1. Introduction
The removal of the mRNA cap by the process of decapping is a critical step during the degradation of eukaryotic mRNAs. Concurrent with transcription, eukaryotic mRNAs are capped on the 5′ end with a 7-methyl-guanosine (m7G) cap. The m7G cap promotes post-transcriptional gene expression at multiple levels [1]. In the nucleus, the m7G cap associates co-transcriptionally with the nuclear cap-binding complex (CBC), which stimulates pre-mRNA splicing and mRNA export [2]. After nuclear export, the CBC is exchanged for translation initiation factor 4F (eIF4F), which plays a critical role in translation initiation of most mRNAs [3]. In addition to serving as a ligand for the cap binding proteins, the m7G cap protects mRNAs from nuclear and cytoplasmic 5′-to-3′ exoribonucleases [4]. Thus decapping of mRNA exposes the mRNA to degradation from the 5′ end and, in the cytoplasm, simultaneously shuts down translation initiation. Depending on the specific pathway, decapping can be the first, an intermediary, or the last step in mRNA decay [5,6,7]. Multiple decapping enzymes have been characterized which differ in their cellular localization and substrate specificities. In this review we will discuss current structural and functional insights into the mechanism and control of one of the key complexes in mRNA decapping, the highly conserved Dcp2 decapping machinery.
2. The Dcp1-Dcp2 decapping complex
The most well-characterized and widely conserved eukaryotic decapping enzyme is Dcp2. Dcp2 was originally identified as a decapping cofactor in the budding yeast Saccharomyces cerevisiae [8]. In subsequent biochemical studies, Dcp2 from human, budding yeast, the nematode Caenorhabditis elegans and the plant Arabidopsis thaliana was shown to have intrinsic decapping activity, releasing m7GDP from m7G-capped RNAs [9,10,11,12]. Consistent with its role in the 5′-to-3′ mRNA decay pathway, purified recombinant Dcp2 is unable to effectively hydrolyze unmethylated cap or free GTP, and shows poor activity on short m7G-capped oligonucleotides [13]. Dcp2 contains a Nudix domain (originally termed MutT) [8,9], which is found in enzymes that hydrolyze nucleoside diphosphates linked to other moieties [14], and is responsible for the catalytic activity of Dcp2 [15,16].
An essential cofactor for Dcp2 in vivo in budding yeast is Dcp1. Dcp1 was originally described as the catalytic decapping protein, and S. cerevisiae strains defective for Dcp1 are highly deficient in decapping [13]. Dcp2 directly interacts with Dcp1 [17,18] and, while recombinant yeast Dcp2 has intrinsic decapping activity in vitro, it is greatly stimulated by Dcp1 [10]. Though the Dcp1-Dcp2 complex is conserved among eukaryotes, metazoans may require additional factors to stimulate Dcp1-Dcp2 interaction [17,18].
2.1 Structural insights into the Dcp1-Dcp2 decapping complex
Dcp2 catalyzes cap hydrolysis in the 5′ to 3′ mRNA decay pathway in a divalent metal ion dependent reaction [19]. Sequence alignment reveals that the N-terminal region of Dcp2 is conserved while its C-terminal region is divergent containing anywhere from ~175 amino acids in human to nearly 700 amino acids in budding yeast [9]. The conserved N-terminal region of Dcp2 is featured by the presence of the Nudix fold which is flanked on both sides by two additional conserved regions known as Box A and Box B [9] (Figure 1A). Box A has been implicated in facilitating the interaction of Dcp2 with Dcp1 while Box B has been shown to be important in RNA binding [20,21]. This has recently been confirmed by NMR chemical shift perturbation experiments in which residues important for RNA binding have been mapped to Box B in the Nudix domain of S. cerevisiae Dcp2 [15].
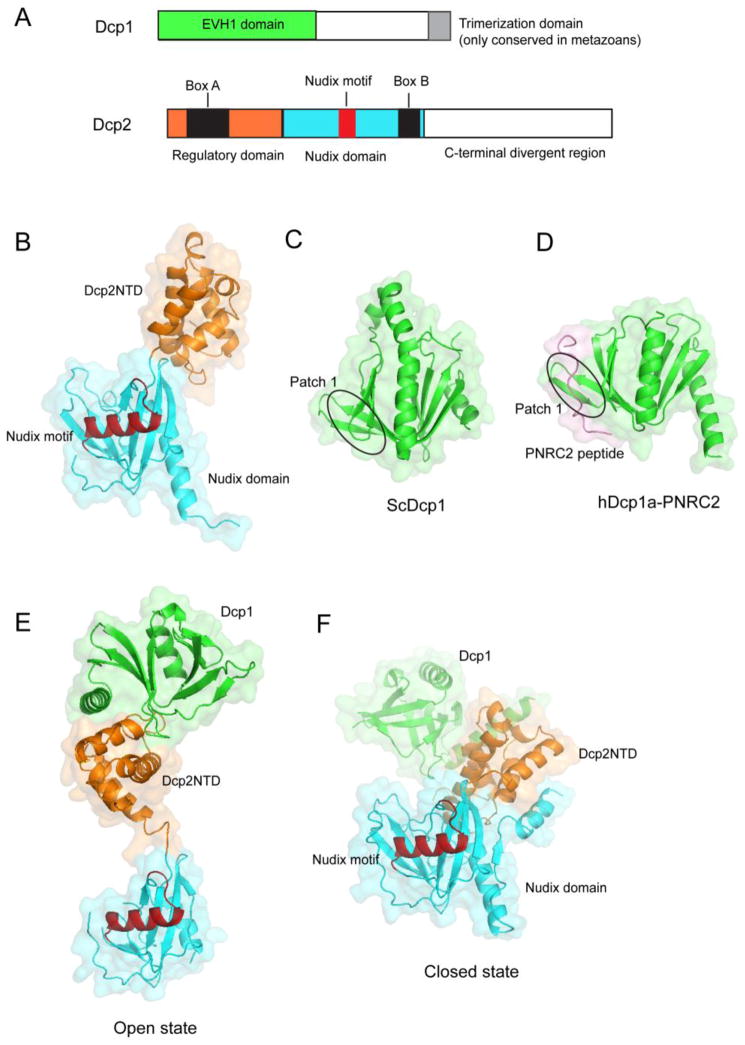
Figure 1. The crystal structures of Dcp1 and Dcp2 proteins.
(A) Schematic diagrams of the domain organization of Dcp1 and Dcp2 proteins. (B) The crystal structure of the S. pombe Dcp2 (residues 1–266) in the apo form. (C) Crystal structure of S. cerevisiae Dcp1 (green) with the patch 1 region marked. (D) Crystal structure of the EVH1 domain of human Dcp1a (green) in complex with the PNRC2 peptide (pink). (E) and (F) The open and closed conformations of the Dcp1-Dcp2 complex with Dcp1 in green, Dcp2 NTD in orange, Nudix domain in cyan and Nudix motif in red.
