidence exists for a targeting or signal sequence that would uniquely direct proteins to the invagination. The most substantiated mechanism of protein localization to the magnetosome membrane appears to be mediated by protein–protein interactions. Interactions, described above between MamB, MamM, and MamE, have been suggested through in vitro work [17••]. Additionally, MamJ, MamA and Mms6 have all been implicated in the recruitment or stability of subsets of other magnetosome proteins [23•,24,25•,26•]. Genetic analyses have shown that MamE, a DegP/HtrA serine protease with putative heme-binding motifs, is required for the localization of a number of proteins to the magnetosome. In its absence, empty magnetosome membranes are formed but a number of proteins such as MamJ, MamI and MamC are mislocalized within the cell [20,27•]. Interestingly, MamE appears to be a dual function protein. When its putative protease or heme-binding residues are mutated MamE is still capable of localizing proteins to the magnetosome [20••]. However, these mutants have a defect in biomineralization and cannot form mature magnetite crystals [20••].
These results support a model where protein sorting depends on a specific network of protein–protein interactions that produce a functional magnetosome. An alternative possibility is that an affinity for established membrane curvature could drive a subset of proteins to the magnetosome; protein affinities for both positive and negative membrane curvature in bacteria have already been demonstrated [28–30]. One could envision that both mechanisms could be at play in localizing magnetosome proteins to the invagination; negative curvature is highly accentuated at the neck of the magnetosome whereas the positive curvature of the magnetosome body could attract an alternate set of proteins.
Magnetosome chain formation
To maximize their magnetic response, MTB align their magnetosomes into one or more chains along the long axis of the cell. Through ECT imaging of two magnetospirilla species, filaments have been observed running parallel to the magnetosome chain. These filaments are proposed to be comprised of MamK, a bacterial actin-like protein encoded by the MAI of all sequenced MTB [5,31,32]. mamK deletions exhibit disorganized magnetosome chains, ectopic chain placement near cell poles, magnetosome clustering, and most tellingly – the absence of filaments near magnetosomes [5••,33•]. In vitro polymerization of MamK into bundles of long filaments further supports the hypothesis of MamK as the building block of the magnetosome cytoskeleton [31].
Recent evidence using fluorescence recovery after photo-bleaching (FRAP) indicates that MamK filaments, like most actin homologs, are dynamic within the cell [34••]. Bleached segments of MamK-GFP filaments were seen to recover fluorescence in a manner dependent on the putative ATPase activity of the protein [34••]. This pattern of recovery is often related to the exchange of unbleached monomers as a result of depolymerization and repolymerization events. However, recent evidence with other families of bacterial actins has shown that entire filaments are motile within the cell raising the possibility that MamK dynamics in the FRAP experiment may also be influenced by such movements [35–37].
MamK dynamics, however, are not an intrinsic property of the protein and are regulated by additional factors as MamK-GFP expressed in a MAI deletion strain fails to recover in FRAP experiments [34••]. A candidate MamK regulator is MamJ, a protein with an acidic repeat domain that is encoded by the mamAB gene cluster. When mamJ is deleted in Magnetospirillum gryphiswaldense MSR-1, magnetosomes cluster in clumps within the cell [38•]. Additionally, in some but not all cases, MamJ and MamK appear to interact in a bacterial two-hybrid assay [23]. In Magnetospirillum magneticum AMB-1, MamJ and its paralog LimJ, are necessary for both chain organization and MamK dynamics in a redundant manner. In the absence of these two regulators, large gaps are apparent within the magnetosome chain and bundles of filaments, presumably composed of MamK, can be seen within these empty spaces. However, additional unknown factors encoded by the MAI are also necessary to regulate the dynamics of MamK in vivo, as expression of either MamJ or LimJ in a ΔMAI strain is unable to rescue MamK filament dynamics [34].
