Abstract
Background
Chronic kidney disease is associated with cardiovascular disease. We tested for evidence of a shared genetic basis to these traits.
Study Design
We conducted two targeted analyses. First, we examined whether known single nucleotide polymorphisms (SNPs) underpinning kidney traits were associated with a series of vascular phenotypes. Additionally, we tested whether vascular SNPs were associated with markers of kidney damage. Significance was set to 1.5 × 10-4 (0.05/325 tests).
Setting & Participants
Vascular outcomes were analyzed in participants from the AortaGen (20,634), CARDIoGRAM (86,995), CHARGE Eye (15,358), CHARGE IMT (31,181), ICBP (69,395) and NeuroCHARGE (12,385) consortia. Tests for kidney outcomes were conducted in up to 67,093 participants from the CKDGen consortium.
Predictor
We used 19 kidney SNPs and 64 vascular SNPs.
Outcomes & Measurements
Vascular outcomes tested were blood pressure, coronary artery disease, carotid intima-media thickness, pulse wave velocity, retinal venular caliber and brain white matter lesions. Kidney outcomes were estimated glomerular filtration rate and albuminuria.
Results
In general, we found that kidney disease variants were not associated with vascular phenotypes (127 of 133 tests were non-significant). The one exception was rs653178 near SH2B3 (SH2B adaptor protein 3), which showed direction-consistent association with systolic (p=9.3E-10) and diastolic (p=1.6E-14) blood pressure and coronary artery disease (p=2.2E-6), all previously reported. Similarly, the 64 SNPs associated with vascular phenotypes were not associated with kidney phenotypes (187 of 192 tests were non-significant), with the exception of 2 high-correlated SNPs at the SH2B3 locus (p=1.06E-07 and p=7.05E-08).
Limitations
Combined effect size of the SNPs for kidney and vascular outcomes may be too low to detect shared genetic associations.
Conclusions
Overall, although we confirmed one locus (SH2B3) as associated with both kidney and cardiovascular disease, our primary findings suggest that there is little overlap between kidney and cardiovascular disease risk variants in the overall population. The reciprocal risks of kidney and cardiovascular disease may not be genetically mediated, but rather a function of the disease milieu itself.
Chronic kidney disease (CKD), expressed by decreased estimated glomerular filtration rate (eGFR) and/or the presence of albuminuria, is a major health issue, affecting almost 18 million adults in the United States.1;2 CKD is associated with a range of serious adverse outcomes, including end-stage renal disease and death.3 However, the major complication of CKD is cardiovascular disease (CVD),4 and the majority of CKD patients die from cardiovascular disease before reaching end-stage kidney disease.5-7 Furthermore, the risk for CVD events increases rapidly as eGFR declines or albuminuria increases,8;9 independent of other common risk factors such as diabetes or hypertension.10
The mechanisms of excess cardiovascular risk in CKD patients are not fully understood. Predictive modeling using traditional cardiovascular risk factors performs poorly in this patient group.11 Novel risk factors for cardiovascular disease, such as vascular calcification, anemia, inflammation or increased oxidative stress may explain some of the excess risk in advanced CKD,12-14 but they fail to fully explain the link between the two diseases.15;16
Recent genome-wide association studies (GWAS) of large European populations have identified novel genetic risk single nucleotide polymorphisms (SNPs) for hypertension,17 coronary artery disease,18 subclinical vascular disease 19-22 as well as kidney functional traits and CKD.23-25 We hypothesized that kidney variants might increase the risk of vascular disease and that vascular variants might be similarly associated with kidney disease traits. We tested this hypothesis by conducting bidirectional cross-trait targeted SNP analyses between variants of eGFR, albuminuria and CVD and well-defined clinical cardiovascular and kidney outcomes. In the first of two targeted SNP analyses, we selected 19 SNPs known to be associated with kidney function 23-25 and investigated their association with the vascular phenotypes of systolic and diastolic blood pressure from the ICBP17 (International Consortium for Blood Pressure) and coronary artery disease from the CARDIoGRAM (Coronary Artery Disease Genome-wide Replication and Meta-Analysis) consortium.18 We also analyzed the association of known kidney SNPs with well-defined subclinical disease phenotypes, including: carotid-femoral pulse wave velocity from the AortaGen consortium,20 retinal venular caliber from the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Eye consortium,21 carotid intima-media thickness (cIMT) from the CHARGE IMT consortium,19 and white matter lesions from the NeuroCHARGE consortium.22 In the second targeted SNP analysis, 64 validated vascular SNPs from a range of cardiovascular phenotypes were tested in eGFR and albuminuria datasets from the CKDGen consortium.23-25
METHODS
Overall Study Design
For all phenotypes, testing for genetic association was performed using existing GWAS meta-analysis datasets in individuals of European descent with self-reported ethnicity. Genetic association testing for the kidney outcomes of eGFR and albuminuria was performed in the CKDGen cohorts using existing meta-analysis results.23;24 Similarly, association tests for vascular outcomes were conducted in most recent meta-analysis results from the AortaGen,20 CARDIoGRAM,18 CHARGE Eye,21 CHARGE IMT,19 ICBP,17 and NeuroCHARGE22 consortia. Heterogeneity tests were conducted in all consortia17-24 (e.g. using Cochran's Q test in CKDGen).
