Abstract
Lithoautotrophic nitrite-oxidizing bacterial populations from moving-bed biofilters of brackish recirculation aquaculture systems (RAS; shrimp and barramundi) were tested for their metabolic activity and phylogenetic diversity. Samples from the biofilters were labeled with 13C-bicarbonate and supplemented with nitrite at concentrations of 0.3, 3 and 10 mM, and incubated at 17 and 28°C, respectively. The biofilm material was analyzed by fatty acid methyl ester - stable isotope probing (FAME-SIP). High portions of up to 45% of Nitrospira-related labeled lipid markers were found confirming that Nitrospira is the major autotrophic nitrite oxidizer in these brackish systems with high nitrogen loads. Other nitrite-oxidizing bacteria such as Nitrobacter or Nitrotoga were functionally not relevant in the investigated biofilters. Nitrospira-related 16S rRNA gene sequences were obtained from the samples with 10 mM nitrite and analyzed by a cloning approach. Sequence studies revealed four different phylogenetic clusters within the marine sublineage IV of Nitrospira, though most sequences clustered with the type strain of Nitrospira marina and with a strain isolated from a marine RAS. Three lipids dominated the whole fatty acid profiles of nitrite-oxidizing marine and brackish enrichments of Nitrospira sublineage IV organisms. The membranes included two marker lipids (16∶1 cis7 and 16∶1 cis11) combined with the non-specific acid 16∶0 as major compounds and confirmed these marker lipids as characteristic for sublineage IV species. The predominant labeling of these characteristic fatty acids and the phylogenetic sequence analyses of the marine Nitrospira sublineage IV identified organisms of this sublineage as main autotrophic nitrite-oxidizers in the investigated brackish biofilter systems.
Introduction
In biofilters of recirculation aquaculture systems (RAS), nitrite oxidation by lithoautotrophic bacteria (NOB) is the most important process to prevent the cultivated organisms from intoxication with nitrite [1]. Nitrite is formed by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) during the first process of aerobic nitrification. Together with heterotrophic bacteria, the nitrifying communities form biofilms on carrier elements in biofilter systems [2]–[3]. Although recirculation aquaculture systems are increasing worldwide [4], we still lack a detailed understanding of the process of bacterial nitrification, and especially nitrite oxidation, in marine RAS [5].
The guild of NOB is phylogenetically heterogeneous and comprises bacteria of at least five different genera: Nitrobacter, Nitrospina, Nitrococcus, Nitrospira and the recently discovered Nitrotoga. Co-existence of two [6]–[10], or three [11]–[12] genera of NOB in the same habitat has previously been reported. NOB grow slowly due to the low energy yield during oxidation of nitrite [13]. Thus the successful cultivation of NOB is a challenging and time consuming task. Consequently, NOB cultures have only rarely been established in the laboratory [14].
Hovanec et al. [15], and Juretschko et al. [16], were first to identify Nitrospira as the dominant NOB in freshwater aquaria and wastewater treatment plants. Later this genus was also detected in marine aquaculture and aquaria systems [2]–[3], [17]. However, only two studies so far have found Nitrospira as the dominant nitrite oxidizer in marine aquacultural biofilters [5], [18]. Here, we aimed to expand the metabolic activity data from Keuter et al. [5] of marine RAS by analyzes of the metabolic activity of NOB in brackish RAS.
Metabolic activity of nitrite-oxidizing populations can be assessed with DNA-based or chemotaxonomic methods. A DNA-based method of assimilation of labeled substrates performed by Radajewski et al. [19] provided the characterization of the ecology and function of methanol-utilizing organisms from forest soil. The incorporation of labeled substrates followed by gas chromatography and combustion isotope ratio spectrometry (GC-C-IRMS) was first used by Boschker et al. [20] for acetate assimilation of sulphate-reducing bacteria. More recently, quadrupole GC-MS (gas chromatography coupled with mass spectrometry) analyses of labeled lipids with 13C-acetate have been conducted [21] to study soil associated bacteria and fungi. The GC-MS analysis is generally less sensitive compared to GC-C-IRMS, but GC-MS systems are readily available to most laboratories [22]. A method based on quadrupole GC-MS for the labeling with 13C-bicarbonate of autotrophic populations in complex environments was described by Knief et al. [23]. Lipid analyses with fatty acid profiling in combination with stable-isotope probing (FAME-SIP) are very sensitive, making this a suitable technique for the quantification of autotrophic 13C-assimilation in nitrite supplemented biofilter material. For the analyses of the nitrite-oxidizing genera Nitrobacter, Nitrococcus, Nitrospira, Candidatus Nitrotoga and Nitrospina [14] by FAME-SIP, reference data sets for marker lipids are available [10]–[11], [24]–[25]. Especially the genus Nitrospira can be differentiated from the other genera of NOB based on characteristic lipid markers. These NOB are characterized by varying combinations of specific major compounds, which are the palmitoleic acid isomers cis-7 and cis-11 [24]. The different content of the characteristic lipids has been proved to be a good indicator for the presence of Nitrospira populations.
To determine the relevant NOB in two moving-bed biofilters of a brackish (17–21 psu (practical salinity units)) recirculation aquaculture system (shrimp and barramundi), we used FAME-SIP after incubation of the nitrifying community with different nitrite concentrations and temperatures. The chemotaxonomic analyses were supplemented by a cloning approach to get in-depth information about the diversity and the taxonomic positions of Nitrospira populations. Further, the fatty acid profiles of enrichment cultures obtained from the biofilter of the shrimp RAS and from marine sediments of the Laptev Sea were determined in order to expand the marker lipids data set of marine Nitrospira species.
Methods
Samples and enrichments
The samples were collected from two brackish recirculation aquaculture systems (RAS) in Strande (Germany (Marifarm; EAP AquaCulture AG)).
One system comprised shrimp (Penaeus vannamei) tanks with a total volume of 189 m3. The moving-bed biofilter of this system has a volume of 11.3 m3 and was started in early 2007 with approx. 3 m3 black and white high density polyethylene (HDPE) biocarriers (type HX09KL, Stöhr, Marktrodach, Germany) with a total surface area of 836 m2/m3 and a protected surface area of 494 m2/m3. The white biocarriers were made of new HDPE material, and the black were made of recycled HDPE material with the addition of 3% carbon black (Figure S1).
