Abstract
Fc receptor-like 5 (FCRL5) regulates BCR signaling and has been reported to bind aggregated IgG. Using surface plasmon resonance, we analyzed the interaction of native IgG samples with FCRL5, revealing a complex binding mechanism, where isotype is just one factor. FCRL5 bound IgG1 and IgG4 with approximately 1 μM KD, while the interaction with IgG3 was a magnitude weaker. However, IgG2 samples displayed a wide range of affinities, indicating that additional factors affect binding. We used a panel of 19 anti-FCRL5 mAbs with defined reactivity to identify domains involved in ligand binding. Six mAbs blocked IgG binding, indicating critical roles of FCRL5 domains 1 and 3, as well as epitopes at the domain 1/2 and domain 2/3 boundaries. We found that only glycosylated IgG containing both Fab arms and the Fc region bound with high affinity. Furthermore, the presence of sialic acid in the IgG carbohydrate altered FCRL5 binding. The interaction of IgG and FCRL5 consisted of two kinetic components, suggesting a complex binding mechanism. We established that the IgG-Fc and IgG-F(ab’)2 fragments bind FCRL5 independently but with low affinity, revealing the mechanism behind the two-step binding of whole IgG. This complex binding mechanism is distinct from that of Fc-receptors, which bind through the Fc. We propose that FCRL5 is a new type of receptor that recognizes intact IgG, possibly enabling B cells to sense immunoglobulin quality. Recognition of undamaged IgG molecules by FCRL5 could allow B cells to engage recently produced antibodies.
INTRODUCTION
The family of Fc receptor-like (FCRL) proteins was discovered in part by searching for Fc-receptor homologues (1-3). Human FCRL1-6 (CD307a-f) are membrane proteins preferentially expressed on B cells (4), while FCRLA and FCRLB reside in the cytoplasm. FCRL1-6 possess cytoplasmic tails with inhibitory ITIM and/or activating ITAM phosphorylation signaling motifs. The signaling potential of FCRL1-5 was established in model systems using either stimulation with Abs (Ab), or chimeric FCRL cytoplasmic tails fused to FcgRIIB extracellular domains. FCRL1 was found to promote B cell activation, whereas FCRL2-5 were each shown to inhibit B cell antigen receptor signaling (5-9). In particular, chimeric FCRL5 recruited SHP1 to two ITIM motifs upon B cell antigen receptor co-stimulation, resulting in diminished calcium influx and protein tyrosine phosphorylation (7). We showed that co-stimulation of FCRL5 and the B cell antigen receptor promotes proliferation and differentiation of naive B cells (10). FCRL5 is expressed on both mature B cells and plasma cells, and is induced by EBV proteins (11,12). FCRL are implicated in human diseases, including cancer and autoimmune conditions (13,14). We and others reported FCRL5 to be overexpressed on malignant B cells of hairy cell leukemia, chronic lymphocytic leukemia, mantle cell lymphoma and multiple myeloma patients (11,14-16). In addition, serum levels of soluble FCRL5 are elevated in patients with several types of B cell tumors (16). A recent study demonstrated the usefulness of FCLR5 as combination biomarker to predict non-response to anti-CD20 therapy in rheumatoid arthritis (17). FCRL5 is a novel target in the treatment of multiple myeloma (18).
Despite substantial progress suggesting physiological roles for FCRL in B cell biology, the identification of FCRL ligands has been lagging. During the last two years, the first ligand candidates emerged. FCRL6, expressed on cytotoxic T cells and NK cells, binds HLA-DR, a MHC class II molecule related to Igs (19). FCRLA in the endoplasmic reticulum binds IgG, IgM and IgA (20,21). FCRL5 has recently been shown to bind aggregated IgG, while FCRL4 binds IgA (22). Specifically, FCRL5 expressed on HEK293T cells bound heat-aggregated IgG1 and IgG2, and bound IgG3 and IgG4 less efficiently. An FCRL5 fragment containing three N-terminal domains was shown capable of binding IgG1 and an Ab reactive to D1-3 inhibited IgG binding. The discovery that FCRL5 is a specific IgG receptor suggests a role for secreted IgG regulating B cells through FCRL5, analogous to FcgRIIB (23).
We sought to further define the IgG ligands of FCRL5 by scrutinizing the interactions of a large panel of native as well as various fragmented and modified IgG samples using surface plasmon resonance, which provides the detailed kinetics of the interactions. Our studies revealed a complex interaction that requires intact IgG molecules. This novel concept will help in understanding the physiological roles of FCRL5 and related proteins.
MATERIALS AND METHODS
Native and modified IgG samples
Intact IgG samples are listed in Table 1. Ig samples were obtained from Athens Research (Athens, GA), Bethyl Laboratories (Montgomery, TX), Sigma-Aldrich (St. Louis, MO), Calbiochem-EMD Millipore (Darmstadt, Germany) and Southern Biotech (Birmingham, AL). Therapeutic mAbs were obtained from the NIH pharmacy (Bethesda, MD) or were a gift. Polyclonal IgG-Fab and polyclonal IgG-Fc were obtained from Athens Research. Polyclonal IgG-F(ab’)2 was from Jackson ImmunoResearch (West Grove, PA). Sample purities were assessed by SDS-PAGE analysis, followed by protein staining. To produce the Fab-Fc fragment, we used the strategy developed by Hambly et al., taking advantage of partial resistance of IgG1 to LysC cleavage upon extended incubation of IgG1 at pH 5.2, due to isomerization of Asp222 (24). Briefly, mIgG1 (#1) in 10 mg/ml in 150 mM NaCl, 10 mM Na-acetate, pH 5.2 was incubated for 3 months at 37°C. Then, limited proteolysis was performed in 0.1 M Tris-HCl, pH 7.5, using 0.1 μg LysC (Pierce/Fisher Scientific, Pittsburgh, PA) per mg IgG1 for 15 minutes at 37°C. The reaction was quenched with 150 mM ammonium acetate, pH 4.7. The Fab-Fc fragment was enriched on two sequential Superdex200 size-exclusion columns (GE Healthcare, Piscataway, NJ), using an AKTA purifier 10 system (GE Healthcare). The final Fab-Fc sample was approximately 95% pure, as determined by SDS-PAGE analysis, with some intact IgG1 remaining.
Table 1.
Characteristics and FCRL5 binding of intact Ig samples by Biacore.
| # | Ig sample 1 | Source 2 | KD (μM) 3 | Binding model 4 |
|---|---|---|---|---|
| 1 | mIgG1κ | Genentech (Trastuzumab) | 1.39 | Two-state |
| 2 5 | mIgG2κ | Athens Research | 1.50 | Two-state |
| 3 | pIgG3 | Athens Research | 18 | Two-state |
| 4 5 | mIgG4κ | Athens Research | 0.87 | Two-state |
| 5 | Intravenous Ig | Talecris (Gamunex) | 2.9 | Two-state |
| 6 | pIgGκ | Bethyl Laboratories | 1.0 | Two-state |
| 7 | pIgGλ | Bethyl Laboratories | 3.1 | Two-state |
| 8 | mIgG1κ | Athens Research | 0.51 | Two-state |
| 9 | pIgG1 | Athens Research | 0.41 | Two-state |
| 10 | mIgG1κ | Genentech (Omalizumab) | 2.0 | Two-state |
| 11 | mIgG2κ | Sigma | 46 | Two-state |
| 12 | mIgG2λ | Sigma | 0.32 | Two-state |
| 13 6 | mIgG2κ | Calbiochem | 85 | 1:1 |
| 14 6 | mIgG2κ | Calbiochem | 0.035 | Two-state |
| 15 | mIgG2κ | Amgen (Panitumumab) | 7.5 | Two-state |
| 16 | mIgG2κ | Amgen (Denosumab) | 205 | 1:1 |
| 17 | mIgG4 | Calbiochem | 0.94 | Two-state |
| 18 | Mouse IgG | Southern Biotech | 75 | Two-state |
Representative Biacore binding curves and KD values are shown on Fig 1 for samples 1-4 or on Fig. S1 for the remaining samples. All Ig human, except #18. m, monoclonal/myeloma; p, polyclonal. Type of light chain is indicated if known.