Study Exposure
In the first targeted SNP analysis, we selected 18 kidney SNPs previously identified by CKDGen in two-stage meta-analysis in association with creatinine-based eGFR (eGFRcr) (15 loci), cystatin-C based eGFR (eGFRcys) (37 loci: CST3 [cystatin C] and SH2B3 [SH2B adaptor protein 3])23 and albuminuria (1 locus, CUBN [cubulin]),24 as well as one SNP yielded by IBC (Institute of Translational Medicine and Therapeutics Broad CARe) candidate-gene SNP association analyses in African Americans from the CARe Renal Consortium (1 locus, KCNQ1 [kidney and cardiac voltage dependent K+ channel).25 The 19 kidney SNPs were tested for association with GWAS results for systolic and diastolic blood pressure from ICBP (n=69,395), coronary artery disease from CARDIoGRAM (n=86,995 with 52,120 cases), as well as with markers of atherosclerosis: aortic pulse wave velocity from AortaGen (n=20,634), retinal venular caliber from CHARGE Eye (n=15,358), cIMT from CHARGE IMT (n=31,181) and brain white matter lesions from NeuroCHARGE (n=12,385) consortia.
For the second targeted SNP analysis, 64 variants associated with blood pressure (29 loci in ICBP), coronary artery disease (25 loci in CARDIoGRAM) or atherosclerosis (1 locus for pulse-wave velocity in AortaGen, 4 loci with retinal venular caliber in CHARGE Eye, 3 loci associated with cIMT in CHARGE IMT and 2 loci with white matter lesions in NeuroCHARGE) were tested for association with two different estimations of GFR (n=67,093 for eGFRcr and n=20,966 for eGFRcys) and albuminuria (n=31,580) in the CKDGen consortium.
Kidney Measures
eGFRcr was estimated using the 4-variable Modification of Diet in Renal Disease Study equation.26 eGFRcys was estimated as eGFRcys = 76.7 × (serum cystatin C)–1.19.27 CKD was defined as eGFRcr < 60 ml / min /1.73 m2, according to National Kidney Foundation guidelines.28 Urinary albumin-creatinine ratio (UACR) was log-transformed for analysis and age and sex specific residuals were determined, as previously described.24 Albuminuria was defined as UACR >17 mg/g for men and >25 mg/g for women.29
Vascular Measures
Hypertension was defined in ICBP consortium as systolic blood pressure ≥ 140mmHg or diastolic blood pressure ≥ 90 mmHg.17 Coronary artery disease was defined in the CARDIoGRAM consortium as symptoms of angina pectoris, previous myocardial infarction or prior cardiac intervention.18 cIMT was measured in the CHARGE IMT consortium in the common carotid artery as the distance between the lumen-intima interface and media-adventitia interface by means of ultrasonography.19 The carotid-femoral pulse wave velocity was assessed by AortaGen using carotid-femoral transit time assessed by tonometry or Doppler flow and transit distance assessed by body surface measurements.20 The retinal venular caliber was measured in CHARGE Eye using in-vivo imaging techniques.21 White matter lesions were detected in NeuroCHARGE by means of magnetic resonance imaging.22
Statistical Methods
For the targeted SNP analyses, we used previously published results of meta-analyses of HapMap [International HapMap Project]-imputed SNPs to test 19 kidney SNPs for association with vascular traits and 64 cardiovascular SNPs in kidney traits. For the targeted SNP analysis of 19 eGFR SNPs in 7 different vascular outcomes (blood pressure, coronary artery disease, cIMT, aortic pulse-wave velocity, retinal venular caliber and white matter lesions) we performed 133 tests. Similarly, for the targeted SNP analysis of 64 vascular SNPs in 3 kidney-related outcomes (eGFRcr, eGFRcys and UACR), we performed 192 tests. We adjusted for multiple testing using a Bonferroni corrected alpha of 1.5e-4 (0.05/325), adjusting for all tests performed (133 + 192).
For each SNP, the results of the targeted meta-analysis included the effect estimator β, its standard error, a two-sided p-value, as well as coded and non-coded allele, frequency of the coded allele and the total sample size. Whenever the requested (lead) SNP was not available, a proxy SNP with an r2 correlation of over 80% was used. In 4 cases, this was not possible, therefore 3 SNPs are missing from the analysis for coronary artery disease and 1 locus is absent in all kidney analyses. Meta-analysis results corrected for genomic control were used for all targeted analyses.
RESULTS
Study Participants
All study participants were of European descent. Study sample characteristics can be found in Table 1. Details on the individual cohorts included are available in the original papers that reported findings for eGFR,23;25 albuminuria,24 blood pressure,17 coronary artery disease,18 cIMT,19 aortic pulse wave velocity,20 retinal venular caliber21 and white matter lesions.22
Table 1.