In the second, smaller experimental system, barramundi (Lates calcarifer) was reared in four tanks of a total volume of 10.8 m3. This system was started in 2006. Here, the biofilter comprised of 10 tanks containing 160 l water each, of which two were filled with 96 l black HX09KL (recycled material) and two with white HX09KL (new material) biocarriers.
Both systems ran at 26 to 29°C, the oxygen concentration was between 6.5 and 7 mg/l and pH was kept between 6.8 and 7. Make up water was taken from the Baltic Sea nearby, filtered and ozonized.
For the whole cell fatty acids profiles, enrichment cultures were started with biocarriers from the biofilter of the shrimp system as inoculum. The carriers were separated in white and black ones; cells were removed by shaking vigorously with glass beads (1.7–2 mm) and incubated for several months in marine mineral salt medium [26] with 70% brackish sea water at 28°C. Consumption of nitrite (1 mM) was regularly monitored with the Griess–Ilosvay spot test [27] and supplemented with NaNO2 (2.5 M stock solution) if necessary. In addition, the marine Nitrospira-enrichment culture, “S11”, which originated from marine sediments of the Laptev Sea in the permafrost region of Siberia [28], was incubated in marine mineral salt medium supplemented with 0.3 mM NaNO2 as described above. This culture was grown at 10°C and 17°C, respectively. Whole cell fatty acid profiles were analyzed from biomass from the two enrichments (see below).
For labeling experiments and subsequent cloning of Nitrospira-specific DNA fragments, biocarriers from the shrimp biofilter and from both biofilter components of the barramundi system (white and black biocarriers, respectively) were taken in October 2008. Nitrite and nitrate concentrations in the shrimp aquaculture system were 97 and 300 µM respectively, as determined by ion pair chromatography (Elite LaChrom System, Hitachi, Krefeld, Germany [29]). We measured the potential nitrite oxidation rate following the protocol by Spieck and Lipski [14]. The maximum nitrification potential, calculated as the average of two parallels, was 400 nmol NO2 *h−1 per biocarrier of the shrimp biofilter. In the barramundi system 1000 µM nitrate was measured, while nitrite could not be detected. The potential nitrite oxidation rates at sampling time were 1000 (white) and 1400 (black) nmol NO2 *h−1 per biocarrier.
Lipid analyses
Whole cell fatty acid profiles
Cells were harvested by centrifugation (10,000× g; 20 min) when dense flocs had developed. Fatty acid methyl esters (FAMEs) were prepared according to Sasser [30]. The analyses of the FAMEs by GC-MS were performed as described previously [24].
Labeling experiments
In 500 ml gastight flasks filled with 100 ml marine mineral medium [26] prepared with 70% seawater (Baltic Sea; Fischland/Darß, Germany), 100 biocarriers from the two barramundi biofilters (black [Bb] or white [Bw] biocarriers) and the biofilter of the shrimp system (mixed black and white biocarriers [Sh]) were incubated at temperatures of 17°C and 28°C with 20 mM NaH13CO3 and NaNO2 concentrations of 0.3, 3 and 10 mM, resulting in a total of 18 different incubations. Nitrite consumption was tested with the Griess–Ilosvay spot test [27] twice a week and initial concentrations were readjusted if necessary. The oxygen content of the gas phase above the suspension was measured regularly by subjecting 10 µl of gas phase samples to GC-MS. The fatty acids from the sample biomass were extracted when one of samples from the same biofilter, incubated with the same temperature, contained less than 5% oxygen in the gas phase above the medium. Then all three flasks with the different nitrite concentrations (0.3 mM, 3 mM and 10 mM) were harvested. This resulted in incubation times of 41 up to 55 days (17°C) and 27 up to 41 days (28°C). We obtained biomass by shaking the HDPE carriers with glass beads (2 mm) in a horizontal shaker at 400 rpm overnight. The suspended biomass was concentrated by centrifugation (10,000× g; 20 min).
Lipids were extracted from biomass and converted to fatty acid methyl esters (FAMEs) as described by Knief et al. [23]. The resulting FAMEs were analyzed by GC-MS, and the degree of labeling was quantified using the SIM-mode (single ion monitoring) as described previously [23]. The natural concentration of 13C in biofilm samples was analyzed from samples without added NaH13CO3 and these values were subtracted from those of 13C-labeled samples.
Significance of labeled amounts of fatty acids was calculated based on a one-tailed Student's t-test. The 13C-content (L) was calculated for unlabeled references from fatty acids of original biofilm samples. Based on these data the t-test was used for the calculation of threshold values (T). Labeling was significant with amounts L>T (p<0.1). The threshold value was calculated for the fatty acids of the biofilm samples with percentage of 3.7% (p<0.1). Therefore, all fatty acids of the biofilters samples with degree of labeling ≥4% were deemed to be labeled.
16S rRNA gene analyses
Primer design
Nitrospira-specific primer NS1036R (5′-GCAGCACCTGAGCTCGCT-3′) was designed by using the PROBE-DESIGN Tool from ARB ([31]; version ssu_jan04.corr_opt.arb [http://www.arb-home.de]). Specificity of the primer was checked with the online databases RDP II with the Probe Match function (Ribosomal Database Project; [32]) and NCBI with the BLAST function (Basic local alignment search tool; [33]). Optimization of the amplification conditions of the new primer NS1036R was performed with DNA from cells of Nitrospira marina 295 by gradient PCR. Under these conditions amplicons were also obtained from Nitrospira moscoviensis M-1 and Candidatus Nitrospira defluvii. As negative controls reverence strains from several phyla were analyzed: Pseudomonas spec. FB1, FB19 and FB22 (acc. nos. AM933495.1; AM933511.1; AM933513.1) for Proteobacteria, Subtercola spec. FB10 (acc. no. AM940948.1) for Actinobacteria and Mucilaginibacter spec. FB 14.2 (acc. no. AM933506.1) for Bacteroidetes. The sequences exhibited 5 or 7 mismatches to the primer sequence of NS1036R.