Therapeutic mAb names are in parenthesis.
KD determined by kinetic analysis for representative runs are shown. Note that the weaker the affinity the more uncertain the actual KD value is.
Data were fitted using two models (1:1; two-state). Two-state fit is shown if the 1:1 fit was poor. For samples 1-4, 1:1 fits are also shown on Fig. S2B.
Samples bound differently depending on FCRL5 density; binding curves obtained at other density are shown on Fig. S2A.
Two lots bound distinctly differently.
Deglycosylated IgG1 and IgG2 were produced using PNGase F to remove N-linked oligosaccharides (25). Briefly, 1 mg IgG in 50 mM Tris, 150 mM NaCl, pH 8.5 was incubated with 1250 units of PNGase F (New England Biolabs, Ipswich, MA) at 37°C for 42 hours. At least 95% of H chain sugar was removed, as verified by assessing the mobility shift of the H chain on reduced SDS-PAGE.
Sialic acid enriched and depleted IVIg were produced as described (25). Briefly, 120 mg IVIg in TBS, pH 7.5 with 0.1 mM CaCl2 was applied to 4 ml agarose-bound Sambucus nigra agglutinin lectin column (Vector Laboratories, Burlingame, CA). Flow through was collected as sialic acid depleted IVIg. Sialic acid enriched IVIg was sequentially eluted with 0.5 M lactose in TBS and 0.5 M lactose in 0.2 M acetic acid, which together comprised 6.0% of the input IVIg. Sialic acid content of samples was assessed by lectin blotting. One μg protein per sample was resolved by reduced SDS-PAGE, transferred to nitrocellulose membrane, which was blocked with 5% BSA in TBST, then blotted with 10 μg/ml biotinylated Sambucus nigra agglutinin (Vector Laboratories), followed by streptavidin-HRP (Southern Biotech). Signal was visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare).
IgG was reduced and alkylated as described (26). IgG at 2 mg/ml in 0.2 M Tris-HCl, pH 8.0 was allowed to react with 10 mM DTT for 30 minutes, followed by 10 minutes treatment with iodoacetamide at 1.1-fold molar excess over cysteins forming interchain disulfide bonds. Samples were assessed by non-reduced SDS-PAGE. Ig samples used in cell binding studies were biotinylated using a kit from Pierce/Fisher Scientific. Comparable biotinylation of Ig samples was verified by Western blot analysis using streptavidin-HRP.
Anti-FCRL5 mAbs
A panel of 19 mAbs against the extracellular portion of membrane FCRL5 was produced as reported, immunizing with plasmid DNA (15,27). mAbs F25, F56 and F119 serve as the reference Abs for FCRL5 (28). The reactivity of all mAbs to FCRL5 was confirmed by flow cytometry (15). Affinities were measured by ELISA, as described (29). FCRL5 domain reactivities were established by ELISA using a series of FCRL5 deletion constructs (30). Topological location of epitopes were established based on the mutual competition of all possible pairs of mAbs for FCRL5 (31). Briefly, ELISA plates were coated with anti-mouse IgG (Jackson Immunoresearch), then incubated overnight with indicator mAb#1. In a separate tube, competitor mAb#2 was incubated with 10 ng/ml of FCRL5-Fc, then added to wells of washed plates and incubated for 1 hour. As standard, 1-10 ng/ml of FCRL5-Fc was used. mAb#2-FCRL5-Fc complexes were captured and probed with Fc-specific goat anti-human IgG-HRP (Jackson Immunoresearch). The pair-wise matrix table of the competition values was analyzed by a cluster analysis for automated grouping of the epitopes.
Surface plasmon resonance
Experiments were performed on Biacore T200 (GE Healthcare). About 12,000 RU anti-His mouse IgG1 mAb (R&D Systems, Minneapolis, MN) was immobilized on CM5 sensors using the amine coupling kit. Recombinant human FCRL5 containing the entire extracellular region (except 7 C-terminal amino acids) with a C-terminal His-tag (R&D Systems) was captured at 2-4 μg/ml for 40-120 s. Recombinant human FCRL3, CD16A and CD32B/C (R&D Systems) containing C-terminal His-tags were used as controls. IgG samples at six concentrations (one of which was run in duplicate), two-fold serially diluted from about 14 μM (2.1 mg/ml) in HBS-P containing 1 mg/ml non-specific binding reducer (GE Healthcare), were injected over FCRL5 for 8 minutes at 20 μl/min and 25°C, then dissociation was monitored for 10 minutes. Bound Ig and FCRL5-His were removed with a 1 minute injection of 10 mM glycine-HCl, pH 1.5. Data were analyzed using the Biacore T200 Evaluation software (GE Healthcare), subtracting the reference surface (immobilized anti-His mouse IgG1 mAb but no captured FCRL5-His) and buffer control signals from each curve. Data were globally fitted by simultaneous numerical integration to the association and dissociation parts of the interaction, using the 1:1 and two-state kinetic analysis models. Steady-state equilibrium was not reached during the association phase with the majority of full IgG samples (except with samples #13 and 16); nevertheless, global fitting produced reproducible kinetic parameters. Experimental Rmax values obtained by global fitting were comparable to theoretical Rmax values calculated from the molecular masses of the interacting proteins and the immobilization level. For interactions where equilibrium was reached, KD was also calculated by equilibrium analysis. Note that the weaker the affinity the more uncertain the actual KD value is.
When using anti-FCRL5 mAb, the analysis was performed similarly, with the following modifications. Anti-FCRL5 mAb at saturating concentration was injected over captured FCRL5-His for 4 minutes, then IgG was injected for 4 minutes, followed by 4 minutes dissociation. Buffer injection, instead of IgG injection, was used as control following every anti-FCRL5 mAb, to correct for mAb dissociation. When calculating percent inhibition by each mAb, buffer controls were subtracted from IgG signals, then % IgG binding was calculated relative to no-Ab sample.
Cell binding studies
HEK293T cells were maintained in DMEM containing 10% FBS. Cells were seeded at 5×106 cells in 10 cm dish 24 hours prior to transfection, then transfected with 5 μg of plasmid encoding transmembrane FCRL5 (15), using Lipofectamine and Plus reagent (Invitrogen, Carlsbad, CA). Forty eight hours later the cells were harvested and incubated with biotinylated IgG and 1 μg anti-FcRL5 F108 for 1 hour on ice. Cells were washed twice with cold PBS with 1% FBS and incubated for 30 minutes on ice with streptavidin-APC and F(ab’)2 anti-mouse Ig-PE (BioSource, Camarillo, CA). Cells were washed twice, fixed with 1% paraformaldehyde, then analyzed on a FACSCalibur (Beckton Dickinson, Franklin Lakes, NJ) using FlowJo (TreeStar, Ashland, OR).