Study sample characteristics for kidney and cardiovascular phenotypes.
| Consortium | Sample sizea | No. of studiesb | Female sex | Age (y) | DM | HTN | Key phenotype | Values corresponding to key phenotype |
|---|---|---|---|---|---|---|---|---|
| CKDGen – eGFR23 | 67,093d | 20 | 72% (48,637) | 59.0 (11.8) | 8% (5,343) | 37% (24,798) | eGFRcr | 86.89 ± 21.62 ml/min/1.73 m2 |
| CKDGen – UACR24 | 31,580 | 12 | 53% (16,723) | 56.4 | 9% (2,818) | 38% (12,001) | UACR | 6.03 [3.52-11.75] mg/gc |
| ICBP17 | 69,395 | 29 | 55% (37,956) | 51.0 (9.9) | NA | 33% (22,573) | SBP/DBP | 129.91 ± 18.38 / 78.54 ± 10.63 mm Hg |
| CARDIoGRAM18 | 86,995 | 14 | 51% (44,505) | 56.0 (9.4) | NA | NA | MI | 52,120 (60) |
| CHARGE IMT19 | 31,181 | 9 | 55% (17,294) | 59.1 (10.3) | 9% (2,661) | 45% (13,996) | cIMT | 0.84 ± 1.6 mm |
| AortaGen20 | 20,634 | 9 | 55% (11,252) | 59.2 (11.3) | NA | NA | Inverse PWV | 125.11 ± 33 ms/m |
| CHARGE Eye21 | 15,358 | 4 | 57% (8,682) | 70.7 (5.8) | NA | NA | RVC | 141.55 ± 14.45 μm |
| NeuroCHARGE22 | 12,385 | 9 | 58% (7,168) | 69.8 (6.3) | 10% (1,250) | 55% (6,833) | WML burden | 4.43 ± 5.35 mL |
Note: Except where otherwise indicated, values for categorical variables are given as number (percentage); values for continuous variables, as mean +/- standard deviation.
DM, diabetes; HTN, hypertension; eGFR, estimated glomerular filtration rate; eGFRcr, creatinine-based eGFR; eGFRcys, cystatin C-based eGFR; UACR, urinary albumin-creatine ratio; CKD, chronic kidney disease; SBP, systolic blood pressure; DBP, diastolic blood pressure; MI, myocardial infarction; IMT, intima-media thickness; cIMT, carotid intima-media thickness; PWV, pulse-wave velocity; RVC, retinal venular caliber; WML, white matter lesion ICBP (International Consortium for Blood Pressure); CARDIoGRAM (Coronary Artery Disease Genome-wide Replication and Meta-Analysis); CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology; NA, not available
Refers to the recruited sample for the respective phenotype, which may differ from the successfully genotyped and analyzed sample.
Details on the individual cohorts included are available in the supplemental materials of the original papers.
Median [interquartile range].
Sample size for cystatin C-based eGFR (eGFRcys) was 20,966.
Analysis of Validated Kidney SNPs in Vascular Traits
Association of kidney SNP variants with blood pressure
Results of associations of kidney SNP variants with blood pressure in the ICBP consortium are presented in Table 2. In general, the variants tested were not associated with blood pressure at α=1.5e-4 (0.05/325) (p-value range, 0.98-0.002). The one exception was rs (reference SNP identifier) 653178 at the SH2B3 gene: each copy of the eGFR risk allele C (lower levels of eGFR) was associated with a higher level of systolic (p=9.3E-10) and diastolic blood pressure (p=1.6E-14). This SNP is in perfect linkage disequilibrium with rs3184504 (r2=1.0), which has been previously reported in association with blood pressure.17
Table 2.
Association results of 19 kidney SNPs with clinical vascular traits in individuals of European descent from the ICBP and CARDIoGRAM consortia.