Sequences of the Nitrospira community
DNA was extracted (PowerSoil™ DNA Isolation Kit; MoBio Laboratories, Carlsbad, CA) from the cell material shaken off the biocarriers which had been incubated with 10 mM nitrite. Nitrospira-specific 16S rRNA gene sequence fragments were obtained with the primer NS1036R, in combination with non-specific general bacterial primer Jur8F [16]. The PCR was performed with 30 cycles in two steps, the first step with 10 cycles at 55°C annealing temperature and the second step with 20 cycles at 45°C. Resulting fragments were cloned using the pGEM®-T vector system (Promega, USA). Clone sequences were analyzed with the online databases RDP II with the “seqmatch” function (Ribosomal Database Project; [32]) and NCBI with the “BLAST” function (Basic local alignment search tool; [33]). Furthermore, the sequences were checked for chimera and sequence anomalies using the Pintail program [34]. The sequences were named according to their origin (Bw: barramundi white biocarrier, Bb: barramundi black biocarrier, Sh: shrimp biocarrier) and the incubation temperature (17 or 28°C). Sequences derived from the gas tight flask incubations were labeled with the appendix OS (acc. nos. HE793388 – HE793423).
In addition to the incubation with NaH13CO3 in gas tight flask, a further set of incubations of the biocarrier samples was prepared in Erlenmeyer flasks with normal caps and without supplemented 13C. Biofilm from the carriers was harvested in a horizontal shaker at 400 rpm overnight. DNA was extracted from the 10 mM nitrite samples and the 16S rRNA genes were cloned and sequenced according to Keuter et al. [5]. The “Sh” DNA extracts were amplified with the primer pair Jur8F/1036R [16], and “Bb” and “Bw” DNA extracts were amplified with the primer pair Jur8F/Ntspa1158R [16], [35]. Sequences resulting from this cloning approach are labeled with the appendix HH (acc. nos. JQ900181-JQ900201; JX028301).
Results
Whole cell fatty acids of enrichments
The enrichment cultures inoculated with black and white biocarriers from the shrimp system (Sh) consisted of 19–26% of the fatty acid 16∶1 cis7 and 14–19% of the lipid 16∶1 cis11. The major fatty acid composition of the enrichment culture from the Laptev Sea (S11) was similar (16∶1 cis7: 32%; 16∶1 cis11: 40%–44%). Additionally, all enrichments showed the lipid 16∶0 as major compound with percentages from 14 up to 39% (Table 1). Data of two previously analyzed Nitrospira cultures, strains Nitrospira marina 295 [24] and Ecomares 2.1 [5], which contained high percentages of the lipid 16∶1 cis7 (30–41%) in combination with 16∶1 cis11 (16–31%), are also presented in Table 1.
Table 1. Whole fatty acid profiles of marine enrichments and Nitrospira marina 295 (fatty acids in %).
| Nitrospira marina 295a | Ecomares 2.1b | Sh (black carrier) | Sh (white carrier) | S11 | |||
| Cultivation temperature | 28°C | 17°C | 28°C | 28°C | 28°C | 10°C | 17°C |
| 12∶0 | 0.6 | - | - | - | - | - | |
| 14∶0 | 1.4 | 1.0 | - | 1.9 | 3.3 | 1.0 | 0.8 |
| 15∶0 iso | 0.8 | - | - | 7.3 | 6.1 | - | - |
| 15∶0 anteiso | - | - | - | 2.4 | 4.1 | - | 1.3 |
| 15∶0 | - | - | - | - | 2.4 | - | - |
| 16∶0 iso | - | - | - | 2.6 | - | 0.7 | 1.7 |
| 16∶1 cis7 | 30.4 | 40.7 | 37.1 | 25.8 | 19.0 | 32.4 | 31.8 |
| 16∶1 cis9 | - | - | - | 2.1 | - | 2.4 | 1.0 |
| 16∶1 cis10 | - | 1.9 | 2.8 | - | - | - | 1.1 |
| 16∶1 cis11 | 15.5 | 30.8 | 19.8 | 18.9 | 13.8 | 44.2 | 40.3 |
| 16∶0 | 36.5 | 24.4 | 35.3 | 29.9 | 39.3 | 14.0 | 15.7 |
| 16∶0 11methyl | 0.8 | - | 2.3 | - | - | - | - |
| 17∶0 iso | 0.8 | - | - | 2.3 | - | - | 0.3 |
| 18∶1 cis9 | 1.8 | - | - | 1.8 | - | 0.8 | 0.7 |
| 18∶1 cis11 | 0.6 | - | - | 2.6 | 5.4 | 3.1 | 1.3 |
| 18∶0 | 8.7 | 2.2 | 2.8 | 2.5 | 3.4 | 1.3 | 1.5 |
| 19∶0 cyclo 9–10 | - | - | - | - | 3.1 | - | 0.6 |
Major fatty acids and labeling of the community
The fatty acid profiles of the original moving-bed biofilter samples from the two barramundi biofilters and the shrimp biofilter were similar (for averages see Figure 1). The profiles were dominated by the fatty acids 16∶0, 16∶1 cis9 and 18∶1 cis11. Highest percentages of the lipid 18∶1 cis11 (up to 27%) were obtained from barramundi biofilter material and the highest percentage of the compound 16∶0 (21%) was obtained from shrimp biofilter material.
Figure 1. Whole fatty acid profile of the three original biofilter samples.
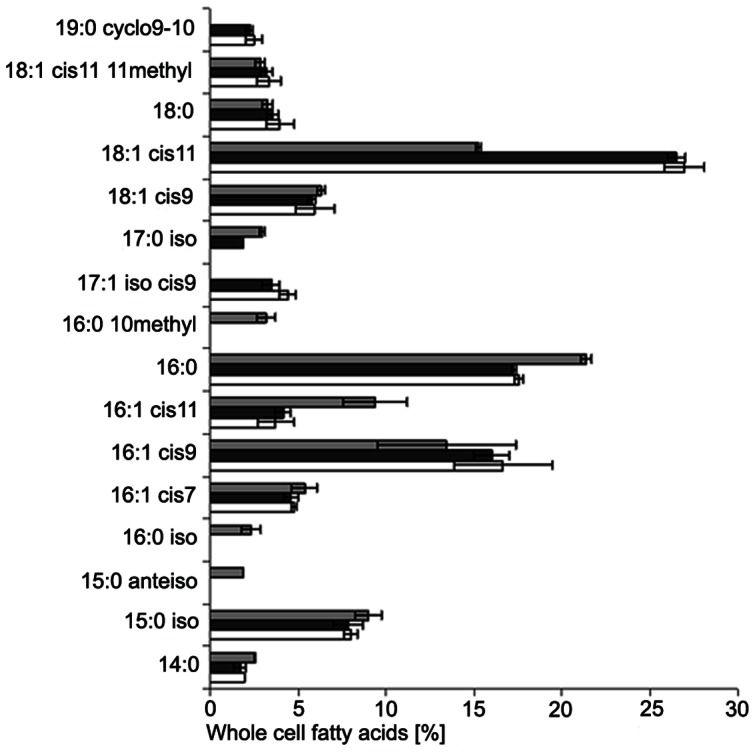
Samples from the moving-bed biofilters were analyzed in parallels (error bars represent the standard deviation of n = 2 samples of each moving-bed system). Data from the barramundi biofilters are shown by white and black bars (white and black biocarriers), and those from the shrimp biofilter by grey bars.