Crystallographic analyses revealed that the conserved N-terminal region of Dcp2 from the fission yeast Schizosaccharomyces pombe forms a bilobed architecture with an N-terminal regulatory domain (NTD) preceding a classic Nudix domain [21] (Figure 1B). Box A lies in the NTD while the Nudix fold together with Box B constitute the Nudix domain characterized by the canonical α/β/α sandwich structure [21]. The Nudix motif forms the catalytic center of Dcp2 by acting through conserved glutamate residues within this motif. These glutamate residues are required for coordination of the divalent Mn2+ orMg2+ ion during the catalytic reaction [10,20]. The NTD alone has no detectable decapping activity in vitro but can affect the decapping efficiency of Dcp2 and is indispensable for decapping in vivo [21]. Moreover, the NTD mediates the binding of Dcp2 to Dcp1 and is required for Dcp1 to stimulate Dcp2 activity.
Dcp2 is an RNA binding protein and prefers longer RNA substrates for efficient decapping [10,16,20]. Structural analysis of the Dcp1-Dcp2 complex reveals that the RNA substrate is bound in a channel on the surface of Dcp2 with the cap structure in the active site and the body of the RNA wrapping across the Nudix domain and a channel along the Box B. The minimum length of the RNA substrate was predicted to be 12 nucleotides for efficient binding to Dcp2 [17]. The requirement for a longer RNA in addition to the cap structure for substrate recognition by Dcp1-Dcp2 has been suggested to prevent accidental decapping of translating mRNAs on which translation initiation complexes are assembled [19].
Dcp1 is a small protein containing an EVH1 domain [22], which generally serves as a protein-protein interaction module [23]. Three conserved patches have been identified on the crystal structure of yeast Dcp1 with patch 1 structurally corresponding to the PRS recognition site of EVH1 domains in other proteins [24,25,26,27] (Figure 1C). Mutations of the conserved residues in patch 1 do not affect either the physical interaction of Dcp1 with Dcp2 or the decapping activity of the Dcp1-Dcp2 complex in vitro [22]. Instead, patch 1 serves as a binding site for a subset of enhancers of decapping (see below). In addition to the well-conserved EVH1 domain, Dcp1 contains a C-terminal trimerization domain which is conserved in metazoans but absent in fungi (Figure 1A). Crystal structures of the Dcp1-trimerization domain from human and Drosophila melanogaster reveal an antiparallel assembly comprised of three kinked alpha-helices [28]. Mutations that disrupt trimerization prevent Dcp1 from being incorporated into active decapping complexes and impairs mRNA decapping in vivo [28]. This finding reveals an unexpected connection and complexity of the mRNA decapping network in metazoans.
The cocrystal structure of the S. pombe Dcp1-Dcp2 complex (Figures 1E and 1F) shows that Dcp2 binds to Dcp1 through the Dcp2 NTD and that the three conserved patches in Dcp1 are not involved in Dcp2 binding [17]. Instead, Dcp1 uses the N-terminal helix α1 and the loop between β5-β6 that are only conserved in yeast species to recognize Dcp2. Consistent with this observation, mutations in S. cerevisiae Dcp1 that affect decapping in vivo are mapped to the N-terminal helix of Dcp1 [29] and yeast two-hybrid assays revealed that mutations in the N-terminus of the yeast Dcp1 disrupt its interaction with Dcp2 and hence decapping [17]. These results explain why stable, direct interactions between Dcp1 and Dcp2 in human or nematode are not observed [12,18] and why additional factors such as Edc4 (also called Hedls or Ge-1, or VARICOSE in Arabidopsis) stimulate the interaction between Dcp1 and Dcp2 in higher eukaryotes [30,31].
The S. pombe Dcp1-Dcp2 complex was shown to exist in open and closed conformations (Figures 1E and 1F) within the crystal, both of which were confirmed to be functionally relevant states as their existence in solution was verified by small angle X-ray scattering (SAXS) [17]. In the open or extended conformation, Dcp2 in the complex resembles the structure of the apo-Dcp2 [21] and adopts a dumbbell conformation. In contrast, in the closed complex Dcp1 and the Dcp2 NTD were shown to be in close proximity to the Nudix domain to form a compact structure. Structural and biochemical data indicated that the closed complex is, or closely resembles, the catalytically more active form of the enzyme [17]. Most recently, NMR spectroscopy [32] showed that Dcp2 exists in a conformational equilibrium in solution between the open and closed states in the absence of ligand, and the open-to-closed conformational switch is mediated by a gatekeeper tryptophan (Trp43). Further enzyme kinetics and NMR experiments [33] revealed that Trp43 promotes formation of the composite active site (see below) and allows Dcp1 and bound coactivators to enhance the catalytic step.
Several lines of evidence suggest that the conformational switch between open and closed states is required for efficient decapping. First, Dcp1 and the NTD of Dcp2 are essential for decapping in budding yeast [21,34]. Second, mutations in prolines of the interdomain linker block closure, hinder decapping in vitro, and cause an accumulation of Dcp2 in P bodies (see section 5) [21]. Third, the NTD of Dcp2 and Dcp1 together contribute a factor of 1,000 to the catalytic step [15,20] but are distinctly located from the active site in the open state of the Dcp1-Dcp2 complex [17]. Kinetic analyses indicate that Dcp1 affects the chemistry of the decapping reaction but not substrate binding [15]. Structural analysis suggests that Dcp1 activates Dcp2 by promoting and/or stabilizing the formation of the closed complex. Mutation of residues that block the formation of the closed complex result in the loss of decapping activity and the inability of Dcp1 to stimulate Dcp2 [17], thus confirming that efficient activation of Dcp2 by Dcp1 requires formation of the closed complex. However, how closure of the Dcp1-Dcp2 complex is linked to efficient cap hydrolysis remains elusive. Recently, Gross and colleagues [33] showed that cap recognition involves both the NTD and Nudix domains of Dcp2. Mutations in the cap-binding site on the NTD block closure and retard the catalytic step by two orders of magnitude whereas Dcp1 enhances the catalytic step by a factor of 10 and promotes closure, likely by stabilizing the closed conformation. These findings suggest that the conversion to the active form of Dcp2 is driven through specific cap recognition by the NTD. The resultant active form of Dcp2 harbors a composite active site with both domains sandwiching the cap.
3. Catalytic enhancers of decapping
The Dcp1-Dcp2 decapping complex is stimulated by multiple enhancers of decapping. The first enhancers of decapping to be described were the S. cerevisiae proteins Enhancer of decapping (Edc) 1 and Edc2, which were identified as high copy suppressors of Dcp1 and Dcp2 mutants, though deletion of Edc1 or Edc2 did not impair in vivo mRNA decay rates of tested reporter mRNAs [35]. Subsequent studies showed that Edc1 and Edc2 bind directly to RNA and can stimulate decapping in vitro [36].