Statistical analysis
Two-sample Student t-test with unequal variances was used, considering P values <0.05 significant.
RESULTS
Recombinant FCRL5 protein binds native IgG
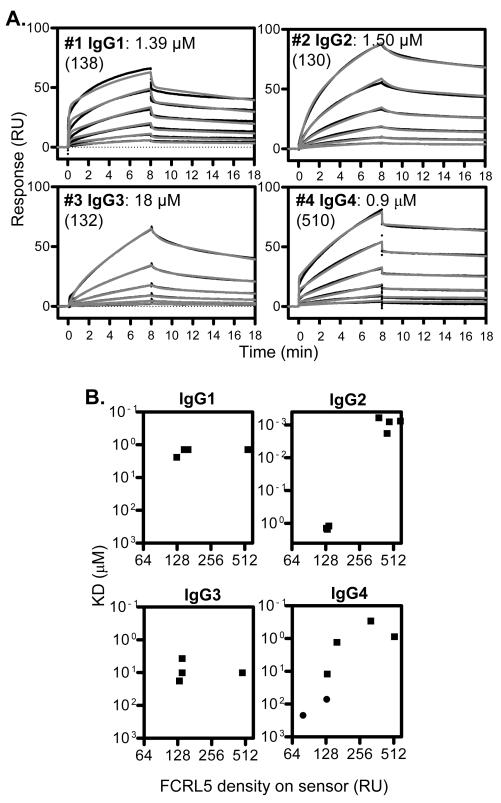
Surface plasmon resonance (Biacore) was used to study the real-time interaction of FCRL5 with native (non-complexed) IgG in a label-free system. Both the association of IgG to FCRL5 and its subsequent dissociation were recorded, from which the association rate constant (ka) and dissociation rate constant (kd) were calculated. The ratio of the rate constants equals the dissociation binding constant (KD), which corresponds to the affinity, equally defined as the IgG concentration at which half of the FCRL5 molecules are in complex at equilibrium. We tested 18 IgG samples (Table 1), representing all human IgG subclasses as well mouse IgG. Trastuzumab and Omalizumab are humanized mAbs containing human sequences except in the CDRs, which are murine derived. Panitumumab and Denosumab are considered fully human IgG2. Three polyclonal IgG samples bound FCRL5 with 1-3 μM KD, while mouse polyclonal IgG bound poorly (Table 1, Fig. 1A and Fig. S1). Four IgG1, including two humanized therapeutic mAbs, bound FCRL5 similarly, displaying 0.4-2 μM KD. Seven IgG2, including two human therapeutic mAbs, displayed a wide range of affinities, from nanomolar to approximately 200 μM KD. Beyond affinities, the kinetics of the IgG2 interactions were markedly different. Two IgG2 (#13 and 16) displayed both association and dissociation rapidly reaching equilibrium, and overall had the weakest affinities. IgG3 had an order of magnitude lower affinity (on average 10.6 μM) than IgG1. Two myeloma IgG4 (#4 and 17) bound similarly, with approximately 1 μM KD and comparable kinetic parameters. Overall, these results indicate significant heterogeneity in FCRL5 binding, in particular for IgG2, suggesting that factors beyond IgG isotype influence the interaction.
Fig. 1. Recombinant FCRL5 binds IgG.
Association and dissociation of soluble Ig to FCRL5 on the sensor were monitored by surface plasmon resonance. (A) Representative binding curves are shown for samples 1-4 (Table 1). Black lines are actual binding curves; overlaid grey lines represent fits obtained by kinetic analysis using two-state binding model. Samples were run at a two-fold dilution series, starting from approximately 14 μM. KD values are shown, in parenthesis indicating FCRL5 densities on the sensor in relative units. (B) KD as a function of FCRL5 density on the sensor is shown. Squares indicate fits were superior using two-state binding; circles indicate fits were excellent using 1:1 binding.
One sample for each IgG subclass (samples 1-4) was used in a detailed comparative analysis of the kinetic parameters of the interactions (Fig. 1). We chose the highest affinity IgG2 for this analysis. Representative Biacore binding curves are shown on Fig. 1A. Binding curves for all IgG produced excellent fits using the two-state binding model, which suggested two linked interactions with separate ka and kd values, while fits from the 1:1 model were poor (Fig. S2A). Indeed, visual inspection of the binding curves, especially those of IgG1 and IgG4, suggested a two-step interaction; rapid initial association was followed by much slower association. Similarly, rapid initial dissociation was followed by slow dissociation. The order of affinities from high to low was: IgG4, IgG2, IgG1, IgG3 (Table 2), although the affinities of IgG1, IgG2 and IgG4 were not statistically different. As negative control, human FCRL3 protein did not bind IgG1, IgG2 or IgG4 at all, while weakly interacted with IgG3 (Table. S1.). In spite of the similar KD values, the primary kinetic parameters were distinct among the IgG subclasses (Table 2). IgG1 and IgG4 displayed an order of magnitude faster initial association (ka1) and initial dissociation (kd1) than IgG2 and IgG3. Remarkably, the secondary interactions were similar among all IgG subclasses, as the ka2 and kd2 values were comparable, suggesting that the secondary interaction is mediated by a region of the molecule that is common among IgG subclasses.
Table 2.
Detailed kinetic analysis of FCRL5 binding of IgG samples #1-4. 1
| Ig sample | KD (μM) | ka1 (1/Ms) | kd1 (1/s) | ka2 (1/s) | kd2 (1/s)×104 |
|---|---|---|---|---|---|
| #1 IgG1 | 1.75 ±0.55 | 6145 ±877 | 0.133 ±0.051 | 0.0057 ±0.0015 | 5.05 ±1.41 |
| #2 IgG2 | 1.37 ±0.15 | 578 ±164 | 0.017 ±0.004 | 0.0068 ±0.0003 | 3.39 ±0.40 |
| #3 IgG3 | 10.6 ±7.1 | 341 ±318 | 0.034 ±0.042 | 0.0070 ±0.0013 | 9.26 ±2.37 |
| #4 IgG4 | 0.82 ±0.51 | 7245 ±4884 | 0.306 ±0.319 | 0.0078 ±0.0017 | 1.98 ±0.70 |
IgG binding was assessed at FCRL5 densities of 120-160 RU, except for IgG4 analyzed at densities of 160-520 RU. Shown are mean ±SD (n=3).
Because our initial results indicated variability, we assessed the correlation of KD and FCRL5 density (relative amount of FCRL5 captured on the sensor). The affinity of the IgG1 and IgG3 interactions did not depend on FCRL5 density (Fig. 1B). However, the affinity of IgG2 increased 1000-fold at approximately 4-fold higher FCRL5 density, from 1.37±0.15 μM to 0.99±0.55 nM, mostly due to slower dissociation (Fig. S2B). In addition, IgG4 binding exhibited strong dependence on FCRL5 density; at low FCRL5 densities (<150 RU) the secondary interaction phase was lost and the affinity dropped 100-fold. We analyzed (as shown on Fig. 1a and Table 2) IgG binding at low FCRL5 densities (120-160 RU), except for IgG4, which was analyzed at higher densities (160-520 RU), because the interaction was lost at lower densities. These protein densities were still lower than those used in recent definitive studies of FcgRs (32-34).
We conclude that IgG binding to FCRL5 consists of two steps, a rapid primary binding event and a secondary binding event with much slower dissociation, contributing to a stable complex. Moreover, variables beyond isotype influence IgG binding.