Fatty acid profiles of the biomass in the three biofilter samples (Bb, Bw and Sh) were analyzed for the presence and isotopic content of selected diagnostic lipids after incubation with 13C-bicarbonate using different nitrite concentrations and temperatures. All major fatty acids of the known nitrite-oxidizing taxa were present in all these samples and showed significant portions of labeled fatty acids. The marker lipids of Nitrospira, fatty acid 16∶1 cis7, 16∶1 cis11 and 16∶0, showed highest 13C assimilation rates of up to 45% (p<0.005). In contrast, the dominating lipids of the genera Nitrobacter (18∶1 cis11), Nitrococcus (16∶1 cis9/18∶1 cis11), Nitrospina (14∶0/16∶1 cis9) and the taxon Nitrotoga (16∶1 cis9) showed only moderate incorporation of 13C in the course of the labeling experiments and never exceeded 13% (p<0.1) (Table 2). The degree of labeling of marker lipids for the genus Nitrospira increased with the nitrite concentration, i.e. the highest assimilation of 13C occurred in the samples with 10 mM nitrite at both temperatures (Figure 2). The samples from the barramundi filter with white biocarriers exhibited highest 13C assimilation rates (except for fatty acid 16∶0 and 16∶1 cis7 at 0.3 mM) (Figure 2) ranging between 32–44% for fatty acid 16∶1 cis7, 26–45% for fatty acid 16∶1 cis11 and 17–29% for fatty acid 16∶0.
Table 2. Labeled amounts of major compounds from nitrite-oxidizing bacteria.
| 14∶0 | 16∶1 cis7 | 16∶1 cis9 | 16∶1 cis11 | 18∶1 cis11 | 16∶0 | |
| Labeled amounts (%) a | 6–13 | 30–44 | 6–13 | 19–45 | 6–10 | 11–29 |
| Significance ( p ) | <0.1 | <0.005 | <0.025 | <0.005 | <0.05 | <0.01 |
| Dominating lipids | ||||||
| Nitrospira IV b | ++ c | +(+) | +(+) | |||
| Nitrobacter | ++ | + | ||||
| Nitrococcus | ++ | ++ | ++ | |||
| Nitrospina | ++ | ++ | + | |||
| Nitrotoga | ++ | ++ |
range of the labeled amounts from the 3 moving bed biofilters (incubations with 10 mM nitrite and both used temperatures), see also Figure 2 for detailed information of Nitrospira marker lipids.
marker lipids of Nitrospira sublineage IV, detailed information see Table 3.
+: marker lipid with percentage ≤20%; ++: marker lipids with >20% of whole fatty acids for the respective taxon.
Figure 2. Degree of labeling of major compounds of the genus Nitrospira.
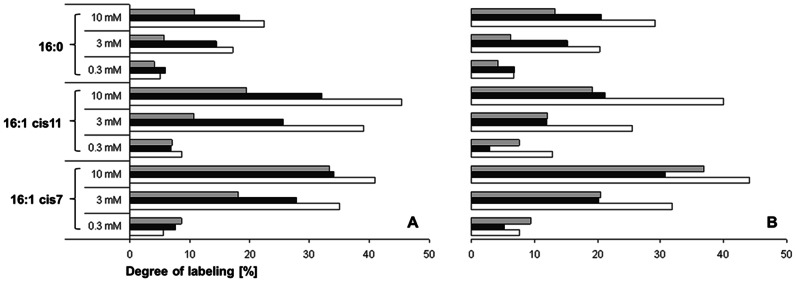
Results from samples of the three moving-bed biofilters incubated at 17°C (A) and 28°C (B). Data from the barramundi biofilters are shown by white and black bars (white and black biocarriers), and those from the shrimp biofilter by grey bars.
Phylogeny of the Nitrospira-community
Phylogenetic analyses were performed on sequences from incubations with 10 mM nitrite in flasks without 13C (appendix HH) and gastight flasks supplemented with 13C-bicarbonat (appendix OS). The consumption rates of supplemented nitrite showed no relevant differences (data not shown) between the two types of incubation.