No homologs of Edc1 and Edc2 have been identified in eukaryotes outside of budding yeast. By contrast, another enhancer of decapping, Edc3, is highly conserved amongst eukaryotes. Edc3 directly interacts with the Dcp1-Dcp2 complex [37]. Yeast Edc3 stimulates the decapping reaction in vitro [38,39] and yeast strains deleted for the Edc3 gene have decreased decapping rates [40]. Another enhancer of decapping, Edc4, appears to be restricted to metazoans [30,41]. The specific mechanism by which Edc4 stimulates decapping is unknown, but human and Arabidopsis Edc4 promotes the association between Dcp1 and Dcp2, and stimulates decapping by Dcp2 in vitro [30,31].
Another enhancer of decapping, Pat1, was initially identified in a yeast mutational screen as a protein that stimulates mRNA decapping [42], and later found to interact directly with Dcp1 and stimulate decapping by recombinant Dcp1-Dcp2 in vitro [39,43]. Pat1, and its orthologs in human and Drosophila, has been found in association with multiple decapping factors [43,44,45]. One of these is another enhancer of decapping, the Lsm1-7 complex [39]. In S. cerevisiae, Lsm1-7 associates with mRNAs that have undergone deadenylation [46], which is generally associated with activation of decapping [47]. Consistent with a role in deadenylation-mediated decapping, lsm1Δ and pat1Δ S. cerevisiae strains show an accumulation of deadenylated mRNA [43]. The purified Lsm1-7-Pat1 complex has intrinsic affinity for the 3′ end of oligoadenylated mRNAs over polyadenylated mRNAs and binds preferentially to deadenylated mRNAs carrying a U-tract at their 3′ terminus over those that do not [48]. Another enhancer of decapping, the DEAD box superfamily 2 helicase Dhh1 (called DDX6 or Rck/p54 in human and Me31B in Drosophila) was first identified as a decapping activator in S. cerevisiae as its depletion leads to the accumulation of capped reporter mRNA decay intermediates [49,50,51]. Dhh1, and its orthologs in other eukaryotes, has been identified in complex with multiple decapping factors, including Dcp2, Edc3 and Pat1 [39,44,45].
An important unresolved question is whether enhancers of decapping are globally involved in cellular decapping, or whether they act in mRNA-specific manners. Microarray studies analyzing mRNA expression in Drosophila S2 cells depleted for Dcp1, Edc3, or Edc4 indicated that Edc4 and Dcp1 generally regulate the same mRNAs, whereas only a subset of those mRNAs were regulated by Edc3 [52]. Thus, some enhancers of decapping might be specific for a subset of decapping substrates, whereas others may act more generally.
3.1 Structural insights into enhancers of decapping
Multiple structural studies have provided insights into the mechanisms by which enhancers of decapping stimulate the Dcp1-Dcp2 complex. The budding yeast decapping coactivators Edc1 and Edc2 were shown to enhance decapping activity of the Dcp1-Dcp2 complex by a 1,000-fold through interaction with the EVH1 domain of Dcp1 [53]. These findings suggest that a crucial function of Dcp1 is to couple the binding of coactivators to substrate recognition and activation of Dcp2. Consistent with this notion, crystal structure of human Dcp1a in complex with proline-rich nuclear receptor coactivator 2 (PNRC2), a protein that stimulates decapping in the nonsense-mediated decay (NMD) pathway, shows that the proline-rich region of PNRC2 is bound to the EVH1 domain of Dcp1a with patch 1 mediating their interaction [54] (Figure 1D). Structural and biochemical analyses of the human Dcp1a-PNRC2 complex [54] showed that both PNRC2 and Dcp1a in isolation stimulate decapping, and that PNRC2 works in synergy with Dcp1a to promote the decapping activity of Dcp2. In contrast to the mode of action of Edc1 and Edc2 [53], which stimulate decapping through Dcp1-mediated recruitment to Dcp2, PNRC2 stimulates decapping through direct binding to Dcp2. These results indicate that PNRC2 is a bona fide decapping coactivator in addition to its adaptor role in NMD.
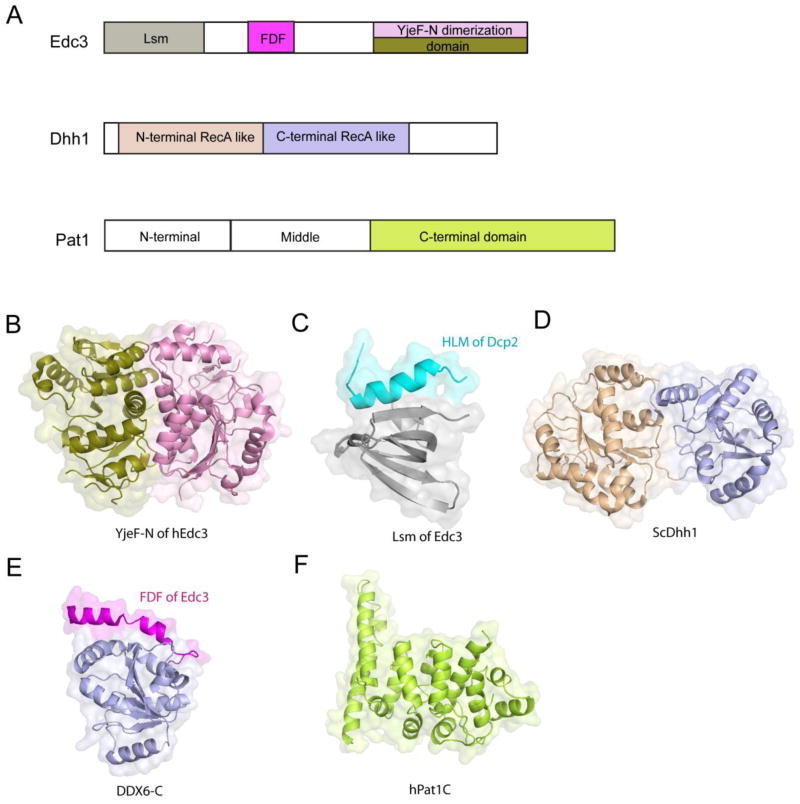
The conserved enhancer of decapping Edc3 consists of an N-terminal divergent Lsm domain, a central FDF domain and a C-terminal YjeF-N domain [55,56] (Figure 2A). The N-terminal Lsm domain of Edc3 adopts a noncanonical Sm fold, which lacks the characteristic N-terminal α helix and remains monomeric in solution [57]. The crystal structure of human Edc3 revealed that the FDF domain is unstructured and the YjeF-N domain adopts a divergent Rossmann fold that forms a dimer [58] (Figure 2B). Residues involved in Edc3 dimerization are highly conserved in eukaryotes. Structure-based mutagenesis demonstrates that Edc3 dimerization is of functional importance for RNA binding, P-body formation and possibly degradation of Rps28b mRNA, a specific substrate of Edc3 [58].