FCRL5 on the cell surface binds native IgG
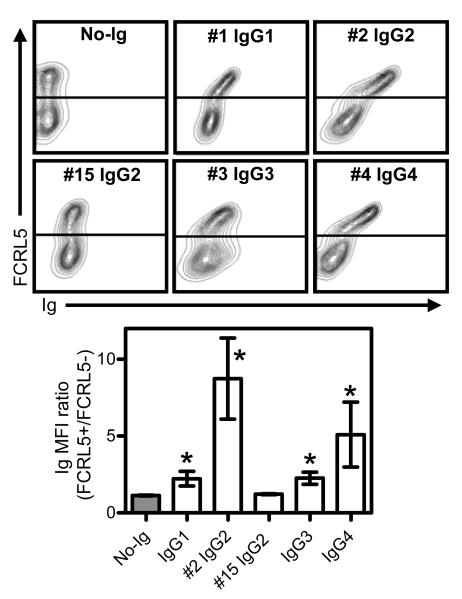
We assessed whether FCRL5 expressed on the cell surface binds native Ig, first using transfected HEK293T cells. The IgG samples were the same as those shown on Fig. 1, allowing direct comparison. One additional IgG2 (#15), which had lower affinity by Biacore, was also tested. Binding of biotinylated IgG at 1.7 μM concentration was monitored by flow cytometry. Co-staining of FCRL5 using a mAb that did not interfere with IgG binding allowed assessment of the correlation of FCRL5 expression and IgG binding. We detected the strongest binding of IgG4 and one of the IgG2 (#2) to FCRL5-positive cells, whereas binding of IgG1 and IgG3 was much weaker (Fig. 2). We could not detect the binding of IgG2 (#15), being tested at a concentration below its KD as measured by Biacore. Intensity of IgG binding correlated positively with FCRL5 expression level. In conclusion, IgG binds FCRL5 on cells, similarly to recombinant FCRL5 on Biacore sensor, although the latter detection method is clearly more sensitive.
Fig. 2. FCRL5 on cells binds IgG.
Binding of selected Ig samples (Table 1) to FCRL5-transfected HEK293T cells at 1.7 μM was monitored by flow cytometry. Cells were co-stained for FCRL5. On top, representative flow patterns are shown. Line(s) indicate gating of FCRL5-positive versus FCRL5-negative cells. Below the bar graphs show the ratios of the Ig signals (MFI) of FCRL5 positive and negative cells (mean±SD; N=3). * denotes p<0.05 compared to no-Ig control.
FCRL5 domains 1 and 3 play key roles in IgG binding
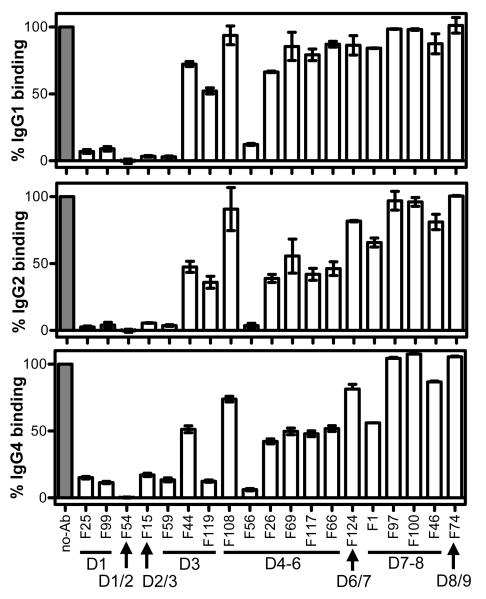
To identify epitopes required for IgG1, IgG2 and IgG4 binding, we used a panel of 19 FCRL5-specific mAbs thoroughly characterized regarding domain and epitope specificity (Table 3). IgG3 was excluded from the analysis due to its lower affinity. Each mAb was allowed to bind FCRL5 at a saturating concentration, and subsequent IgG binding was monitored by Biacore (Fig. 3). IgG1, IgG2 and IgG4 binding was fully blocked by the following six mAbs, five of which bind in the D1-3 region. F25 and F99 (epitopes on D1); F54 (D1/D2 boundary, as it does not bind isolated D1 or D2, but binds tandem D1-2); F59 (D3); F15 (D2/D3 boundary); F56 (D4-6 fragment). An additional six mAbs inhibited IgG2 and IgG4 binding at least 50%: F44 and F119 (D3); F26, F69, F117 and F66 (all bind epitopes on the D4-6 fragment). The same mAbs that partially blocked IgG2 and IgG4 binding had less effect on IgG1 binding, suggesting subtle differences exist between IgG subclass recognition by FCRL5. Partial inhibition of IgG binding may also be due to conformational effects or steric hindrance, even acting across FCRL5 domains, and may not be the result of the proximity of the mAb’s epitope and the IgG-binding region on FCRL5. In conclusion, epitopes on D1, the D1/D2 boundary, the D2/D3 boundary, D3 and one epitope on D4-6 are required for both IgG1 and IgG2 binding.
Table 3.
Properties of anti-FCRL5 mAbs used in the studies.
| mAb | KD 1 (nM) |
Epitope 2 |
React- ivity 3 |
Binding to FCRL5 domains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | D2 | D3 | D1-2 | D1-3 | D4-6 | D7-9 | D4-9 | D1-8 | ||||
| F25 | 1.2 | 1 | D1 | + | − | − | + | + | − | − | − | + |
| F99 | 1.6 | 1 | D1 | + | − | − | + | + | − | − | − | + |
| F54 | 6.1 | 2 | D1/2 | − | − | − | + | + | − | − | − | + |
| F15 | 7.6 | 3 4 | D2/3 | − | − | − | − | + | − | − | − | + |
| F59 | 6.0 | 3 4 | D3 | − | − | + | − | + | − | − | − | + |
| F44 | 4.6 | 4 | D3 | − | − | + | − | + | − | − | − | + |
| F119 | 5.4 | 4 | D3 | − | − | + | − | + | − | − | − | + |
| F108 | 1.9 | 5 | D4-6 | − | − | − | − | − | + | − | + | + |
| F56 | 4.4 | 6 | D4-6 | − | − | − | − | − | + | − | + | + |
| F26 | 2.7 | 7 | D4-6 | − | − | − | − | − | + | − | + | + |
| F69 | 4.1 | 7 | D4-6 | − | − | − | − | − | + | − | + | + |
| F117 | 3.5 | 7 | D4-6 | − | − | − | − | − | + | − | + | + |
| F66 | 7.5 | 7 | D4-6 | − | − | − | − | − | + | − | + | + |
| F124 | 10.6 | 8 | D6/7 | − | − | − | − | − | − | − | + | + |
| F1 | 5.7 | 9 | D7-8 | − | − | − | − | − | − | + | + | + |
| F97 | 7.8 | 9 | D7-8 | − | − | − | − | − | − | + | + | + |
| F100 | 3.6 | 10 | D7-8 | − | − | − | − | − | − | + | + | + |
| F46 | 6.6 | 10 | D7-8 | − | − | − | − | − | − | + | + | + |
| F74 | 4.4 | 11 | D8/9 | − | − | − | − | − | − | + | + | − |
KD for an FCRL5 D1-9 fragment was measured by ELISA.
Topographical epitope assignments, based on mAb cross-competition.
Overall reactivity, based on binding to FCRL5 domains, as shown on the right. Slash indicates that mAb binds at the boundary of two domains.
Epitope assignment was the same, however domain reactivities differed, likely reflecting conformational effect or steric hindrance.