| Kidney SNP | Nearest Gene | Coded (risk) allele | SBP | DBP | CAD | |||
|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | OR (CI) | p-value | |||
| rs10109414 | STC1 | T | -0.054 | 0.6 | 0.07 | 0.3 | 0.99 [0.97 - 1.02] | 0.9 |
| rs11959928 | DAB2 | A | 0.009 | 0.9 | -0.009 | 0.9 | 1.01 [0.98 - 1.04] | 0.3 |
| rs12460876 | SLC7A9 | T | 0.169 | 0.1 | 0.074 | 0.2 | 1.01 [0.97 - 1.03] | 0.6 |
| rs1260326 | GCKR | C | -0.102 | 0.3 | -0.019 | 0.8 | 0.97 [0.94 - 1.00] | 0.1 |
| rs12917707 | UMOD | G | 0.353 | 0.005 | 0.244 | 0.002 | 1.01 [0.97 - 1.051 | 0.4 |
| rs13538 | ALMS1 | A | -0.096 | 0.4 | -0.13 | 0.1 | 0.95 [0.91 - 0.981 | 0.004 |
| rs1394125 | UBE2Q2 | A | 0.112 | 0.3 | 0.021 | 0.8 | NA** | NA** |
| rs17319721 | SHROOM3 | A | -0.143 | 0.1 | -0.042 | 0.5 | 1.01 [0.98 - 1.03] | 0.6 |
| rs1801239 | CUBN | T | -0.017 | 0.9 | 0.038 | 0.7 | 1.01 [0.95 - 1.05] | 0.8 |
| rs267734 | ANXA9 | T | -0.047 | 0.7 | -0.028 | 0.7 | 0.99 [0.96 - 1.03] | 0.9 |
| rs347685 | TFDP2 | A | 0.035 | 0.7 | -0.088 | 0.2 | 1.02 [0.99 - 1.04] | 0.2 |
| rs4744712 | PIP5K1B | A | -0.002 | 0.98 | 0.035 | 0.6 | 1.01 [0.98 - 1.04] | 0.3 |
| rs626277 | DACH1 | A | -0.038 | 0.7 | -0.022 | 0.7 | 1.01 [0.96 - 1.03] | 0.9 |
| rs6420094 | SLC34A1 | G | 0.086 | 0.5 | 0.097 | 0.2 | NA** | NA** |
| rs653178 | SH2B3 | C | 0.605 | 9.3E-10*** | 0.48 | 1.64E-14*** | 1.08 [1.04 - 1.11] | 2.2E-06*** |
| rs7805747 | PRKAG2 | A | 0.359 | 0.02 | 0.178 | 0.1 | NA** | NA** |
| rs81204* | KCNQ1 | C | -0.032 | 0.8 | 0.04 | 0.6 | 1.01 [0.98 - 1.05] | 0.4 |
| rs881858 | VEGFA | A | 0.009 | 0.9 | 0.172 | 0.02 | 1.01 [0.96 - 1.03] | 0.9 |
| rs911119 | CST3 | T | -0.177 | 0.1 | -0.107 | 0.2 | 0.99 [0.96 - 1.02] | 0.8 |
NOTE: The clinical vascular traits tested were SBP, DBP, and CAD. Results are presented for the trait-decreasing eGFR allele.
CAD, coronary artery disease; DBP, diastolic blood pressure; rs, reference NSP identifier; SBP, systolic blood pressure; SNP, single nucleotide polymorphism; ICBP (International Consortium for Blood Pressure); CARDIoGRAM (Coronary Artery Disease Genome-wide Replication and Meta-Analysis); OR, odds ratio; CI, confidence interval; eGFR, estimated glomerular filtration rate; NA, not available.
The rs163160 SNP was used as a proxy for SBP, DBP and CAD (r2=0.81).
These SNPs were not available in the imputed CARDIoGRAM meta-analysis dataset and an adequate proxy was not found.
Significant associations after Bonferroni correction for multiple testing (p ≤ 1.5e-4)
Association of kidney SNP variants with coronary artery disease
Results of associations of kidney SNP variants with coronary artery disease in the CARDIoGRAM consortium are presented in Table 2. The majority of tests were negative at α=1.5e-4, with p-values ranging from 0.96 to 0.004. The one exception was rs653178 in the SH2B3 gene, whose risk allele C was associated with a higher risk for coronary artery disease (odds ratio [OR], 1.08; 95% confidence interval [CI], 1.04-1.11; p=2.2E-6). This SNP has been previously reported by CKDGen in association with cystatin C-based eGFR (eGFRcys).18
Association of kidney SNP variants with subclinical markers of atherosclerosis
The results of the association between kidney SNPs and cIMT, pulse wave velocity, retinal venular caliber and white matter lesions are presented in Table 3. Similar to what we observed for blood pressure and coronary artery disease, these variants were not associated with these phenotypes at α=1.5e-4 (p-value range, 0.99-0.001), with the exception of rs653178 near SH2B3, which was associated with high levels of retinal venular caliber (p=2.9E-11). This SNP is in high linkage disequilibrium (r2=0.96) with another SNP (rs10774625) in the same locus, previously reported by NeuroCHARGE in association with retinal venular caliber.21
Table 3.
Association results of 19 kidney SNPs with subclinical vascular traits in individuals of European descent from the CHARGE IMT, AortaGen, CHARGE Eye and NeuroCHARGE consortia.