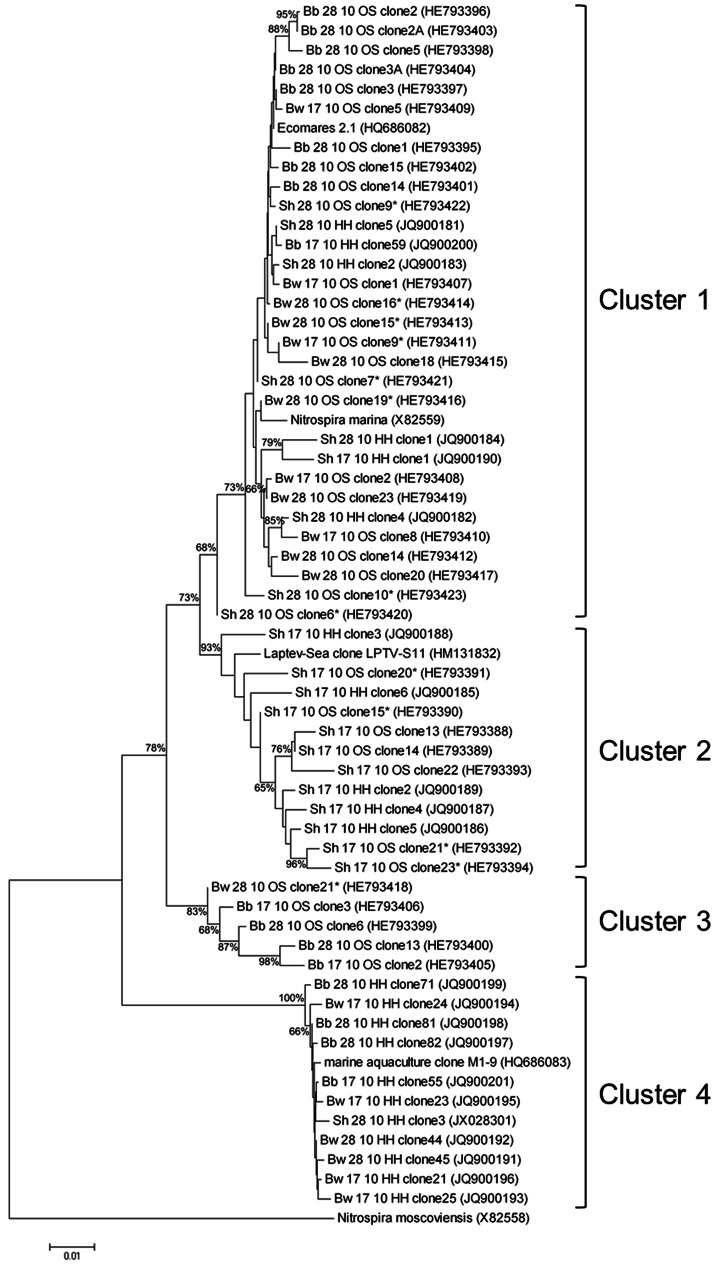
The cloning approach revealed four different clusters within sublineage IV of Nitrospira (Figure 3); cluster 1 comprised sequences of almost all analyzed biofilters affiliated with Nitrospira marina 295 (acc. no. X82559) and Ecomares 2.1, isolated from a marine RAS (acc. no. HQ686082) analyzed previously [5]. Cluster 2 included the sequence of the Laptev Sea sediment clone LPTV S11 (acc. no. HM131832), and comprised only cloned sequences of the Sh17 incubation, i.e. biocarriers from the shrimp biofilter with an incubation temperature of 17°C. Clusters 3 and 4 consisted of sequences from samples incubated in gas tight flasks (OS) or in open flasks (HH), respectively. Cluster 4 included a sequence of the enrichment culture M1–9 from a marine RAS (acc. no. HQ686083; [5]). Clusters 3 and 4 sequences differed from all sequences of cluster 1 and 2 in the nucleotides on position 482–487 (C–ACT in cluster 1 and 2, and T–GCC in cluster 3 and 4), according to the E. coli numbering [36].
Figure 3. Phylogenetic tree based on 16S rRNA gene sequences.
The tree was constructed by neighbor-joining algorithm showing four different clusters (1–4). Nodes ≥65% supported by bootstrap values (based on 1000 iterations). The tree shows sequences of the marine Nitrospira sublineage IV with clones of the brackish RAS biofilters. The sequence of Nitrospira moscoviensis from sublineage II was defined as outgroup. The sequences obtained from analyzed biofilters were named after their origin (Bw: barramundi white biocarrier; Bb: barramundi black biocarrier; Sh: shrimp biocarrier), the incubation temperature (17 or 28°C), as well as the appendix OS (acc. nos. HE793388 – HE793423) and HH (acc. nos. JQ900181-JQ900201; JX028301) for different cloning approaches. Sequences ≤800 bp was marked with “*”. Scale bar = 1% sequence divergence.
Discussion
Metabolically active NOB in the biofilters
Members of the genus Nitrospira often co-exist with other NOB, or even dominate the nitrite oxidation in many engineered ecosystems such as wastewater treatment plants, aquaria and aquaculture biofilters, as indicated by several nucleic acid-based studies [2]–[3], [15]–[18], [37]. Fluorescence in situ hybridization (FISH) with rRNA-targeted probes is well practical for analyses of the microbial structure but is insufficient to determine the metabolic activity of cells. More detailed information on activity of a defined group of bacteria can be obtained by combining FISH with microradiography (MAR-FISH) [38]. However, in contrast to FAME-SIP the target sequences of autotrophic organisms must be known to obtain MAR-FISH signals from metabolic active groups. Because FAME-SIP is independent from available sequence information, it represents an appropriate method to analyze autotrophic communities in environmental samples. The results of the present study extended DNA-based findings for aerobic biofilters in brackish aquaculture biofilters of 17–21 psu salinity. Moreover, the labeling approach with FAME-SIP allows assessment of the metabolic activity of the whole autotrophic, nitrite-oxidizing community in the analyzed brackish aquaculture systems.
The straight chain fatty acids 16∶0, 16∶1 cis9 and 18∶1 cis11 continuously dominated the lipid profile, while other groups of fatty acids, polyunsaturated and iso-/anteiso-branched fatty acids, were detected in lower percentages for all samples. All these lipids occur mainly in heterotrophic cells. The lipid 18∶1 cis9, a compound of different heterotrophic cells, showed no significant assimilation of 13C in the course of the labeling experiments. The degree of labeling never exceeded 3.8% (data not shown). It is assumed that only active autotrophic cells, but not heterotrophic cells, show a considerable incorporation of label under the conditions we used [23].
The high degree of labeled Nitrospira-specific compounds, the cis-7 and cis-11 isomer of hexadecenoic acid, reflects the major role of Nitrospira cells in the biofilters. Marker lipids of the other nitrite-oxidizing autotrophic organisms, Nitrobacter, Nitrospina, Nitrococcus and Candidatus Nitrotoga were only moderately labeled (Table 2). The low degree of labeled Nitrobacter marker fatty acids at 10 mM nitrite contrasts the expectation that this NOB is often found as dominant NOB in habitats with high substrate availability [39]–[41], outcompeting Nitrospira which has high substrate affinity at low nitrite concentration [42]. However, Nitrobacter- and Nitrotoga-like bacteria were detected by specific PCR in samples from the shrimp (both) and the barramundi (only Nitrotoga) filters (data not shown), but no relevant activity could be assigned to these genera by the labeling approach.
In contrast to Nitrobacter (Alphaproteobacteria), Nitrospira-like bacteria of the deep-branching phylum Nitrospirae [43] grow slowly, and seem to be better adapted to low oxygen concentrations [44]. Their high nitrite affinities led to the assumption, that Nitrospira are K-strategists, which thrive at low nitrite concentrations [44]–[45]. The high metabolic activity of Nitrospira populations, especially at a concentration of 10 mM nitrite (Figure 2) is remarkable. In line with our finding, Maixner et al. [35] suggested a broader differentiation of ecotypes within the genus Nitrospira on an imaginary scale reaching from K-strategist to r-strategist. The Nitrospira in the studied brackish RAS hence might have been r-strategists, or/and bear currently unidentified features that are advantageous compared to other NOB.