Fig. 3. Domains and epitopes of FCRL5 involved in IgG binding.
Using Biacore, anti-FCRL5 mAbs at saturating concentrations were allowed to bind FCRL5, then binding of IgG1 (#8), IgG2 (#2) or IgG4 (#4) at 6.7 μM, was assessed. Binding levels were corrected to buffer controls and expressed as a percent of no-Ab control. Mean and range of two or three independent experiments are shown. Domain (D) specificities of mAbs (Table 3) are shown; slash indicates binding at domain boundaries.
Only intact IgG binds FCRL5 with high affinity
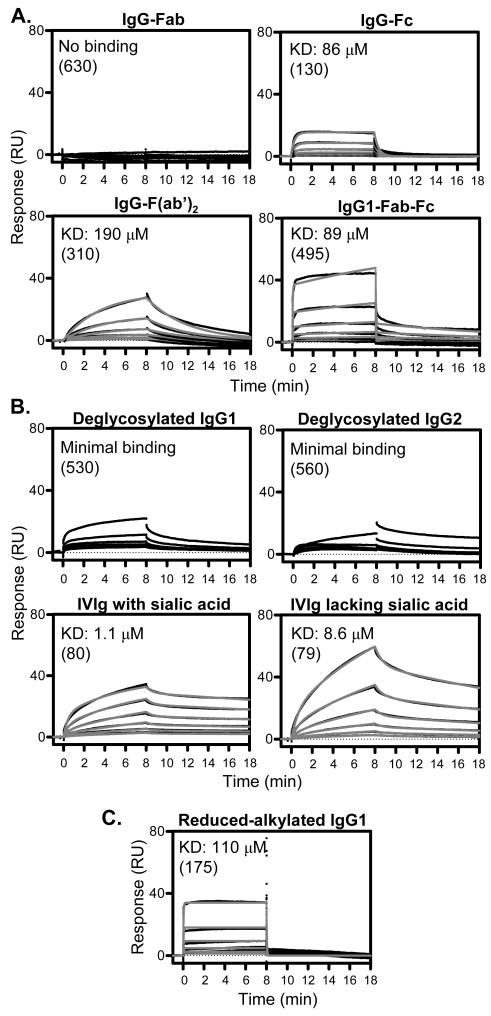
FcgRs bind IgG through the Fc portion of the molecule and display similar affinities for intact IgG and Fc fragment (32). We first assessed whether FCRL5 binds to the Fc and/or Fab fragments of polyclonal IgG. The purity of the samples, assessed by non-reduced and reduced SDS-PAGE, is shown on Fig. S3A and B. IgG-Fab did not bind FCRL5, whereas IgG-Fc displayed weak (86±27 μM KD) 1:1 binding, using kinetic analysis (Fig. 4A). The KD of IgG-Fc, alternatively calculated using steady state analysis was 89±8 μM. Notably, the IgG-Fc fragment bound visibly differently than most intact IgG, with both association and dissociation rapidly reaching equilibrium, resembling the kinetics of IgG binding to low affinity FcgRs (32,33). The binding of IgG-F(ab’)2 to FCRL5 was detectable albeit weak, and displayed clearly different kinetics than IgG-Fc, with slow association and dissociation. For an IgG1-Fab-Fc fragment, which contained one Fab arm and the Fc region, we detected weak (89 μM KD) and close to 1:1 binding, with some residual secondary interaction component. Therefore, the IgG-Fc region and both Fab arms are required for high affinity FCRL5 binding. Two FcgR proteins (CD16A and CD32B/C), bound intact IgG1 and IgG-Fc fragment similarly, but not IgG-F(ab’)2 (Table. S1.), as expected. We propose, based on kinetic considerations, that the Fc and F(ab’)2 regions, each mediate one of the two interaction steps observed for full IgG.
Fig. 4. Molecular determinants of IgG playing roles in FCRL5 binding.
Biacore was performed as indicated in Fig. 1. Representative binding curves are shown. KD values are shown, in parenthesis indicating FCRL5 densities on the sensor in relative units. Note that the weaker the affinity the more uncertain the actual KD value is. (A) Fab and Fc fragments were run from 30 μM. Fab-Fc and F(ab’)2 were run from 15 μM and 20 μM, respectively. Fits were obtained by kinetic analysis (1:1 binding for Fc and F(ab’)2; two-state binding for Fab-Fc). (B) Samples were obtained as described in the Materials and Methods section, and were run from 15 μM. Fits for IVIg with or without sialic acid were obtained by kinetic analysis (two-state binding). (C) Interchain disulfide bonds were reduced and alkylated under mild conditions. Reduced IgG1 was run from 14 μM. Fits were obtained by kinetic analysis (1:1 binding).
Next, the role of IgG glycosylation in FCRL5 binding was investigated. Deglycosylation of IgG1 (#1) and a high affinity IgG2 (#2) greatly diminished their binding to FCRL5, displaying only minimal residual binding likely due to some remaining intact IgG (Fig. 4B). Therefore, glycosylation of the IgG heavy chains is required for the interaction, similar to FcgRs (35). We further assessed the contribution of one component of the IgG glycan, sialic acid. IVIg, representing polyclonal IgG, was fractionated on Sambucus nigra lectin column, which separates IgG glycoforms based on the presence of sialic acid on the Fab regions (36-38). Enrichment of the samples was assessed by lectin blotting (Fig. S3C). We found that IVIg with sialic acid on the Fab interacted with FCRL5 differently than IVIg depleted of sialic acid (Fig. 4B). IVIg with sialic acid displayed significantly higher affinity (1.72±0.61 μM KD) than IVIg lacking sialic acid (8.83±1.10 μM KD). Additionally, IVIg with sialic acid appeared to bind only a subset of FCRL5 proteins, as suggested by a saturation level (Rmax) that was approximately 1/3 of that expected based on the molecular masses and the immobilization level. As controls, two human FcgRs (CD16A and CD32B/C) displayed comparable affinities for IVIg with or without sialic acid (Table S1).
We next assessed the contribution of interchain disulfide bonds. The more accessible interchain disulfide bonds in IgG1 (#1) were reduced and alkylated, resulting in IgG molecules still held together by non-covalent interactions (26). Reduced IgG1 bound FCRL5 with low affinity due to apparent loss of the secondary interaction component, in a manner comparable to that of Fc (Fig. 4C). Although we do not have structural data verifying that reduced-alkylated IgG1 folded correctly, its binding to FCRL5 suggested that it retained the interaction component related to properly folded Fc. However, the Fab regions might be unfolded. We conclude that interchain disulfide bond integrity is likely critical for IgG1 binding.
In summary, we established that strong FCRL5 binding requires an intact IgG molecule, and the Fc and F(ab’)2 regions apparently mediate distinct phases of the interaction.
DISCUSSION
We established that FCRL5 binds intact IgG. A detailed quantitative analysis of IgG binding revealed a complex interaction, engaging several domains of both the IgG and FCRL5 molecules, resulting in two-step binding mediated by different regions of the IgG molecule.