| Kidney SNP | Nearest Gene | Coded (risk) allele | cIMT | PWV | RVC | WMLs | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | z-score | p-value | |||
| rs10109414 | STC1 | T | 0.001 | 0.7 | 0.007 | 0.5 | 0.068 | 0.77 | -0.444 | 0.7 |
| rs11959928 | DAB2 | A | -0.002 | 0.1 | 0.005 | 0.6 | 0.042 | 0.86 | -0.18 | 0.9 |
| rs12460876 | SLC7A9 | T | -0.002 | 0.2 | -0.003 | 0.8 | 0.29 | 0.22 | 0.785 | 0.4 |
| rs1260326 | GCKR | C | 0.002 | 0.3 | -0.036 | 0.001 | -0.022 | 0.93 | -0.004 | 0.9 |
| rs12917707 | UMOD | G | -0.003 | 0.1 | 0.019 | 0.2 | -0.415 | 0.16 | 1.334 | 0.2 |
| rs13538 | ALMS1 | A | 0.001 | 0.4 | -0.007 | 0.6 | -0.419 | 0.13 | 0.454 | 0.6 |
| rs1394125 | UBE2Q2 | A | -0.002 | 0.3 | -0.024 | 0.1 | -0.83 | 0.001 | 0.164 | 0.9 |
| rs17319721 | SHROOM3 | A | -0.001 | 0.5 | -0.002 | 0.8 | -0.319 | 0.17 | -1.387 | 0.2 |
| rs1801239 | CUBN | T | 0.001 | 0.8 | 0.001 | 0.99 | -0.14 | 0.72 | 2.907 | 0.004 |
| rs267734 | ANXA9 | T | -0.001 | 0.8 | 0.001 | 0.9 | 0.251 | 0.37 | -0.476 | 0.6 |
| rs347685 | TFDP2 | A | 0.002 | 0.1 | 0.001 | 0.99 | 0.419 | 0.09 | 0.038 | 0.9 |
| rs4744712 | PIP5K1B | A | 0.002 | 0.1 | -0.001 | 0.99 | 0.375 | 0.11 | -0.029 | 0.9 |
| rs626277 | DACH1 | A | 0.002 | 0.3 | 0.017 | 0.1 | 0.289 | 0.21 | 0.079 | 0.9 |
| rs6420094 | SLC34A1 | G | 0.002 | 0.2 | 0.008 | 0.6 | 0.085 | 0.75 | NA** | NA** |
| rs653178 | SH2B3 | C | -0.001 | 0.6 | 0.017 | 0.1 | 1.529 | 2.9E-11*** | -0.318 | 0.8 |
| rs7805747 | PRKAG2 | A | 0.003 | 0.2 | 0.012 | 0.4 | -0.166 | 0.62 | NA** | NA** |
| rs81204 | KCNQ1 | C | 0.003 | 0.3 | 0.02 | 0.2 | NA** | NA | NA** | NA** |
| rs881858 | VEGFA | A | 0.0001 | 0.9 | -0.029 | 0.03 | 0.822 | 0.002 | 0.519 | 0.6 |
| rs911119 | CST3 | T | 0.001 | 0.7 | -0.034 | 0.01 | -0.047 | 0.87 | -0.337 | 0.7 |
Note: The subclinical vascular traits tested were cIMT, PWV, RVC, and WMLs. Results are presented for the trait-decreasing eGFR allele.
CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology); PWV, pulse-wave velocity; RVC, retinal venular caliber; WMLs, white mater lesions; carotid intima-media thickness (cIMT), eGFR, estimated glomerular filtration rate; SNP, single nucleotide polymorphism; NA, not available
These SNPs were not available in the imputed meta-analysis dataset and an adequate proxy was not found.
Significant associations after Bonferroni correction for multiple testing (p ≤ 1.5e-4)
Analysis of Validated Vascular SNPs in Kidney Traits
Association of blood pressure variants with kidney traits
Association tests of 29 known blood pressure SNPs with eGFR and albuminuria (UACR) in the CKDGen consortium are presented in Table 4. We observed one significant association with low eGFR at α=1.5e-4: rs3184504 at the SH2B3 locus (p=1.06E-07). This SNP is perfectly correlated with rs653178 (r2=1.0), which has been previously reported by CKDGen in association with eGFRcys.23 All other association tests for eGFR and albuminuria were not statistically significant (p-value range, 0.9-0.001).
Table 4.
Association of 29 blood pressure SNPs with kidney traits in individuals of European descent from the CKDGen consortium.