Nitrobacter has rarely been reported to be a major NOB in marine habitats [8], [46]. Rather the marine genera Nitrococcus or Nitrospina would be theoretically more suitable as putative co-inhabitants or competitors in brackish or marine RAS. Both genera are tolerant to high nitrite concentrations [47] and Nitrospina is thought to be the most abundant nitrite oxidizer in the oceans [48], though sound data on the composition and distribution of NOB in the oceans are still lacking [49]. In contrast to the theory, no specific fatty acids of either of these NOB were labeled during our experiments and there was no hint for their relevance in the analyzed biofilters.
The similar pattern of labeled fatty acids at different nitrite concentrations suggests that only one dominant NOB was active under the different conditions. The amounts of the specific labeled acids of Nitrospira correlated positively with the nitrite concentrations. Both moving-bed-systems ran at 26 to 29°C, but the metabolic activity was not influenced by the incubation temperatures of 17 and 28°C. We detected similar activities at the original temperature of the system and the lower temperature conditions suggesting the autotrophic organisms are very resistant to a decrease in temperature. While the temperature difference of 9°C did not seem to have any influence on the activity of the NOB the type of the biocarriers did affect the activity of the NOB: In almost all incubations, the bacterial community on the white biocarriers of the barramundi biofilter incorporated more 13C-bicarbonate than the black carriers of the second barramundi biofilter, even though both biofilters were supplied by the same effluent water from the barramundi tanks. This is in accordance with results from a range of activity tests on black and white biocarriers from the shrimp biofilter between June 2008 and June 2009; the nitrite-oxidizing bacteria on the white biocarriers were more active than those on the black biocarriers (Figure S2). These results are interesting, since there is no difference in shape, structure or size of the white and black biocarriers. The only difference is that white biocarrier consisted of new HDPE material while the black HDPE biocarriers were made of recycled material pigmented with black carbon. Reasons for that are unclear and would require further studies.
Phylogenetic relationship in sublineage IV
We assessed micro-diversity of Nitrospira-strains in the biofilters of the brackish RAS, by constructing a clone library using semi-specific Nitrospira primer pairs with DNA from the 10 mM nitrite incubations at both temperatures.
We assumed that the bacteria in the biofilters would be halotolerant or halophilic, since the systems were run with water from the Kiel Bight, a part of the Baltic Sea which can reach salinities of up to 30 psu. Our phylogenetic analyses showed that sequences from the analyzed biofilters belong to the marine Nitrospira sublineage IV, and some were highly similar to the few known marine cultures.
So far six sublineages of Nitrospira-strains are known, mostly containing only one described strain, each of which might be habitat specific [25], [37]. For instance, species of sublineage I (Candidatus N. defluvii-lineage), are found in wastewater treatment plants, while species of the sublineage VI (N. calida-lineage) seem to be restricted to hot springs. Sequences of sublineage IV have mainly been derived from marine habitats.
In various engineered ecosystems Nitrospira-related strains, from either different or the same sublineages, co-exist [18], [50]. Some authors associate population shifts or differences by physical or chemical factors like nitrite concentration, oxygen content or temperature [35], [51]–[52]. Brown et al. [53] discovered 16S rRNA gene sequences related to Nitrospira marina-like organisms and also sequences related to Nitrospira moscoviensis-like organisms in biofilters of a shrimp RAS. Quantification by qPCR showed higher abundances of Nitrospira marina-like organisms from sublineage IV than Nitrospira moscoviensis-like organisms from sublineage II. In the current study we found only sequences of Nitrospira from sublineage IV grouped into four different clusters using clone libraries of the barramundi and shrimp biocarriers.
Cluster 1 comprised the largest number of sequences originating from all incubations and included the strains Nitrospira marina and Ecomares 2.1. The latter was isolated from the moving-bed biofilter of a marine RAS in Büsum, Germany (North Sea water) [5]. However, a range of sequences confirmed three further clusters within the marine Nitrospira sublineage. Sequences from the colder incubation of the shrimp biocarriers were found clustering together with culture S11 (cluster 2). The culture S11 was observed to convert moderate nitrite concentrations (34–36 µM/day) irrespectively of the temperature (range between: 10–28°C) [28]. Population shift experiments with varying nitrite concentrations [35] revealed shifts within a few days, which is a relatively short time for nitrifiers. Therefore, the incubation at 17°C for approx. 55 days might have favored growth of bacteria of cluster 2.
Except for the colder incubation of biocarriers from the shrimps filter, cluster 4 comprised sequences from all incubation variants together with the sequence of enrichment culture M1-marine, a Nitrospira coexisting with the Ecomares 2.1 in the marine RAS run with North Sea water [5]. No representative of cluster III has been cultivated so far.
The genotypic information of marine and brackish habitats revealed high similarities of the sequences within the marine Nitrospira sublineage. This indicates that there are indeed different ecotypes in Nitrospira sublineage IV, although no grouping into marine and brackish strains could be observed. The marine sublineage IV is the only sublineage of Nitrospira with several isolated or highly enriched and physiologically studied representatives. Experiments on the strains S11 [28], Ecomares 2.1 [5], and N. marina 295 [26] revealed that members of this sublineage, even very close relatives such as N. marina 295 and Ecomares 2.1, can differ immensely in their substrate tolerances, temperature optima or substrate conversion rates. Such physiological diversity, a prerequisite for niche differentiation, might therefore explain the high phylogenetic micro-diversity. Micro-scale conditions in biofilms of biofilter systems can vary extremely over time and/or space [35], [44], enabling the co-existence of strains with differing physiological preferences for resources.
New data sets of fatty acid profiles in sublineage IV
Nitrospira-typical fatty acids are used as biomarker molecules for the in situ detection of this NOB in natural environments. Moreover, different sublineages can be identified by certain combination of these acids. For instance, two cultures in the same sublineage, N. calida and GaII (sublineage VI), exhibited considerable differences in their fatty acids profile [25].