FCRL5 binding of 18 IgG samples was analyzed, revealing a surprising complexity, where isotype may not be the major determinant of the interaction. IgG2 samples displayed a wide range of affinities as weak as 200 μM. The molecular signatures affecting IgG2 binding are unknown, but might be related to flexibility of the Fab regions or differences in glycosylation, which affect IgG function and structure (39,40). On the other hand, the four IgG1 we tested bound similarly, while polyclonal IgG3 had a magnitude weaker affinity. Two IgG4 had comparable affinities at higher FCRL5 densities on the sensor, but binding was almost lost at FCRL5 densities below a sharp threshold. Similarly, one particular IgG2 displayed 1000-fold different affinities, depending on FCRL5 density. IgG2 is able to rearrange disulfide bonds to form distinct isoforms and covalent dimers (41-43), while the disulfide bonds between the IgG4 heavy chains are unstable and isomerize to allow formation of half molecules (44). IgG2 dimers at higher densities could contact two FCRL5 molecules, although this possibility was not supported by SDS-PAGE analysis, which indicated only minor amounts of dimers. We speculate that the strong dependence of both IgG2 and IgG4 binding on FCRL5 density might reflect a role for disulfide bond isomerization. We note that during Biacore analysis, the anti-His mAb on the sensor surface, a mouse IgG1, might compete with soluble IgG for the captured receptor. However, the impact of immobilized mouse IgG1 on Biacore assay performance appears negligible, because FcgRs bound IgG as expected, and no IgG concentration dependent interference was observed with FCRL5.
IgG2 (#2) and IgG4 bound more strongly to FCRL5 on cells than IgG1 and IgG3. However, binding of a lower affinity IgG2 could not be detected. Biacore affinities obtained at higher FCRL5 densities correlated better with binding to FCRL5 on cells, suggesting that IgG2 and IgG4 binding to membrane FCRL5 may be driven not only by the affinity of one-to-one protein interactions, but also by protein clustering or other mechanisms. Our results obtained using HEK293T cells partly agree with a previous report by Wilson et al., which found all IgG subclasses binding FCRL5, IgG1 and IgG2 being superior (22). The paper differs from our study in testing heat-aggregated IgG and using a different set of samples, which together could explain the different isotype rankings.
Compared to CD32B, the sole FcgR expressed on B cells, FCRL5 displays approximately an order of magnitude higher affinity for IgG1 and IgG4, whereas it has two-times lower affinity for IgG3 (33). The majority of IgG2 appears to have higher affinity for FCRL5 than CD32B, which binds IgG2 with approximately 50 μM KD. Beyond affinities, IgG binds FCRL5 and CD32B with distinctly different kinetics. All IgG subclasses bind CD32B with both association and dissociation reaching equilibrium in seconds, resulting in a dynamic interaction that is only stable if polyvalent. In contrast, IgG binding to FCRL5 takes minutes to reach equilibrium, but on the other hand is much more stable than the interaction with CD32B. Under physiological conditions, FCRL5 might bind IgG as part of an immune complex, in which the antigen component could drive the initial contact with the B cell. Based on affinities alone, FCRL5 would preferentially engage the IgG component over CD32B, with the exception of IgG3. Nevertheless, the molecular events taking place on the B cell membrane would also be influenced by additional factors, including the relative localization of the various receptors involved and the kinetics of the interactions.
FCRL5 engages several of its Ig-domains to bind IgG, as established using anti-FCRL5 mAbs with defined reactivity (Fig. 5). Our data implicate D1, D3 and one epitope on the D4-6 fragment in IgG binding, whereas the role of D2 is unknown, as none of the anti-FCRL5 mAbs was D2 specific. The roles of D1-3, each distinct and displaying homology to the three Ig domain types present in FcgRs, are not surprising. FCRL5 domains 4-9, however, are more similar to each other than to domains found in FcgRs (4). A previous study showed that D1-3 of FCRL5, when expressed on cells, were sufficient to bind aggregated IgG (22), thus the significance of the epitope on D4-6 fragment requires further investigation. The observation that mAbs F54 and F15, which recognize epitopes spanning FCRL5 D1/D2 and D2/D3 boundaries, blocked the interaction suggests that bending of the FCRL5 molecule at domain boundaries is critical for IgG binding. Flexibility of both FCRL5 and IgG (45) may be important in aligning multiple domains during the interaction.
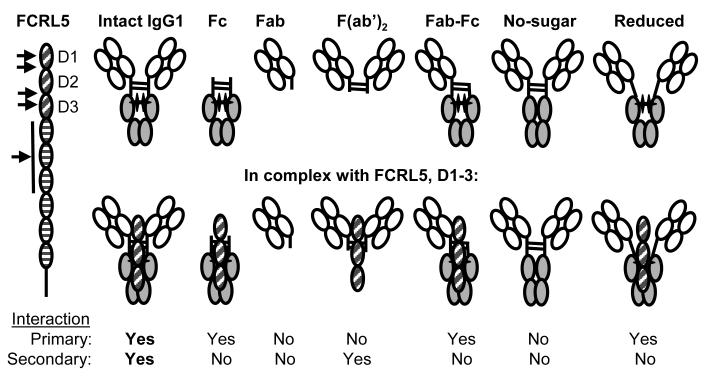
Fig. 5. The interaction of IgG1 and FCRL5.
IgG1, for which most complete data are available, interacts with FCRL5 in a complex fashion. On the left, Ig-domain structure of FCRL5 is shown. Arrows point to domains (D) implicated in intact IgG1 binding, based on inhibition by anti-FCRL5 mAbs. Representations of intact IgG1 as well as various deficient molecules lacking protein domains or other components are shown alone (top) and in a proposed complex with D1-3 of FCRL5 (bottom). The images reflect that the IgG1 sugar pushes the CH2 domains apart (40). Sugar is represented by black cross. The orientation of FCRL5 D1-3 relative to IgG1 is unknown. The interaction of intact IgG1 and FCRL5 consists of two components. In case of the primary interaction, both association and dissociation are rapid, while for the secondary interaction both are slow. We propose based on kinetic considerations that the Fc region mediates the primary interaction, whereas the F(ab’)2 the secondary interaction. Lack of either IgG1 sugar or interchain disulfide bonds diminish the interaction. As a result, only intact IgG1 binds with high affinity.
IgG clearly employs multiple regions to bind FCRL5 (Fig. 5). Although IgG-Fc fragment bound FCRL5, its affinity was greatly reduced and its binding lacked the secondary interaction component. Remarkably, the rapid association and dissociation of IgG-Fc resembled that of the primary interaction of intact IgG, suggesting that the Fc mediates this interaction phase. The kinetic parameters of the primary interactions were different among the IgG subclasses and could partly be mediated by the hinge, where subclasses differ most. The IgG-Fab fragment did not bind FCRL5, while IgG-F(ab’)2 bound with low affinity and slow kinetics, resembling that of the secondary interaction of full IgG. Furthermore, the affinity of IgG1-Fab-Fc fragment was comparable to that of the Fc fragment and similarly lacked most of the secondary interaction component observed with full IgG. Therefore, the secondary interaction phase requires both Fab arms. The upper hinge sequences, present in F(ab’)2, are different among the four IgG subclasses, whereas all IgG subclasses showed similar secondary interactions, arguing against the role of the upper hinge in FCRL5 binding. Therefore, the secondary interaction phase of full IgG is likely mediated by a region located within the Fab and common among IgG subclasses, perhaps on the CH1 domain or the L-chain. Deglycosylated IgG lost its ability to bind FCRL5, implicating the CH2 domains where the carbohydrates are located. Importance of the sugar suggests either direct FCRL5 contacts with carbohydrate moieties, or structural requirements, as the carbohydrate alters the steric arrangement of the CH2 domains by pushing them apart (39,40). IgG lacking sugar did not bind FCRL5 in spite of containing the F(ab’)2, which alone bound, perhaps as a result of steric inhibition due to the closely-spaced CH2 domains. IgG containing sialic acid as component of sugars present on the Fab bound FCRL5 with 5-times higher affinity, supporting the involvement of IgG regions located outside the Fc portion and suggesting that sialylation of the Fab modifies the interaction with FCRL5. IgG1 lacking interchain disulfide bonds kept the fast first interaction but lost the slow secondary interaction component, likely reflecting the importance of the proximity of the two Fab arms, which together could form one interaction surface. In conclusion, the interaction of IgG with FCRL5 is profoundly different from that with FcgRs. While FcgR binding is a one-step, rapid interaction mediated by the Fc (32,33), FCRL5 binding consists of two components, one dependent on the Fc, the other on the F(ab’)2 region. Nevertheless, additional studies including solving the crystal structure of the receptor-ligand complex are required to fully elucidate the structural bases of the interaction.