| BP SNP | Nearest Gene | Coded (risk) allele | UACR | eGFRcr | eGFRcys | |||
|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | |||
| rs17367504 | MTHFR-NPPB | A | 0.003 | 0.8 | 0.001 | 0.5 | 0.002 | 0.5 |
| rs2932538 | MOV10 | G | 0.002 | 0.9 | 0.002 | 0.2 | -0.002 | 0.5 |
| rs13082711 | SLC4A7 | C | 0.008 | 0.5 | 0.003 | 0.1 | 0.003 | 0.3 |
| rs3774372 | ULK4 | C | -0.014 | 0.2 | 0.001 | 0.6 | 0.009 | 0.004 |
| rs419076 | MECOM | T | 0.005 | 0.6 | -0.001 | 0.3 | -0.003 | 0.2 |
| rs1458038 | FGF5 | T | -0.019 | 0.1 | 0.004 | 0.005 | -0.001 | 0.6 |
| rs13107325 | SLC39A8 | C | -0.02 | 0.3 | 0.005 | 0.048 | 0.002 | 0.7 |
| rs13139571 | GUCY1A3-GUCY1B3 | C | 0.002 | 0.9 | -0.004 | 0.03 | -0.007 | 0.006 |
| rs1173771 | NPR3-C5orf23 | G | -0.004 | 0.6 | -0.001 | 0.7 | 0.003 | 0.2 |
| rs11953630 | EBF1 | C | 0.014 | 0.1 | 0.002 | 0.2 | 0.002 | 0.5 |
| rs1799945 | HFE(H63D) | G | 0.003 | 0.8 | 0.006 | 0.001 | -0.004 | 0.2 |
| rs805303 | BAT2-BAT5 | G | -0.003 | 0.8 | 0.004 | 0.007 | 0.0004 | 0.9 |
| rs4373814 | CACNB2(5′) | C | 0.008 | 0.4 | -0.001 | 0.7 | -0.001 | 0.7 |
| rs1813353 | CACNB2(3′) | T | 0.019 | 0.047 | -0.002 | 0.3 | 0.003 | 0.3 |
| rs4590817 | C10orf107 | G | 0.005 | 0.7 | -0.0004 | 0.8 | 0.005 | 0.1 |
| rs932764 | PLCE1 | G | -0.004 | 0.7 | 0.001 | 0.6 | -0.001 | 0.8 |
| rs11191548 | CYP17A1-NT5C2 | T | -0.04 | 0.01 | 0.004 | 0.1 | 0.002 | 0.6 |
| rs7129220 | ADM | A | 0.012 | 0.4 | -0.002 | 0.4 | -0.006 | 0.1 |
| rs381815 | PLEKHA7 | T | 0.015 | 0.1 | -0.003 | 0.1 | -0.002 | 0.4 |
| rs633185 | FLJ32810-TMEM133 | C | -0.008 | 0.4 | 0.003 | 0.1 | -0.002 | 0.5 |
| rs17249754 | ATP2B1 | G | 0.011 | 0.4 | -0.004 | 0.02 | -0.007 | 0.02 |
| rs3184504 | SH2B3 | T | -0.008 | 0.4 | -0.003 | 0.01 | -0.012 | 1.06E-07* |
| rs10850411 | TBX5-TBX3 | T | 0.008 | 0.4 | 0.001 | 0.7 | 0.003 | 0.2 |
| rs1378942 | CYP1A1-ULK3 | C | -0.005 | 0.6 | -0.001 | 0.3 | -0.002 | 0.5 |
| rs2521501 | FURIN-FES | T | -0.016 | 0.3 | -0.002 | 0.4 | 0.005 | 0.1 |
| rs17608766 | GOSR2 | C | 0.015 | 0.3 | -0.003 | 0.2 | -0.005 | 0.2 |
| rs12940887 | ZNF652 | T | 0.007 | 0.5 | -0.001 | 0.5 | -0.001 | 0.6 |
| rs1327235 | JAG1 | G | 0.021 | 0.02 | -0.0001 | 0.9 | 0.004 | 0.1 |
| rs6015450 | GNAS-EDN3 | G | 0.006 | 0.7 | -0.004 | 0.04 | 0.005 | 0.1 |
Note: The kidney traits tested are UACR, eGFRcr, and eGFRcys. Results are presented in the trait-increasing allele for BP. BP, blood pressure; SNP, single nucleotide polymorphism; eGFR, estimated glomerular filtration rate; eGFRcr, creatinine-based eGFR; eGFRcys, cystatin C-based eGFR; UACR, urinary albumin-creatine ratio; CKD, chronic kidney disease
Significant associations after Bonferroni correction for multiple testing (p ≤ 1.5e-4).
Association of coronary artery disease variants with kidney traits
Association tests of 25 coronary artery disease variants with kidney traits in the CKDGen consortium are presented in Table 5. In general, they did not associate with either eGFR or albuminuria at α=1.5e-4 (0.05/325), with p-values ranging from 0.9 to 0.002. The exception was rs3184504 at SH2B3, whose risk allele T for coronary artery disease was associated with lower levels of eGFR.
Table 5.
Association of 25 CAD SNPs with kidney traits in individuals of European descent from the CKDGen consortium.