Figure 2. The structures of the enhancers of decapping Edc3, Dhh1 and Pat1.
(A) Schematic diagrams showing the domain organization of Edc3, Dhh1 and Pat. (B) The crystal structure of YjeF-N dimerization domain from human Edc3 with each subunit colored in pink and deep olive, respectively. (C) The solution structure of the Lsm domain of Edc3 (gray) in complex with the helical leucine-rich motif (HLM) of Dcp2 (cyan). (D) The crystal structure of yeast Dhh1 showing two Rec-A like domains with the N-terminal domain in brown and the C-terminal domain in light blue. (E) The crystal structure of DDX6 C-terminal Rec-A like domain (the ortholog of yeast Dhh1, shown in light blue) in complex with the FDF motif of Edc3 (magenta). (F) The crystal structure of human Pat1 C-terminal α-α superhelical domain.
S. cerevisiae Scd6 (Lsm13), vertebrate RAP55 (Lsm14), D. melanogaster Trailer Hitch (Lsm15 or Tral), and C. elegans CAR-1 belong to the Scd6 family of proteins and share a number of sequence and functional features with Edc3 [55,56,57,59,60]. However, in contrast to Edc3, Scd6 shows only mild stimulation of the Dcp1-Dcp2 complex in vitro [37,61]. More recently, the structure of yeast Edc3 Lsm domain in complex with a short helical leucine-rich motif (HLM) from Dcp2 was determined [37] (Figure 2C). The HLM motif is located close to the C-terminal end of the Nudix domain and identification of its interaction with Edc3 highlights the regulatory role of Edc3 in decapping. Structure based sequence analysis of the C-terminal unstructured extension of S. cerevisiae Dcp2 reveals additional HLMs, which can also interact with Edc3 and are essential for the localization of the Dcp1-Dcp2 complex to P-bodies [37]. Not surprisingly, the Scd6 Lsm domain competes with Edc3 for the interaction with these HLMs [37]. Unexpectedly, in contrast to yeast, the HLM that mediates Edc3 binding is present in metazoan Dcp1 but absent from Dcp2 [37].
Crystal structure of S. cerevisiae Dhh1 (Figure 2D) shows that it consists of two RecA-like α/β domains with a unique arrangement in such a way that the two domains are linked through the interactions of the canonical helicase motifs [62]. Electrostatic potential mapping combined with mutagenesis revealed that motifs I, V, and VI are involved in RNA binding. Limited proteolysis of Dhh1 suggested that ATP binding enhances an RNA-induced conformational change. However, how the conformational change of Dhh1 upon ATP and RNA binding affects the decapping activity of the Dcp1-Dcp2 complex remains elusive. One possibility is that Dhh1 utilizes the energy of ATP hydrolysis to unwind RNA with secondary structures and/or to displace bound translation factors, thereby facilitating access of the Dcp1-Dcp2 complex to the cap structure of the mRNA substrate.
A previous study showed that fragments of Edc3 and Tral proteins covering the FDF motifs can coimmunoprecipitate with the C-terminal RecA-like domain of Me31B, the Drosophila ortholog of S. cerevisiae Dhh1 [60]. The crystal structure of the C-terminal RecA-like domain of DDX6 (Figure 2E), the human ortholog of Dhh1, in complex with the FDF motif of Edc3 [59] shows that the FDF motif adopts an α-helical conformation upon binding to DDX6, occupying a shallow groove opposite to the DDX6 surface involved in RNA binding and ATP hydrolysis. The interaction of DDX6 with the Edc3 FDF motif is required for its P body localization and its role in translation repression [59] (see below). Similarly, Tral also employs its FDF motif to interact with Me31B, thus competing the interaction of Edc3 with Me31B in a mutually exclusive manner [59]. The biological significance of this competitive interaction remains to be characterized. Conceivably, the competition between Tral and Edc3 to associate with DDX6/Me31B may facilitate the formation of distinct decapping complexes that have different functions and/or target distinct sets of mRNAs.
The Lsm1-7-Pat1 complex plays important roles in mRNA decapping. Recently, a crystal structure of the C-terminal domain of human Pat1 (Pat1-C), which is the only structured domain of Pat1, was determined. Pat1-C folds into an α-α superhelix [63] (Figure 2F). Structure-based mutagenesis shows that a conserved and basic surface patch of Pat1-C is required for its binding to RNA, Dcp2, Edc4 and Lsm1-7 [63]. While the Lsm1-7 complex is predicted to form a heteroheptameric ring [43,64,65,66,67], its subunits may form various subcomplexes. A crystal structure of Lsm3 from S. cerevisiae shows that it forms an octameric ring structure [68]. Recent crystallographic analysis combined with analytical ultracentrifugation of Lsm3, Lsm4 and a Lsm5/6/7 subcomplex from S. pombe indicate that these exist in solution as a heptamer, a monomer and a hexamer, respectively [69,70]. Moreover, like Lsm3 alone, the S. pombe Lsm2/3 subcomplex is also in a heptameric state in solution. Given that subcomplexes or individual subunits of Lsm1-7 proteins have a propensity to form oligomers [68,69,70], it is unclear how the seven Lsm proteins assemble into the ring structure and whether subcomplexes exist in vivo as functional units. In addition, how Pat1 interacts with the Lsm1-7 complex and how this hetero-octameric complex modulates decapping remains to be understood.
3.3 Stimulation of mRNA decapping through mRNP remodeling leading to repression of translation initiation
The mRNA m7G cap is generally not free, but bound in the nucleus by the CBC and in the cytoplasm by the eIF4F complex. The eIF4E cap-binding subunit of eIF4F can inhibit decapping by yeast Dcp2 in vitro [71,72], and eIF4G, which stimulates eIF4E binding to the cap, can further decrease the in vitro decapping rate [73]. This suggests that the translation initiation machinery can protect mRNA from decapping and that a competition between the translation initiation complex and decapping exists. Evidence supporting this can be seen in vivo with yeast strains harboring defective translation initiation factors. Expression of mutant translation initiation factors, including eIF4E and eIF4G, which slow the rate of translation, generally increase the rate of mRNA decay [74]. Thus, an important mechanism for enhancing decapping likely involves remodeling of the translation initiation complex associated with the mRNA 5′ end to allow access for Dcp2 to the m7G cap. Indeed, a subset of enhancers of decapping also act as repressors of translation.