We showed that FCRL5 is not a bona fide FcR, as the Fc was insufficient for the interaction. We propose that FCRL5 is a receptor for intact IgG, because it displayed reduced affinity for IgG molecules that are fragmented, lack glycosylation or have improper interchain disulfide structure. What might be the physiological relevance of IgG binding to FCRL5 be restricted to intact molecules? One distinction may be that damaged IgG molecules do not compete for recognition, allowing FCRL5 to function as a receptor of newly secreted, intact IgG molecules, which were not subjected to enzymatic or structural alterations. Preferential sensing of recently produced IgG molecules by FCRL5 could be part of a mechanism to focus the immune response on emerging infections. In addition, the ability of B and plasma cells to sense and discriminate intact IgG could provide quality control at multiple levels. Mature B cells could specifically be regulated by intact but not fragmented or otherwise altered IgG. Plasma cells, which express high levels of FCRL5, might be regulated by their secreted IgG in an autologous manner. Finally, if discriminating intact IgG is a property shared with FCRLA (20,21), this could entail quality control of newly synthesized IgG in the endoplasmic reticulum. Nevertheless, the physiological relevance of the complex interaction requiring intact IgG and multiple FCRL5 domains remains to be established.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Lute and Michael Murphy for advice, Milos Dokmanovic, Mei-ying Yu and Wen Jin Wu for reagents.
Abbreviations used
- D
domain
- FCRL
Fc receptor-like
Footnotes
Grant support: This work was supported by the Intramural Research Program of CDER/FDA. A.F. and H.L. were supported through the Research Fellowship Program for CDER administered by the Oak Ridge Associated Universities. B.D. was supported by the FDA Commissioner’s Fellowship Program. S.N. and T.I. were supported by Leukemia Research Foundation, CLL Global Research Foundation and a NIH COBRE grant (1P20RR024219-01A2).
REFERENCES
- 1.Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9772–9777. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatzivassiliou G, Miller I, Takizawa J, Palanisamy N, Rao PH, Iida S, Tagawa S, Taniwaki M, Russo J, Neri A, Cattoretti G, Clynes R, Mendelsohn C, Chaganti RS, la-Favera R. IRTA1 and IRTA2, novel immunoglobulin superfamily receptors expressed in B cells and involved in chromosome 1q21 abnormalities in B cell malignancy. Immunity. 2001;14:277–289. doi: 10.1016/s1074-7613(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 3.Xu MJ, Zhao R, Cao H, Zhao ZJ. SPAP2, an Ig family receptor containing both ITIMs and ITAMs. Biochem. Biophys. Res. Commun. 2002;293:1037–1046. doi: 10.1016/S0006-291X(02)00332-7. [DOI] [PubMed] [Google Scholar]
- 4.Davis RS. Fc receptor-like molecules. Annu. Rev. Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 5.Leu CM, Davis RS, Gartland LA, Fine WD, Cooper MD. FcRH1: an activation coreceptor on human B cells. Blood. 2005;105:1121–1126. doi: 10.1182/blood-2004-06-2344. [DOI] [PubMed] [Google Scholar]
- 6.Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD. The inhibitory potential of Fc receptor homolog 4 on memory B cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13489–13494. doi: 10.1073/pnas.1935944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haga CL, Ehrhardt GR, Boohaker RJ, Davis RS, Cooper MD. Fc receptor-like 5 inhibits B cell activation via SHP-1 tyrosine phosphatase recruitment. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9770–9775. doi: 10.1073/pnas.0703354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kochi Y, Myouzen K, Yamada R, Suzuki A, Kurosaki T, Nakamura Y, Yamamoto K. FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J. Immunol. 2009;183:5502–5510. doi: 10.4049/jimmunol.0901982. [DOI] [PubMed] [Google Scholar]
- 9.Jackson TA, Haga CL, Ehrhardt GR, Davis RS, Cooper MD. FcR-like 2 Inhibition of B cell receptor-mediated activation of B cells. J. Immunol. 2010;185:7405–7412. doi: 10.4049/jimmunol.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dement-Brown J, Newton CS, Ise T, Damdinsuren B, Nagata S, Tolnay M. Fc receptor-like 5 promotes B cell proliferation and drives the development of cells displaying switched isotypes. J Leukoc. Biol. 2012;91:59–67. doi: 10.1189/jlb.0211096. [DOI] [PubMed] [Google Scholar]
- 11.Polson AG, Zheng B, Elkins K, Chang W, Du C, Dowd P, Yen L, Tan C, Hongo JA, Koeppen H, Ebens A. Expression pattern of the human FcRH/IRTA receptors in normal tissue and in B-chronic lymphocytic leukemia. Int. Immunol. 2006;18:1363–1373. doi: 10.1093/intimm/dxl069. [DOI] [PubMed] [Google Scholar]
- 12.Mohan J, Dement-Brown J, Maier S, Ise T, Kempkes B, Tolnay M. Epstein-Barr virus nuclear antigen 2 induces FcRH5 expression through CBF1. Blood. 2006;107:4433–4439. doi: 10.1182/blood-2005-09-3815. [DOI] [PubMed] [Google Scholar]
- 13.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae SC, Tokuhiro S, Chang X, Sekine A, Takahashi A, Tsunoda T, Ohnishi Y, Kaufman KM, Kang CP, Kang C, Otsubo S, Yumura W, Mimori A, Koike T, Nakamura Y, Sasazuki T, Yamamoto K. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat. Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li FJ, Ding S, Pan J, Shakhmatov MA, Kashentseva E, Wu J, Li Y, Soong SJ, Chiorazzi N, Davis RS. FCRL2 expression predicts IGHV mutation status and clinical progression in chronic lymphocytic leukemia. Blood. 2008;112:179–187. doi: 10.1182/blood-2008-01-131359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ise T, Maeda H, Santora K, Xiang L, Kreitman RJ, Pastan I, Nagata S. Immunoglobulin superfamily receptor translocation associated 2 protein on lymphoma cell lines and hairy cell leukemia cells detected by novel monoclonal antibodies. Clin. Cancer Res. 2005;11:87–96. [PubMed] [Google Scholar]
- 16.Ise T, Nagata S, Kreitman RJ, Wilson WH, Wayne AS, Stetler-Stevenson M, Bishop MR, Scheinberg DA, Rassenti L, Kipps TJ, Kyle RA, Jelinek DF, Pastan I. Elevation of soluble CD307 (IRTA2/FcRH5) protein in the blood and expression on malignant cells of patients with multiple myeloma, chronic lymphocytic leukemia, and mantle cell lymphoma. Leukemia. 2007;21:169–174. doi: 10.1038/sj.leu.2404445. [DOI] [PubMed] [Google Scholar]