| CAD SNP | Nearest Gene | Coded (risk) allele | UACR | eGFRcr | eGFRcys | |||
|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | |||
| rs11206510 | PCSK9 | T | -0.012 | 0.3 | -0.001 | 0.5 | -0.003 | 0.4 |
| rs599839 | SORT1 | A | 0.018 | 0.1 | 0.003 | 0.1 | 0.001 | 0.6 |
| rs17465637 | MIA3 | C | -0.009 | 0.7 | -0.006 | 0.4 | -0.005 | 0.4 |
| rs6725887 | WDR12 | C | -0.024 | 0.1 | -0.004 | 0.04 | -0.007 | 0.04 |
| rs2306374 | MRAS | C | 0.012 | 0.3 | -0.001 | 0.5 | 0.003 | 0.3 |
| rs12526453 | PHACTR1 | C | 0.003 | 0.8 | 0.0002 | 0.9 | 0.002 | 0.4 |
| rs3798220 | LPA | C | NA* | NA* | NA* | NA* | NA* | NA* |
| rs4977574 | CDKN2A, CDKN2B | G | 0.013 | 0.1 | -0.002 | 0.2 | 0.003 | 0.2 |
| rs1746048 | CXCL12 | C | -0.018 | 0.2 | -0.001 | 0.7 | 0.001 | 0.9 |
| rs3184504 | SH2B3 | T | -0.008 | 0.4 | -0.003 | 0.01 | -0.012 | 1.06E-07** |
| rs1122608 | LDLR | G | -0.016 | 0.1 | -0.001 | 0.5 | -0.002 | 0.5 |
| rs9982601 | MRPS6 | T | 0.006 | 0.7 | -0.001 | 0.6 | 0.005 | 0.2 |
| rs17114036 | PPAP2B | A | -0.018 | 0.2 | -0.003 | 0.2 | 0.001 | 0.8 |
| rs17609940 | ANKS1A | G | -0.008 | 0.5 | -0.001 | 0.5 | 0.0003 | 0.9 |
| rs12190287 | TCF21 | C | 0.014 | 0.2 | 0.002 | 0.1 | -0.004 | 0.1 |
| rs11556924 | ZC3HC1 | C | -0.015 | 0.1 | -0.001 | 0.6 | -0.002 | 0.5 |
| rs579459 | ABO | C | -0.018 | 0.1 | 0.002 | 0.3 | -0.004 | 0.1 |
| rs12413409 | CYP17A1 | G | -0.038 | 0.017 | 0.003 | 0.1 | -0.0002 | 0.9 |
| rs964184 | ZNF259 | G | 0.008 | 0.6 | -0.002 | 0.3 | -0.01 | 0.002 |
| rs4773144 | COL4A1 | G | -0.022 | 0.02 | -0.002 | 0.1 | 0.001 | 0.7 |
| rs2895811 | HHIPL1 | C | 0.006 | 0.6 | -0.001 | 0.4 | -0.001 | 0.7 |
| rs3825807 | ADAMTS7 | A | 0.002 | 0.8 | 0.001 | 0.5 | 0.001 | 0.6 |
| rs216172 | SMG6 | C | -0.008 | 0.6 | 0.002 | 0.4 | 0.002 | 0.5 |
| rs12936587 | RASD1 | G | 0.02 | 0.04 | 0.003 | 0.02 | 0.004 | 0.1 |
| rs46522 | UBE2Z | T | 0.018 | 0.046 | 0.001 | 0.7 | 0.002 | 0.5 |
Note: The kidney traits tested are UACR, eGFRcr, and eGFRcys. Results are presented in the trait-increasing allele for CAD. CAD, coronary artery disease; SNP, single nucleotide polymorphism; eGFR, estimated glomerular filtration rate; eGFRcr, creatinine-based eGFR; eGFRcys, cystatin C-based eGFR; UACR, urinary albumin-creatine ratio; CKD, chronic kidney disease; NA, not available.
This SNP was not available in the imputed CKDGen meta-analysis dataset and an adequate proxy was not found.
Significant associations after Bonferroni correction for multiple testing (p ≤ 1.5e-4)
Association of subclinical cardiovascular SNP variants with kidney traits
The association results for 10 known subclinical vascular SNP variants (3 loci for cIMT, 1 locus for pulse-wave velocity, 4 loci for retinal venular caliber and 2 loci for white matter lesions) with kidney traits in the CKDGen consortium are reported in Table 6. The majority of tests were not significant at α=1.5e-4 (p-value range, 0.9-0.0005), with the exception of rs10774625 at SH2B3, which was associated with lower levels of eGFR (p=7.05E-08). This SNP was previously noted in association with retinal venular caliber21 and is in strong linkage disequilibrium (r2=0.96) with the other 2 SNPs in the same locus, which have previously been reported.17;18;23
Table 6.
Associations of 10 subclinical atherosclerosis SNPs with kidney traits in individuals of European descent from the CKDGen consortium.
| CVD SNP | Nearest Gene | Coded (risk) allele | CVD phenotype | UACR | eGFRcr | eGFRcys | |||
|---|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | ||||
| rs11781551 | ZHX2 | G | cIMT | -0.005 | 0.6 | -0.0001 | 0.9 | -0.001 | 0.6 |
| rs445925 | APOC1 | G | cIMT | -0.01 | 0.8 | -0.007 | 0.1 | -0.001 | 0.9 |
| rs6601530 | PINX1 | G | cIMT | -0.017 | 0.1 | 0.003 | 0.03 | 0.001 | 0.7 |
| rs7152623 | 3′-BCL11B | G | PWV | -0.007 | 0.4 | -0.001 | 0.5 | 0.001 | 0.8 |
| rs2287921 | RASIP1 | C | RVC | 0.009 | 0.3 | -0.003 | 0.1 | -0.006 | 0.01 |
| rs225717 | VTA1 | T | RVC | 0.009 | 0.4 | 0.004 | 0.02 | 0.001 | 0.7 |
| rs10774625 | SH2B3 | A | RVC | -0.008 | 0.4 | -0.003 | 0.03 | -0.013 | 7.05E-08* |
| rs17421627 | MEF2C | G | RVC | 0.002 | 0.9 | 0.004 | 0.1 | 0.001 | 0.8 |
| rs3744028 | TRIM65 | C | WML | -0.01 | 0.4 | 0.006 | 0.0005 | -0.0003 | 0.9 |
| rs1055129 | TRIM47 | G | WML | -0.011 | 0.3 | 0.005 | 0.002 | 0.001 | 0.9 |
Note: The subclinical CVD SNPs (atherosclerosis SNPs) were previously associated with cIMT, PWV, RVC, and WMLs. The kidney traits tested are UACR, eGFRcr, and eGFRcys. Results are presented in the trait-increasing allele for the respective subclinical CVD phenotype.