Despite these advances in understanding the in vivo and in vitro properties of MamK, its specific function and the manner by which it contributes to chain organization are still unclear. MamK might act as a guide to establish the magnetosome chain by moving new magnetosomes into a preexisting chain. Such a model has been suggested for MSR-1, in which both MamK filaments and magnetic interactions between adjacent magnetosomes seem to be required for chain organization [38•,39•]. Alternatively, MamK may act to maintain the chain after it has already been formed. Finally, as discussed below, MamK may act during cell division to ensure the proper segregation of the magnetosome chain.
Cell cycle and magnetosome formation
Once the cell has formed its magnetosome membranes, properly sorted proteins to the compartment to promote biomineralization, and aligned the magnetosomes in a chain, it faces the additional challenge of cell division. In MTB, initial EM studies suggested that the magnetosome chain is divided evenly between the two daughter cells [40,41•]. For the population to maintain its magnetic properties throughout multiple rounds of growth, each daughter cell must synthesize and incorporate new magnetosomes into the existing chain.
To determine how this process occurs requires investigation of both the timing of magnetosome formation and the mechanisms involved in magnetosome maturation. The time needed for a magnetosome to invaginate from the inner membrane and form a 50 nm wide spherical compartment is currently unknown. Related to this issue is the outstanding question of the time frame during the division cycle in which new magnetosomes are formed and incorporated into the existing chain. Inner membrane invaginations could simply be synthesized continually throughout growth or there could be discrete portions of the cell cycle in which MTB are primed for magnetosome membrane synthesis.
The actual cell division event in MTB requires not only successful division plane formation in between two segregated chromosomes but also bisection of the magnetosome chain (Figure 2). Intriguingly, in MSR-1 cell division appears to proceed asymmetrically from one lateral edge of the cell, which may provide the force necessary to segregate the magnetosome chain [42••]. Preferential localization of the chain spanning the future division site at midcell primes equal distribution of magnetosomes to the daughter cells. In MSR-1, chain halves rapidly relocalize from the poles to the new future division site in a process that is mediated by MamK filaments [42••]. In a broad sense this process is reminiscent of the segregation of the proteinaceous carbon-fixation microcompartments of cyanobacteria: carboxysomes. These organelles are linearly arranged in the cell and their alignment and equitable division to daughter cells is dependent upon with the action of ParA cytoskeletal filaments [43].
The coordination of the development of polar organelles such as flagella, stalks, and pili with the progression of the cell cycle has been extensively investigated in Alphaproteobacteria related to the magnetospirilla. These processes are controlled through the CtrA regulatory network, and it was hypothesized that the biogenesis of magnetosomes in the context of the cell cycle could be similarly regulated by components of this pathway, most of which are conserved in MTB. While CtrA and other members of its pathway are essential for viability and cell cycle progression in some Alphaproteobacteria, they were recently shown to be dispensable in AMB-1 [44]. Mutants lacking ctrA had no discernible cell cycle defects and produced functional magnetosome chains [44]. Thus, the existence and identity of elements regulating the cell cycle in MTB and coordinating the formation of magnetosomes in its context remain elusive.
Conclusion
Over the past decade significant progress has been made in the discovery of magnetosome genes and the elucidation of a basic pathway for the assembly of this bacterial organelle. The next challenge is to understand the mechanisms by which these factors act and to define the coordination of these processes in the context of the cell cycle. Further investigations of bacterial organelles using magnetosomes and other compartments as model systems will continue to illuminate similarities and differences in how bacterial and eukaryotic cells solve the common problems associated with organelle biogenesis. As is the case for biological tasks across the entire tree of life, some solutions share an ancient derivation, whereas others may prove to have arisen independently and convergently. Regardless of their evolutionary histories, the mechanisms MTB have evolved to generate and organize intracellular compartments are proving to be elegant, intricate, and intriguing.