One such decapping factor that functions as a translational repressor is Pat1. Budding yeast strains lacking Pat1 are unable to effectively inhibit translation in response to glucose deprivation, and overexpression of Pat1 leads to general translation repression [75]. These observations suggest that in addition to playing a possible role in directly recruiting or activating the decapping complex, Pat1 may promote decapping by stimulating remodeling or release of translation initiation components. Interestingly, in budding yeast Pat1 shows RNA-dependent association with the translation initiation factors eIF4E and eIF4G, as well as with the poly(A)-binding protein Pab1 [46]. By contrast, the Lsm1-7 complex fails to associate with eIF4E and eIF4G, even in strains lacking Dcp1, Dcp2, or Pat1 [46]. This is consistent with the idea that the association of the Lsm1-7 complex with mRNPs is accompanied by an eIF4F release step distinct from the catalytic activity of Dcp2. Recent observations that the human [45,76] and Drosophila [44] homologs of Pat1 also exists in complex with decapping factors, including Dcp2 and the Lsm1-7 complex, and stimulate decay of a tethered reporter mRNA, shows that some of these functions have been conserved throughout evolution. Human and Drosophila Pat1, unlike the budding yeast homolog, also interact with deadenylation factors in an RNA-independent manner, suggesting a function in both deadenylation and decapping [44,45]. Collectively, these observations suggest a possible role of the Pat1-Lsm1-7 complex in the release of translation initiation factors in preparation for decapping. However, given their link to deadenylation, an alternative possibility is that the release of eIF4F occurs as a secondary consequence of removal of the poly(A)-binding protein, which is known to stimulate cap-binding by the eIF4F complex [77].
Another enhancer of decapping, the DEAD box helicase Dhh1, may play an important role in mRNP remodeling. Dhh1 deletion and overexpression in budding yeast has similar effectsas those observed for Pat1, namely decreased and increased translational repression, respectively [75], and Dhh1 inhibits translation in vitro [75]. The exact mechanism by which Dhh1 represses translation and how this relates to enhancement of decapping is not fully understood. In yeast, the effect of Dhh1 on translation may be to inhibit formation of a stable 48S translation preinitiation complex [75], or it may function to slow the movement of ribosomes along the mRNA [78]. Surprisingly, tethered Dhh1 is able to repress translation and stimulate decay in yeast even when its ATPase activity is abolished, suggesting a conformational change may not be necessary for these activities [79]. In vitro studies have shown that mutant DDX6, the human ortholog of Dhh1, deficient in ATP hydrolysis can relax structured RNA in the presence of ATP but not ADP, suggesting that ATP hydrolysis may trigger a switch from an active to an inactive state [80]. DDX6 function may also depend on its ability to oligomerize on mRNA, which has been observed in cells from both human and the frog Xenopus laevis [80,81]. In yeast, decapping of long non-coding (lnc) RNAs, which are capped and polyadenylated, is mediated by Dcp2 but unaffected by deletion of Dhh1 or Lsm1 suggesting these cofactors are not needed for decapping of these non-translated RNA species [82].
A number of decapping enhancers can also be found in complexes that appear to promote translation repression in the absence of decapping. For example, in D. melanogaster, C. elegans, and X. laevis the Dhh1 orthologs Me31B, CGH-1, and Xp54, respectively, play important roles in translational repression and storage of mRNAs during development of the oocyte [81,83,84]. In D. melanogaster this involves a complex that also contains the eIF4E-binding protein CUP [83], and competitive binding of Me31B by either Trailer Hitch (Tral) or Edc3 may determine if an mRNA is translationally repressed or decapped [60]. Contrasting the activity of Dhh1 in yeast, Xp54 ATPase mutants stimulate the translation of a tethered reporter in X. laevis oocytes [85]. Homologs of these proteins have also been characterized in complex with decapping factors in other eukaryotes (see section 3.1), but what dictates which complexes form on mRNPs under different conditions remains unclear.
Thus, multiple lines of evidence support that decapping complexes interfere with translation initiation. However, while decapping is generally in competition with translation initiation, evidence suggests that decapping can occur while mRNAs are still engaged with elongating ribosomes [86,87]. The specific mechanism by which decapping factors promote remodeling of translation initiation complexes, and what dictates whether complexes containing decapping factors repress translation only, or also activate decapping, remains to be answered.
4. Cis-elements and trans-factors that promote mRNA-specific decapping by Dcp2
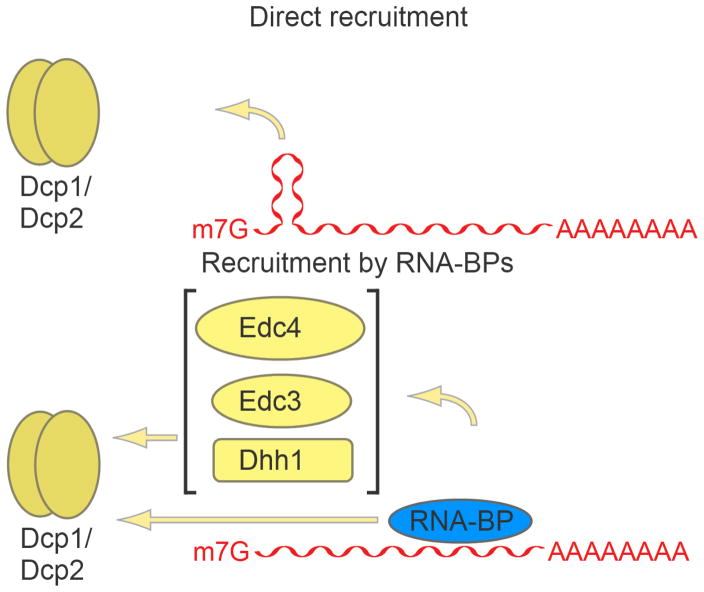
Though decapping factors can stimulate the function of Dcp2 to initiate decapping and eventual 5′-to-3′ decay of mRNA substrates, regulation of Dcp2 is important to control which transcripts are decapped and degraded. One mechanism by which decapping by Dcp2 is thought to be regulated is through substrate-dependent recruitment. Several ways by which Dcp2 can be enlisted to decap specific substrates have been uncovered, including direct recruitment of Dcp2 to mRNA cis-elements, and recruitment via RNA binding proteins that interact with Dcp2 or other members of the decapping complex (Figure 3).
Figure 3. General principles of decapping complex recruitment.
RNA cis-elements, such as the hairpin structure in Rrp41 mRNA, can recruit Dcp2 directly. RNA binding proteins (RNA-BPs) can stimulate decapping by recruiting the Dcp1-Dcp2 complex directly or through decapping enhancers.