- 17.Owczarczyk K, Lal P, Abbas AR, Wolslegel K, Holweg CT, Dummer W, Kelman A, Brunetta P, Lewin-Koh N, Sorani M, Leong D, Fielder P, Yocum D, Ho C, Ortmann W, Townsend MJ, Behrens TW. A plasmablast biomarker for nonresponse to antibody therapy to CD20 in rheumatoid arthritis. Sci Transl. Med. 2011;3:101ra92. doi: 10.1126/scitranslmed.3002432. [DOI] [PubMed] [Google Scholar]
- 18.Elkins K, Zheng B, Go M, Slaga D, Du C, Scales SJ, Yu SF, McBride J, de TR, Rawstron A, Jack AS, Ebens A, Polson AG. FcRL5 as a Target of Antibody-Drug Conjugates for the Treatment of Multiple Myeloma. Mol. Cancer Ther. 2012;11:2222–2232. doi: 10.1158/1535-7163.MCT-12-0087. [DOI] [PubMed] [Google Scholar]
- 19.Schreeder DM, Cannon JP, Wu J, Li R, Shakhmatov MA, Davis RS. Cutting edge: FcR-like 6 is an MHC class II receptor. J. Immunol. 2010;185:23–27. doi: 10.4049/jimmunol.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson TJ, Gilfillan S, Colonna M. Fc receptor-like A associates with intracellular IgG and IgM but is dispensable for antigen-specific immune responses. J Immunol. 2010;185:2960–2967. doi: 10.4049/jimmunol.1001428. [DOI] [PubMed] [Google Scholar]
- 21.Santiago T, Kulemzin SV, Reshetnikova ES, Chikaev NA, Volkova OY, Mechetina LV, Zhao M, Davis RS, Taranin AV, Najakshin AM, Hendershot LM, Burrows PD. FCRLA is a resident endoplasmic reticulum protein that associates with intracellular Igs, IgM, IgG and IgA. Int Immunol. 2011;23:43–53. doi: 10.1093/intimm/dxq456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson TJ, Fuchs A, Colonna M. Cutting edge: Human FcRL4 and FcRL5 are receptors for IgA and IgG. J Immunol. 2012;188:4741–4745. doi: 10.4049/jimmunol.1102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 24.Hambly DM, Banks DD, Scavezze JL, Siska CC, Gadgil HS. Detection and quantitation of IgG 1 hinge aspartate isomerization: a rapid degradation in stressed stability studies. Anal. Chem. 2009;81:7454–7459. doi: 10.1021/ac901258g. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 26.Stevenson GT, Dorrington KJ. The recombination of dimers of immunoglobulin peptide chains. Biochem. J. 1970;118:703–712. doi: 10.1042/bj1180703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata S, Salvatore G, Pastan I. DNA immunization followed by a single boost with cells: a protein-free immunization protocol for production of monoclonal antibodies against the native form of membrane proteins. J Immunol Methods. 2003;280:59–72. doi: 10.1016/s0022-1759(03)00192-3. [DOI] [PubMed] [Google Scholar]
- 28.Vidal-Laliena M, Romero X, March S, Requena V, Petriz J, Engel P. Characterization of antibodies submitted to the B cell section of the 8th Human Leukocyte Differentiation Antigens Workshop by flow cytometry and immunohistochemistry. Cell Immunol. 2005;236:6–16. doi: 10.1016/j.cellimm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 30.Ise T, Kreitman RJ, Pastan I, Nagata S. Sandwich ELISAs for soluble immunoglobulin superfamily receptor translocation-associated 2 (IRTA2)/FcRH5 (CD307) proteins in human sera. Clin Chem. Lab. Med. 2006;44:594–602. doi: 10.1515/CCLM.2006.115. [DOI] [PubMed] [Google Scholar]
- 31.Nagata S, Numata Y, Onda M, Ise T, Hahn Y, Lee B, Pastan I. Rapid grouping of monoclonal antibodies based on their topographical epitopes by a label-free competitive immunoassay. J Immunol Methods. 2004;292:141–155. doi: 10.1016/j.jim.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Maenaka K, van der Merwe PA, Stuart DI, Jones EY, Sondermann P. The human low affinity Fcgamma receptors IIa, IIb, and III bind IgG with fast kinetics and distinct thermodynamic properties. J. Biol. Chem. 2001;276:44898–44904. doi: 10.1074/jbc.M106819200. [DOI] [PubMed] [Google Scholar]
- 33.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 34.Ferrara C, Stuart F, Sondermann P, Brunker P, Umana P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J. Biol. Chem. 2006;281:5032–5036. doi: 10.1074/jbc.M510171200. [DOI] [PubMed] [Google Scholar]
- 35.Mimura Y, Sondermann P, Ghirlando R, Lund J, Young SP, Goodall M, Jefferis R. Role of oligosaccharide residues of IgG1-Fc in Fc gamma RIIb binding. J Biol. Chem. 2001;276:45539–45547. doi: 10.1074/jbc.M107478200. [DOI] [PubMed] [Google Scholar]
- 36.Stadlmann J, Weber A, Pabst M, Anderle H, Kunert R, Ehrlich HJ, Peter SH, Altmann F. A close look at human IgG sialylation and subclass distribution after lectin fractionation. Proteomics. 2009;9:4143–4153. doi: 10.1002/pmic.200800931. [DOI] [PubMed] [Google Scholar]
- 37.Guhr T, Bloem J, Derksen NI, Wuhrer M, Koenderman AH, Aalberse RC, Rispens T. Enrichment of sialylated IgG by lectin fractionation does not enhance the efficacy of immunoglobulin G in a murine model of immune thrombocytopenia. PLoS One. 2011;6:e21246. doi: 10.1371/journal.pone.0021246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasermann F, Boerema DJ, Ruegsegger M, Hofmann A, Wymann S, Zuercher AW, Miescher S. Analysis and functional consequences of increased Fab-sialylation of intravenous immunoglobulin (IVIG) after lectin fractionation. PLoS One. 2012;7:e37243. doi: 10.1371/journal.pone.0037243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: properties of a series of truncated glycoforms. Mol. Immunol. 2000;37:697–706. doi: 10.1016/s0161-5890(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 40.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 41.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, Bondarenko PV, Ricci MS, Dillon TM, Balland A. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol. Chem. 2008;283:16194–16205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu YD, Chen X, Enk JZ, Plant M, Dillon TM, Flynn GC. Human IgG2 antibody disulfide rearrangement in vivo. J Biol. Chem. 2008;283:29266–29272. doi: 10.1074/jbc.M804787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo EM, Wims LA, Chan LA, Morrison SL. Human IgG2 can form covalent dimers. J Immunol. 2003;170:3134–3138. doi: 10.4049/jimmunol.170.6.3134. [DOI] [PubMed] [Google Scholar]
- 44.van der Neut KM, Schuurman J, Losen M, Bleeker WK, Martinez-Martinez P, Vermeulen E, den Bleker TH, Wiegman L, Vink T, Aarden LA, De Baets MH, van de Winkel JG, Aalberse RC, Parren PW. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science. 2007;317:1554–1557. doi: 10.1126/science.1144603. [DOI] [PubMed] [Google Scholar]
- 45.Saphire EO, Stanfield RL, Crispin MD, Parren PW, Rudd PM, Dwek RA, Burton DR, Wilson IA. Contrasting IgG structures reveal extreme asymmetry and flexibility. J Mol. Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.