Acknowledgments
Arash Komeili is supported by the NIH (NIGMS R01GM084122) and by a David and Lucille Packard Foundation Fellowship in Science and Engineering. Shannon Greene is supported by the NIH Genetics Training Grant GM07127.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Mobius KA. Das Sterben der einzelligen und der vielzelligen Tiere. Vergleichend betrachtet. Biologisches Centralblatt. 1884;4:389–392. [Google Scholar]
- 2.Murat D, Byrne M, Komeili A. Cell biology of prokaryotic organelles. Cold Spring Harbor Perspect Biol. 2010:2. doi: 10.1101/cshperspect.a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vats P, Yu J, Rothfield L. The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci. 2009;66:3353–3362. doi: 10.1007/s00018-009-0092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorby YA, Beveridge TJ, Blakemore RP. Characterization of the bacterial magnetosome membrane. J Bacteriol. 1988;170:834–841. doi: 10.1128/jb.170.2.834-841.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. In this study electron cryotomography was used to show that magnetosomes are permanent invaginations of the inner membrane that are flanked by cytoskeletal filaments. These filaments were probably composed of the bacterial actin-like protein, MamK, which plays a central role in the organization of the magnetosome chain. [DOI] [PubMed] [Google Scholar]
- 6.Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2011;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünberg K, Muller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünberg K, Wawer C, Tebo BM, Schuler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics. 2006;6:5234–5247. doi: 10.1002/pmic.200500887. [DOI] [PubMed] [Google Scholar]
- 11.Richter M, Kube M, Bazylinski DA, Lombardot T, Glockner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the stepwise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA. 2010;107:5593–5598. doi: 10.1073/pnas.0914439107. A genetic dissection of the Magnetosome Island identified genes involved in the step-wise formation of magnetosomes, including magnetosome membrane formation, protein sorting, and biomineralization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Lohbe A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schuler D. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS ONE. 2011;6:e25561. doi: 10.1371/journal.pone.0025561. Deletions of large regions of the Magnetosome Island suggests that the mamAB gene cluster is sufficient for magnetosome formation. Other gene clusters such as the mamXY were identified as integral for biomineralization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 15.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 16.Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch Microbiol. 2005;183:9–18. doi: 10.1007/s00203-004-0739-4. [DOI] [PubMed] [Google Scholar]
- 17••.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Pósfai M, et al. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol. 2011;82:818–835. doi: 10.1111/j.1365-2958.2011.07863.x. In this study, the cation diffusion superfamily proteins MamB and MamM were shown to be essential for magnetosome membrane formation and biomineralization, respectively. Furthermore, these proteins were shown to interact with one another and additional magnetosome proteins such as MamE. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Arakaki A, Matsunaga T. Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Mol Microbiol. 2010;76:480–488. doi: 10.1111/j.1365-2958.2010.07117.x. [DOI] [PubMed] [Google Scholar]
- 19.Scheffel A, Gardes A, Grunberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Quinlan A, Murat D, Vali H, Komeili A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol. 2011;80:1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x. The serine protease MamE was shown to be a bifunctional protein, with a role in biomineralization dependent on both protease activity and putative heme-binding, and a secondary role in protein sorting to the magnetosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J Biol Chem. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- 22.Amemiya Y, Arakaki A, Staniland SS, Tanaka T, Matsunaga T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials. 2007;28:5381–5389. doi: 10.1016/j.biomaterials.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 23•.Scheffel A, Schüler D. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J Bacteriol. 2007;189:6437–6446. doi: 10.1128/JB.00421-07. Surprisingly, the distinctive acidic repeat domain of MamJ is not required for magnetosome chain assembly. In this study, the authors demonstrate ex vivo interactions between MamJ and MamK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto D, Taoka A, Uchihashi T, Sasaki H, Watanabe H, Ando T, Fukumori Y. Visualization and structural analysis of the bacterial magnetic organelle magnetosome using atomic force microscopy. Proc Natl Acad Sci USA. 2010;107:9382–9387. doi: 10.1073/pnas.1001870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Zeytuni N, Ozyamak E, Ben-Harush K, Davidov G, Levin M, Gat Y, Moyal T, Brik A, Komeili A, Zarivach R. Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc Natl Acad Sci USA. 2011;108:E480–E487. doi: 10.1073/pnas.1103367108. The TPR-protein MamA forms oligomeric globular complexes in vitro and structural analysis predicts potential protein interactions sites, which could contribute to the formation of a multi-protein complex shell at the magnetosome membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Tanaka M, Mazuyama E, Arakaki A, Matsunaga T. Mms6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J Biol Chem. 2011;286:6386–6392. doi: 10.1074/jbc.M110.183434. A deletion of mms6 results in smaller and elongated crystals in M. magneticum AMB-1, and also leads to altered protein profiles at the magnetosome, including decreases of Mms5, Mms7 and Mms13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Yang W, Li R, Peng T, Zhang Y, Jiang W, Li Y, Li J. mamO and mamE genes are essential for magnetosome crystal biomineralization in Magnetospirillum gryphiswaldense MSR-1. Res Microbiol. 2010;161:701–705. doi: 10.1016/j.resmic.2010.07.002. MamE and MamO were identified as essential for magnetite crystal formation in M. gryphiswaldense through a transposon mutagenesis study. [DOI] [PubMed] [Google Scholar]
- 28.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 29.Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taoka A, Asada R, Wu L-F, Fukumori Y. Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J Bacteriol. 2007;189:8737–8740. doi: 10.1128/JB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rioux J-B, Philippe N, Pereira S, Pignol D, Wu L-F, Ginet N. A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic Magnetosome Island. PLoS ONE. 2010;5:e9151. doi: 10.1371/journal.pone.0009151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol. 2010;77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. A deletion of mamK in M. gryphiswaldense MSR-1 leads to magnetosome clumping and ectopic magnetosome chain placement. However, MamK in MSR-1 is not strictly required for chain formation as short chains are still observed. [DOI] [PubMed] [Google Scholar]
- 34••.Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, Komeili A. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82:342–354. doi: 10.1111/j.1365-2958.2011.07815.x. Using fluorescence recovery after photobleaching (FRAP), the authors demonstrate that MamK, a bacterial actin, forms filaments in vivo and that these filaments exhibit dynamic behavior dependent on ATP hydrolysis and the redundant actions of MamJ and its paralog LimJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 37.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. Using electron cryotomography, the authors demonstrate that the acidic repeat-containing protein MamJ is essential for magnetosome chain formation; its deletion exhibits severe clustering of magnetosomes with empty vesicles scattered throughout the cytoplasm. [DOI] [PubMed] [Google Scholar]
- 39•.Klumpp S, Faivre D. Interplay of magnetic interactions and active movements in the formation of magnetosome chains. PLoS ONE. 2012;7:e33562. doi: 10.1371/journal.pone.0033562. Computational simulations of magnetic interactions and modeled active transport mechanisms, involving cytoskeletal filaments of MamK, predict that both features are necessary for the formation and stability of magnetosome chains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Posfai M, Kasama T, Dunin-Borkowski R. Characterization of bacterial magnetic nanostructures using high-resolution transmission electron microscopy and off-axis electron holography. Microbiol Monogr. 2006:198–225. [Google Scholar]
- 41•.Staniland SS, Moisescu C, Benning LG. Cell division in magnetotactic bacteria splits magnetosome chain in half. J Basic Microbiol. 2010;50:392–396. doi: 10.1002/jobm.200900408. Magnetosome chains are divided symmetrically at the midcell upon cell division, and quantitative analysis of magnetosome numbers versus cell length suggests against a burst of magnetosome synthesis just before septation. [DOI] [PubMed] [Google Scholar]
- 42••.Katzmann E, Müller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schüler D. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol. 2011;82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. The authors show that MamK filaments probably mediate the recruitment of magnetosome chains to the midcell before cell division. Using cryoelectron tomopgraphy, they also demonstrate that septation in M. gryphiswaldense MSR-1 proceeds asymmetrically from one lateral edge of the cell. [DOI] [PubMed] [Google Scholar]
- 43.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 44.Greene SE, Brilli M, Biondi E, Komeili A. Analysis of the CtrA pathway in magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol. 2012;194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]