The observation that Dcp2 has a preference for capped RNA substrates over free cap raised the possibility that Dcp2 has a preference for specific RNA sequences [20]. Consistent with this, the mRNA encoding the human exosome component Rrp41 was found to contain an element in the 5′UTR which stimulates Dcp2 binding and rapid decapping when in close proximity to the mRNA cap [5]. This region was predicted to have a stem-loop structure and compensatory mutational analysis suggested that the secondary structure is important for Dcp2 recruitment and activation [88]. An interesting question is if this direct recruitment of Dcp2 to an mRNA is a widespread mechanism of mRNA decay regulation. A genome-wide analysis predicted hundreds of human mRNAs that could harbor similar stem loop structure in their 5′UTRs, and several of these showed enhanced decapping in vitro by Dcp2 [88]. Contrasting this, in C. elegans 70% of mRNA transcripts contain a 22-nucleotide splice leader sequence, which reduces their in vitro decapping rate compared to transcripts without the splice leader [12]. The sequence is not predicted to have a strong secondary structure and the mechanism by which decapping is inhibited is unknown. This is independent of the non-canonical trimethylated m2,2,7G cap found on trans-spliced C. elegans mRNAs, which is effectively hydrolyzed by budding yeast, human, and nematode Dcp2 [12].
In addition to direct RNA recruitment, the decapping machinery can be recruited to target mRNAs through RNA binding proteins. For example, as discussed in section 3.1, evidence suggests that the Lsm1-7-Pat1 complex serves a general role in recruiting the decapping complex to deadenylated mRNAs, in what is thought to be a major pathway of mRNA decay in eukaryotes [47]. More recent studies have also presented evidence that the Lsm1-7 complex serves to recruit decapping factors to RNAs, including histone mRNAs, that have undergone tailing by unconventional terminal ribonucleotidyltransferases [89,90].
Additionally, multiple examples exist for mRNA-specific RNA binding proteins that activate the decapping complex. For example, in S. cerevisiae, ribosomal protein (rp) S28b autoregulates its own mRNA by binding to the 3′ UTR and activating decapping independently of deadenylation, through interaction of rpS28b with Edc3 and Dcp1 [91]. Another example in budding yeast is the Puf5 protein, a member of the PUF family of proteins that generally target substrate mRNAs for translational repression and/or mRNA decay [92,93]. Puf5 associates with the deadenylase Caf1/Pop2 and the decapping factor Dcp1 [94], and stimulates the deadenylation and decapping of target mRNAs [95].
In mammalian cells, multiple trans-factors are involved in degrading mRNAs containing destabilizing 3′ UTR AU-rich elements (AREs) [96]. One such protein is tristetraprolin (TTP), which interacts with multiple mRNA decay factors [97,98,99], including Edc3 and Dcp2 components of the decapping complex, and stimulates the decapping activity of Dcp2 in vitro on ARE-containing RNA [30]. There is also evidence to suggest that decapping plays a role in mRNA silencing by micro (mi)RNAs. Experiments in Drosophila S2 cells indicated that depletion of Dcp2 resulted in an increase in both steady state miRNA-target reporter mRNA levels and translation [100,101]. In murine embryonic fibroblast cells hypomorphic for Dcp2, no distinguishable defect in silencing of a miRNA reporter mRNA was observed, unless the cells were also depleted for another cytoplasmic decapping enzyme, Nudt16 [102]. In human cells, DDX6 interacts with the miRNA effectors Ago1 and Ago2 in an RNA independent manner, and depletion of DDX6 diminishes repression of a miRNA target reporter [103]. Decapping is also activated in the nonsense mediated decay (NMD) pathway. The central factor in NMD, Upf1, interacts with Dcp2 and activates decapping [18,104]. In yeast, depletion of the decapping factor Dcp1 leads to a major defect in NMD [105,106]. In human cells, the interaction of Upf1 with Dcp2 is bridged by the protein PNRC2 [107], and appears to serve as one of many mechanisms by which Upf1 can initiate mRNA degradation, which also includes endonucleolytic cleavage and deadenylation [108,109].
While a number of examples of recruitment of the decapping machinery by trans-factors have been documented, in most cases it remains to be established how the trans-factors communicate with decapping complexes (Figure 3). Do they all communicate with a common decapping holoenzyme complex, or are there multiple inroads into the decapping machinery, for example through alternative decapping sub-complexes all leading to decapping by Dcp2? Moreover, in most cases, how critical decapping is as compared to the two other mechanisms of initiation of mRNA decay, deadenylation and endonucleolytic cleavage, remains poorly understood. Finally, could mechanisms other than mRNA-specific recruitment contribute to substrate specificity by the decapping complex, such as mRNP remodeling events that expose the mRNA cap?
5. mRNA Decapping and P Bodies
Dcp2 and many of its enhancers and cofactors can be seen to accumulate in discrete foci in the cytoplasm termed P bodies [110], which are conserved across eukaryotes [111,112]. Accumulating evidence suggests that P bodies are self-assembling RNA granules that primarily assemble from RNA decay intermediates that accumulate with decapping factors [111] (Figure 4). Consistent with this, the depletion of factors that activate mRNA decay prior to the decapping step, such as deadenylases, reduces the number of visible P bodies in cells, while depleting cells of factors, such as Dcp1 or Xrn1, that enhance decapping or the subsequent 5′-to-3′ decay step generally increases the size or number of detectable P bodies [75,110,113]. Moreover, mRNA decay intermediates have been localized to P bodies, and RNase treatment disrupts P bodies, suggesting that RNA is a necessary component for P body integrity [113,114,115,116]. In budding yeast, the decapping stimulator Edc3 and the Lsm4 subunit of the Lsm1-7 complex play critical roles in assembling mRNPs into P bodies, likely by promoting intermolecular contacts between mRNPs assembled with decapping complexes [49,117]. Ribosomal proteins, poly(A)-binding protein, and translation initiation factors, with the exception of eIF4E, are absent from P bodies [118,119], and translation elongation inhibitors generally rapidly disassemble P bodies [120], suggesting that mRNPs that assemble into P bodies need to be free of ribosomes. While P bodies may primarily consist of intermediates of mRNA decay, pulse transcribed reporter mRNAs that accumulate in P bodies during glucose starvation in budding yeast have been observed to reenter translation after glucose restoration, suggesting that decay is not the only fate for mRNAs that accumulate in P bodies [118]. Despite ongoing interest and research on P bodies, and despite their conservation in all tested eukaryotes, their function remains unclear, and there is currently little evidence that the assembly of RNA decay intermediates into P bodies plays a rate-limiting role in mRNA degradation.
Figure 4. P body dynamics.