Beside N. marina and its close relative Ecomares 2.1, the culture S11 from the Laptev Sea is the third culture of the marine Nitrospira-sublineage IV, of which a fatty acids profile has been generated. This profile and profiles of the two enrichment cultures from the shrimp biofilter (black and white biocarriers) were similar to the profiles of the other two Nitrospira strains originating from marine environments. The specific fatty acids of Nitrospira sublineage IV consisted of the two marker lipids 16∶1 cis7 and 16∶0 cis11 in combination with the non-specific fatty acid 16∶0 as major compounds in the membranes (Table 1). This characteristic pattern (Table 3) is distinguishable from the lipid profiles of the sublineages I, II, V and VI [10], [24]–[25]. The lipid pattern of sublineage III is not yet determined due to the lack of enrichment cultures for this group. The marker lipid analyses confirmed the possibility of differentiation of marine Nitrospira strains of sublineage IV from other Nitrospira sublineages using this method.
Table 3. Marker lipids for Nitrospira species from whole fatty acid profiles.
| 16∶1 cis7 | 16∶1 cis11 | 16∶0 | 16∶0 11methyl | ||
| sublineage I | Candidatus Nitrospira defluviia | ++ e | + | ||
| sublineage II | Nitrospira moscoviensis b | ++ | + | ++ | |
| sublineage IV | Nitrospira marina b | ++ | + | ++ | |
| Enrichment Ecomares 2.1c | ++ | ++ | + | ||
| Enrichment S11 | ++ | ++ | + | ||
| Enrichment Sh black | ++ | + | ++ | ||
| Enrichment Sh white | + | + | ++ | ||
| sublineage V | Candidatus Nitrospira bockianab | + | ++ | ++ | |
| sublingeage VI | Nitrospira calida d | + | ++ | ++ | |
| Enrichment Ga IId | ++ | ++ |
Conclusion
The labeling approach with 13C as substrate indicated the presence and the metabolic activity of a Nitrospira-related population as the dominant nitrite-oxidizer in analyzed brackish moving-bed filter systems. The abundance of Nitrospira-like organism was shown under various incubation parameters. The sequencing approach of the 16S rRNA genes revealed four Nitrospira phylotypes, but only the second cluster was restricted to the specific incubation conditions of low temperature. Our study could also confirm that whole fatty acid profiles of currently known Nitrospira sublineage IV organisms from marine habitats always consist of the two marker lipids 16∶1 cis7 and 16∶1 cis11 combined with the non-specific acid 16∶0 as major compounds in the membranes.
Supporting Information
New high density polyethylene biocarries. Biocarriers of the type HX09KL (Stöhr, Marktrodach, Germany) made of new material (white) and of recycled material (black color, due to the addition of 3% carbon black). Scale of the ruler in centimeter (cm).
(TIF)
Nitrite oxidizing potentials of black and white biocarriers from the shrimp biofilter. 10 biocarriers were shaken in 50 ml mineral medium spiked with 1 mM nitrite. Bars right axis: nitrite-oxidizing potentials (in nmol substrate per hour) of NOB on 1 recycled (stripes) or new (dots) HDPE biocarrier. Left axis; nitrate concentrations (black line) of the biofilter water indicating the N load of the system over the sampling period of one year.
(TIF)
Acknowledgments
We thank Wiebke Schütt for proofreading the manuscript.
Funding Statement
This research was funded by the German Research Foundation (DFG; www.dfg.de (Project: LI 1624/1-1 and LI 1624/1-2)) and by the Deutsche Bundesstiftung Umwelt (DBU; www.dbu.de (Project: AZ 23821/01-03)). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lewis WM, Morris DP (1986) Toxicity of nitrite to fish: a review. Trans Am Fish Soc 115: 183–195. [Google Scholar]
- 2. Itoi S, Ebihara N, Washio S, Sugita H (2007) Nitrite oxidizing bacteria, Nitrospira, distribution in the outer layer of the biofilm from filter materials of a recirculating water system for the goldfish Carassius auratus . Aquaculture 264: 297–308. [Google Scholar]
- 3. Sugita H, Nakamura H, Shimada T (2005) Microbial communities associated with filter materials in recalculating aquaculture systems of freshwater fish. Aquaculture 243: 403–409. [Google Scholar]
- 4. Losordo TM, Hobbs AO (2000) Using computer spreadsheets for water flow and biofilter sizing in recirculating aquaculture production systems. Aquac Eng 23: 95–102. [Google Scholar]
- 5. Keuter S, Kruse M, Lipski A, Spieck E (2011) Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ Microbiol 13 (9) 2536–2547. [DOI] [PubMed] [Google Scholar]
- 6. Attard E, Poly F, Commeaux C, Laurent F, Terada A, et al. (2010) Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol 12: 315–326. [DOI] [PubMed] [Google Scholar]
- 7. Cébron A, Garnier J (2005) Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: Detection, quantification and growth along the lower Seine River (France). Water Res 39: 4979–4992. [DOI] [PubMed] [Google Scholar]
- 8. Kumar VJ, Joseph V, Philip R, Bright Singh IS (2010) Nitrification in brackish water recirculating aquaculture system integrated with activated packed bed bioreactor. Water Sci Technol 61: 797–805. [DOI] [PubMed] [Google Scholar]
- 9. Schramm A, de Beer D, Gieseke A, Amann R (2000) Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ Microbiol 2: 680–686. [DOI] [PubMed] [Google Scholar]
- 10. Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, et al. (2006) Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol 8: 405–415. [DOI] [PubMed] [Google Scholar]
- 11. Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E (2007) Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1: 256–264. [DOI] [PubMed] [Google Scholar]
- 12. Xiao Y, Zeng GM, Yang ZH, Liu YS, Ma YH, et al. (2009) Coexistence of nitrifiers, denitrifiers and Anammox bacteria in a sequencing batch biofilm reactor as revealed by PCR-DGGE. J Appl Microbiol 106: 496–505. [DOI] [PubMed] [Google Scholar]
- 13. Thauer RK, Jungermann K, Decker K (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41: 100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spieck E, Lipski A (2011) Cultivation, growth physiology and chemotaxonomy of nitrite-oxidizing bacteria. In: Klotz MG, editor. Methods of Enzymology. Vol. 486, Part A: Research on nitrification and related processes. Oxford: Academic Press/Elsevier. pp. 109–130. [DOI] [PubMed]
- 15. Hovanec TA, Taylor LT, Blakis A, de Long EF (1998) Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol 64: 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Röser A, et al. (1998) Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64: 3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tal Y, Watts JEM, Schreier SB, Sowers KR, Schreier HJ (2003) Characterization of the microbial community and nitrogen transformation processes associated with moving bed bioreactors in a closed recirculated mariculture system. Aquaculture 215: 187–202. [Google Scholar]
- 18. Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, et al. (2007) Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63: 192–204. [DOI] [PubMed] [Google Scholar]
- 19. Radajewski S, Ineson P, Parekh NR, Murell JC (2000) Stable-isotope probing as a tool in microbial ecology. Nature 403: 646–649. [DOI] [PubMed] [Google Scholar]
- 20. Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, et al. (1998) Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392: 801–805. [Google Scholar]
- 21. Arao T (1999) In situ detection of changes in soil bacterial and fungal activities by measuring 13C incorporation into soil phospholipid fatty acids from 13C acetate. Soil Biol Biochem 31: 1015–1020. [Google Scholar]
- 22.Lipski A (2006) Detection of autotrophic sulphur- and iron-oxidizing bacteria using labelled fatty acid methyl esters (FAMEs). In: Cooper JE, Rao JR, editors. Molecular approaches to soil, rhizosphere and plant microorganism analysis. Oxfordshire: CAB International. pp. 132–145.