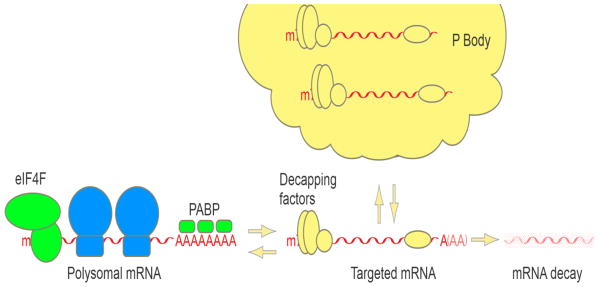
P bodies assemble from translationally repressed, ribosome-free, mRNPs associated with decapping factors. mRNPs assembled into P bodies are either targets of mRNA decay, or can return to translation.
6. Additional decapping enzymes
In addition to Dcp2, there are other eukaryotic proteins capable of hydrolyzing the mRNA cap. While Dcp2 was believed to be the only factor that catalyzed decapping in the 5′-to-3′ mRNA decay pathway, a hypomorphic Dcp2 mouse showed no reduction in mRNA decay rates in cells where Dcp2 was no longer detectable [121]. By testing the ability of other mammalian Nudix domain-containing proteins to hydrolyze capped RNA, Nudt16 was discovered to have decapping activity similar to Dcp2 [121]. Nudt16 is the mammalian ortholog of the X. laevis protein X29, which was initially described to hydrolyze the m7G cap of the U8 snoRNA and may function in the turnover of other nuclear capped RNAs [122]. Interestingly, unlike its Xenopus homolog, mammalian Nudt16 appears to be primarily cytoplasmic [121]. Further studies in mammalian cells suggest that Nudt16 and Dcp2 may be responsible for regulating distinct subsets of mRNAs with some shared targets [102].
In human cell lysates, a decapping activity was found to cosediment with the 3′-to-5′ degradation machinery, the exosome. This activity, termed DcpS (Dcp Scavenger), is able to hydrolyze free m7GpppG cap, which is the product of a complete mRNA degradation reaction by the exosome [123]. By contrast, DcpS shows no activity towards unmethylated GpppG. Contrasting with the Dcp2 reaction, which has a preference for long capped substrates and releases m7GDP, the product of the scavenger decapping reaction is m7GMP and DcpS is most effective in hydrolyzing free cap, though it shows some decapping activity on substrates of up to ten nucleotides [7].
Further decapping enzymes exist that may perform specialized functions rather than act as part of general mRNA decay pathways. The yeast factor Rai1, which exists in complex with the nuclear 5′-to-3′ exonuclease Rat1 [124], is able to remove unmethylated GpppG from RNA substrates in vitro, and Rai1Δ strains accumulate aberrantly capped mRNA substrates under certain stress conditions [125], suggesting that Rai1 has a role in a nuclear capping quality control mechanism. Recently, a yeast homolog of Rai1 has been identified that appears to have both decapping and exonucleolytic activity and was named Dxo1 [126]. Rai1Δ/Dxo1Δ double deletion mutants show an increase in endogenous aberrantly capped mRNA, but normal levels of properly capped mRNA, suggesting that like Rai1, Dxo1 might function in capping quality control [126]. Recent work in humans has provided evidence that also Dcp2 has a role in nuclear decapping, in which it in complex with Xrn2, the human ortholog of Rat1, promotes premature transcription termination by activating 5′-to-3′ degradation of nascent transcripts [127].
7. Looking forward at decapping
Biochemical, cellular, and structural analyses have led to the identification and characterization of many of the central components involved in mRNA decapping allowing for the understanding of the basic principles of cytoplasmic decapping. While a lot has been learned in recent years about the mechanisms and regulation of these factors, there are many interesting open questions that need to be addressed. For example, although the structures of S. pombe Dcp2 in the apo form and in complex with Dcp1 have been determined, how the 5′ cap of the mRNA substrate is recognized and cleaved by Dcp2 still remains unclear. Because cap recognition occurs during the catalytic step and requires the presence of the RNA body, a high resolution crystal structure of Dcp2, or the Dcp1-Dcp2 complex, with a bound capped RNA would be required to elucidate the mechanism of cap recognition. In addition, what are the mechanisms by which the enhancers of decapping stimulate the decapping activity of Dcp2? Since the assembly of the decapping complex is a dynamic and complex process and decapping enhancers may use different mechanisms to bind Dcp2 and activate decapping, an important area of future study will be directed towards understanding the interactions of various decapping enhancers with Dcp2 and their functional significance for regulating mRNA decapping. In addition to these structural questions, there are a number of important functional questions for future study. For example, studies in D. melanogaster, C. elegans, and X. laevis have identified mRNPs containing decapping factors that appear to repress translation and store mRNA, rather than targeting them for decay. What dictates whether an mRNA is stored or degraded and how are decapping factors involved in both of these seemingly contrasting functions? There is substantial evidence that suggests translation initiation and decapping are in competition. How is the mRNP remodeled to allow for translational repression and access to the 5′ cap by Dcp2? While decapping was long thought to be an irreversible process that triggered the rapid decay of mRNA, the recent identification of a cytoplasmic capping complex has challenged this view [128,129]. Howwidespread is cytoplasmic capping and is cap homeostasis an important mechanism for translation regulation? P bodies are conserved cytoplasmic foci containing many components of the decapping machinery as well as mRNA in various states of decay. What role do P bodies play in translational repression, mRNA decapping and decay, and what determines their composition and formation? Nudt16 has been identified as a nuclear and cytoplasmic factor capable of decapping and its function appears to at least partially overlap with Dcp2. What role does Nudt16 play in mRNA decapping and what factors regulate its function? Insights into these and other questions regarding decapping will allow us to better understand this important mechanism of eukaryotic post-transcriptional gene regulation.
Highlights.
Decapping inactivates translation initiation and initiates 5′-to-3′ decay of mRNA.
The Dcp1-Dcp2 decapping complex is widely conserved among eukaryotes.
An open-to-closed transition controls the activity of the Dcp1-Dcp2 complex.
Multiple enhancers of decapping promote decapping by different mechanisms.
mRNAs that accumulate with decapping complexes can assemble into P bodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell. 2010;39:950–62. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–9. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 3.McKendrick L, Pain VM, Morley SJ. Translation initiation factor 4E. Int J Biochem Cell Biol. 1999;31:31–5. doi: 10.1016/s1357-2725(98)00129-0. [DOI] [PubMed] [Google Scholar]
- 4.Stevens A. An exoribonuclease from Saccharomyces cerevisiae: effect of modifications of 5′ end groups on the hydrolysis of substrates to 5′ mononucleotides. Biochem Biophys Res Commun. 1978;81:656–61. doi: 10.1016/0006-291x(78)91586-3. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Song M-G, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol. 2008;28:939–48. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–22. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–8. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–8. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki S, Takeda A, Motose H, Watanabe Y. Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 2007;581:2455–9. doi: 10.1016/j.febslet.2007.04.051. [DOI] [PubMed] [Google Scholar]
- 12.Cohen LS, Mikhli C, Jiao X, Kiledjian M, Kunkel G, Davis RE. Dcp2 Decaps m2,2,7GpppN-capped RNAs, and its activity is sequence and context dependent. Mol Cell Biol. 2005;25:8779–91. doi: 10.1128/MCB.25.20.8779-8791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaGrandeur TE, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–96. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–62. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 15.Deshmukh MV, Jones BN, Quang-Dang D-U, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell. 2008;29:324–36. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 16.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–24. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell. 2008;29:337–49. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–21. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–90. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 20.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–47. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She M, Decker CJ, Chen N, Tumati S, Parker R, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol. 2004;11:249–56. doi: 10.1038/nsmb730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: structure, function and interactions. FEBS Lett. 2002;513:45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- 24.Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143–54. doi: 10.1016/s0896-6273(00)81145-9. [DOI] [PubMed] [Google Scholar]
- 25.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–80. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 26.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 27.Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA. Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich Syndrome. Cell. 2002;111:565–76. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- 28.Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, Weichenrieder O. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc Natl Acad Sci U S A. 2009;106:21591–6. doi: 10.1073/pnas.0909871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–85. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple Processing Body Factors and the ARE Binding Protein TTP Activate mRNA Decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Yang J-Y, Niu Q-W, Chua N-H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. The Plant Cell. 2006;18:3386–98. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floor SN, Borja MS, Gross JD. Interdomain dynamics and coactivation of the mRNA decapping enzyme Dcp2 are mediated by a gatekeeper tryptophan. Proc Natl Acad Sci U S A. 2012;109:2872–7. doi: 10.1073/pnas.1113620109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Floor SN, Jones BN, Hernandez GA, Gross JD. A split active site couples cap recognition by Dcp2 to activation. Nat Struct Mol Biol. 2010;17:1096–101. doi: 10.1038/nsmb.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–6. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 35.Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–51. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012;31:279–90. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol. 2010;30:1446–56. doi: 10.1128/MCB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–83. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–39. doi: 10.1093/genetics/166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu JH, Yang W-H, Gulick T, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–8. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–8. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 44.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b Connects Deadenylation with mRNA Decapping and Controls the Assembly of Processing Bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tharun S, Parker R. Targeting an mRNA for Decapping: Displacement of Translation Factors and Association of the Lsm1p 7p Complex on Deadenylated Yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 47.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–7. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 48.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–49. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–97. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–27. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang S-F, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–70. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borja MS, Piotukh K, Freund C, Gross JD. Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA. 2011;17:278–90. doi: 10.1261/rna.2382011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai T, Cho H, Liu Z, Bowler MW, Piao R, Parker S, Kim YK, Song H. Structural basis of PNRC2-mediated link between mRNA surveillance and decapping. Structure. 2012;20:2025–2037. doi: 10.1016/j.str.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Albrecht M, Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 56.Anantharaman V, Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 2007;27:8600–11. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ling SHM, Decker CJ, Walsh MA, She M, Parker R, Song H. Crystal structure of human Edc3 and its functional implications. Mol Cell Biol. 2008;28:5965–76. doi: 10.1128/MCB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–8. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 60.Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol. 2008;28:6695–708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J-C, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci U S A. 2008;105:5821–6. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–70. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun JE, Tritschler F, Haas G, Igreja C, Truffault V, Weichenrieder O, Izaurralde E. The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 2010;29:2368–80. doi: 10.1038/emboj.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005;33:433–8. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- 65.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–71. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol. 2000;12:346–50. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 67.Zaric B, Chami M, Rémigy H, Engel A, Ballmer-Hofer K, Winkler FK, Kambach C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem. 2005;280:16066–75. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]
- 68.Naidoo N, Harrop SJ, Sobti M, Haynes PA, Szymczyna BR, Williamson JR, Curmi PMG, Mabbutt BC. Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment. J Mol Biol. 2008;377:1357–71. doi: 10.1016/j.jmb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 69.Mund M, Neu A, Ullmann J, Neu U, Sprangers R. Structure of the LSm657 complex: an assembly intermediate of the LSm1-7 and LSm2-8 rings. J Mol Biol. 2011;414:165–76. doi: 10.1016/j.jmb.2011.09.051. [DOI] [PubMed] [Google Scholar]
- 70.Wu D, Jiang S, Bowler MW, Song H. Crystal structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe. PloS One. 2012;7:e36768. doi: 10.1371/journal.pone.0036768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–42. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramirez CV, Vilela C, Berthelot K, McCarthy JEG. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol. 2002;318:951–62. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- 73.Vilela C, Velasco C, Ptushkina M, McCarthy JE. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 2000;19:4372–82. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–56. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–86. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Totaro A, Renzi F, La Fata G, Mattioli C, Raabe M, Urlaub H, Achsel T. The human Pat1b protein: a novel mRNA deadenylation factor identified by a new immunoprecipitation technique. Nucleic Acids Res. 2011;39:635–47. doi: 10.1093/nar/gkq797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou M-N, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–13. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sweet T, Kovalak C, Coller J. The DEAD-Box Protein Dhh1 Promotes Decapping by Slowing Ribosome Movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–37. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Bénard M, Andrey P, Pierron G, Kress M, Standart N, le Cam E, Weil D. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–1715. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–34. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45:279–91. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 84.Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol. 2008;182:543–57. doi: 10.1083/jcb.200801183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell. 2009;20:2464–72. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–9. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–7. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y, Ho ES, Gunderson SI, Kiledjian M. Mutational analysis of a Dcp2-binding element reveals general enhancement of decapping by 5′-end stem-loop structures. Nucleic Acids Res. 2009;37:2227–37. doi: 10.1093/nar/gkp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song M-G, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–65. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 92.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–7. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 93.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–9. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 95.Blewett NH, Goldstrohm AC. An eIF4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol. 2012 doi: 10.1128/MCB.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 97.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–61. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–66. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sandler H, Kreth J, Timmers HTM, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–86. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–7. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Y, Song M, Kiledjian M. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA. 2011;17:419–28. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chu C, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–54. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 105.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–81. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 106.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 107.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 108.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 109.Mühlemann O, Lykke-Andersen J. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 2010;7:28–32. doi: 10.4161/rna.7.1.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–15. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–35. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–72. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–9. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–27. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song M-G, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ghosh T, Peterson B, Tomasevic N, Peculis BA. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol Cell. 2004;13:817–28. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- 123.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–62. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 124.Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–15. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–11. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chang JH, Jiao X, Chiba K, Oh C, Martin CE, Kiledjian M, Tong L. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat Struct Mol Biol. 2012;19:1011–7. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–24. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29:2155–67. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR. Identification of Cytoplasmic Capping Targets Reveals a Role for Cap Homeostasis in Translation and mRNA Stability. Cell Rep. 2012;2:674–84. doi: 10.1016/j.celrep.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]