- 23. Knief C, Altendorf K, Lipski A (2003) Linking autotrophic activity in environmental samples with specific bacterial taxa by detection of 13C-labeled fatty acids. Environ Microbiol 5: 1155–1167. [DOI] [PubMed] [Google Scholar]
- 24. Lipski A, Spieck E, Makolla A, Altendorf K (2001) Fatty acid profiles of nitrite-oxidizing bacteria reflect their phylogenetic heterogeneity. Syst Appl Microbiol 24: 377–384. [DOI] [PubMed] [Google Scholar]
- 25. Lebedeva EV, Off S, Zumbrägel S, Kruse M, Shagzhina A, et al. (2011) Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol Ecol 75: 195–204. [DOI] [PubMed] [Google Scholar]
- 26. Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U (1986) Nitrospira marina gen. Nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol 144: 1–7. [Google Scholar]
- 27.Schmidt EL, Belser LW (1994) Autotrophic Nitrifying Bacteria. In: Weaver RW, Angle JS, Bottomley PJ, editors. Methods of Soil Analysis. Part 2-Microbiological and Biochemical Properties. Madison: Soil Science Society of America. pp.159–177.
- 28.Alawi M (2007) Diversity of nitrite-oxidizing bacteria in soils of the North Siberian permafrost and sediments of the Laptev Sea (Diversität Nitrit oxidierender Bakterien in Böden des nordsibirischen Permafrostes und Sedimenten der Laptev-See). Dissertation. Hamburg, Germany: University of Hamburg.
- 29. Meincke M, Bock E, Kastrau D, Kroneck PMH (1992) Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch Microbiol 158: 127–131. [Google Scholar]
- 30.Sasser M (1990) Identification of bacteria through fatty acids analysis. In: Klement Z, Rudolph K, Sands DC, editors. Methods on Phytobacteriology. Budapest: Akademiai Kiado. pp. 199–204.
- 31. Ludwig W, Strunk O, Westram R, Richter L, Meier, et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, et al. (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 34. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, et al. (2006) Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8: 1487–1495. [DOI] [PubMed] [Google Scholar]
- 36. Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli . Proc Natl Acad Sci 75: 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (2001) In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67: 5273–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagner M, Horn M, Daims H (2003) Fluorescence in situ hybridization for the identification and characterization of prokaryotes. Curr Opin Microbiol 6: 302–309. [DOI] [PubMed] [Google Scholar]
- 39. Degrange V, Bardin R (1995) Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol 6: 2093–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim DJ, Kim SH (2006) Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res 40: 887–894. [DOI] [PubMed] [Google Scholar]
- 41. Wagner M, Rath G, Koops HP, Flood J, Amann R (1996) In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol 34: 237–244. [Google Scholar]
- 42. Nogueira R, Melo LF (2006) Competition between Nitrospira spp. and Nitrobacter spp. in nitrite-oxidizing bioreactors. Biotechnol Bioeng 95: 169–175. [DOI] [PubMed] [Google Scholar]
- 43. Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E (1995) A new obligately chemolithoautotrophic, nitrite oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol 164: 16–23. [DOI] [PubMed] [Google Scholar]
- 44. Schramm A, de Beer D, Wagner M, Amann R (1998) Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol 64: 3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blackburne R, Vadivelu VM, Yuan Z, Keller J (2007) Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter . Water Res 41: 3033–3042. [DOI] [PubMed] [Google Scholar]
- 46. Ward BB, Carlucci AF (1985) Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol 50: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watson SW, Waterbury JB (1971) Characteristics of two nitrite oxidizing bacteria Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Microbiol 77: 203–230. [Google Scholar]
- 48. Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, et al. (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9: 1162–75. [DOI] [PubMed] [Google Scholar]
- 49.Ward BB (2010) Nitrification in the Ocean. In: Ward BB, Arp DJ, Klotz MG, editors. Nitrification. Washington DC: ASM Press. pp. 325–346.
- 50. Zhu P, Ye Y, Pei F, Lu K (2012) Characterizing the structural diversity of a bacterial community associated with filter materials in recirculating aquaculture systems of Scortum barcoo . Can J Microbiol 58 (3) 303–310. [DOI] [PubMed] [Google Scholar]
- 51. Park HD, Noguera DR (2008) Nitrospira community composition in nitrifying reactors operated with two different dissolved oxygen levels. J Microbiol Biotechnol 18 (8) 1470–1474. [PubMed] [Google Scholar]
- 52. Siripong S, Rittmann BE (2007) Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res 41: 1110–1120. [DOI] [PubMed] [Google Scholar]
- 53. Brown MN, Briones A, Diana J, Raskin L (2012) Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol 83: 17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
New high density polyethylene biocarries. Biocarriers of the type HX09KL (Stöhr, Marktrodach, Germany) made of new material (white) and of recycled material (black color, due to the addition of 3% carbon black). Scale of the ruler in centimeter (cm).
(TIF)
Nitrite oxidizing potentials of black and white biocarriers from the shrimp biofilter. 10 biocarriers were shaken in 50 ml mineral medium spiked with 1 mM nitrite. Bars right axis: nitrite-oxidizing potentials (in nmol substrate per hour) of NOB on 1 recycled (stripes) or new (dots) HDPE biocarrier. Left axis; nitrate concentrations (black line) of the biofilter water indicating the N load of the system over the sampling period of one year.
(TIF)