CVD, cardiovascular disease; eGFR, estimated glomerular filtration rate; eGFRcr, creatinine-based eGFR; eGFRcys, cystatin C-based eGFR; UACR, urinary albumin-creatine ratio; CKD, chronic kidney disease;; cIMT, carotid intima-media thickness; PWV, pulse-wave velocity; RVC, retinal venular caliber; WML, white matter lesion; SNP, single nucleotide polymorphism
Significant associations after Bonferroni correction for multiple testing (p ≤ 1.5e-4).
Concordance Between Vascular SNP Variants and eGFR
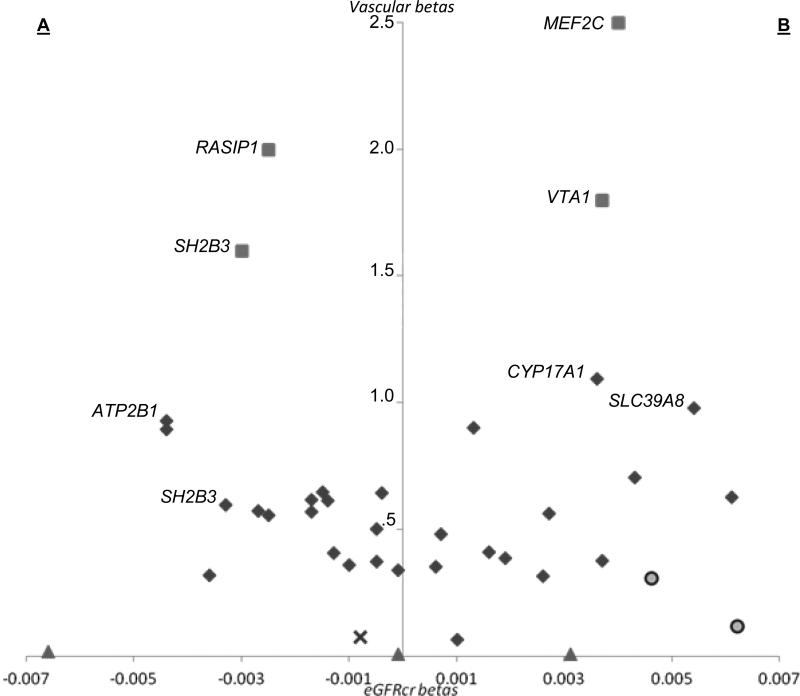
In the CKDGen Consortium, only 96 of 189 CVD risk variants tested were in the expected direction, (i.e. were associated with lower eGFR) whereas 93 were in the opposite direction (p=0.9) (Fig. 1). Similar patterns of discordance were observed in all vascular consortia, where only 70 out of 127 tested variants showed association in the hypothesized direction (p=0.3).
Figure 1.
Scatter plot of SNP effect sizes (beta) for creatinine-based eGFR (eGFRcr) and vascular phenotypes (systolic blood pressure [filled diamonds], carotid-intima medial thickness [filled triangles], pulse-wave velocity [×], retinal venular caliber [filled squares], and white matter lesions [filled circles]. The side labeled A (lower eGFRcr effect size, higher vascular effect size) represents associations consistent with the study hypothesis; the side labeled B (higher eGFRcr effect size, higher vascular effect size) represents associations inconsistent with the hypothesis. The results show no significant correlation between eGFRcr and vascular SNP variants.
DISCUSSION
We observed minimal overlap in genetic association of well-validated SNPs across a variety of kidney and vascular traits. The exception was SH2B3, which has previously been reported as associated with several traits. The absence of association observed suggests that there is little overlap between kidney disease and CVD risk variants.
The general lack of association seen in our analyses is consistent with recent findings of the ICBP consortium, where investigators constructed a genetic risk score for the 29 SNPs related to blood pressure and found evidence of genetic overlap with left ventricular wall thickness, incident heart failure, stroke and coronary artery disease, but not with kidney disease or kidney function.17 Our study advances our understanding in this area, by demonstrating a similar lack of a shared genetic basis to a broad range of well-characterized cardiovascular and kidney traits.
The SH2B3 locus is the one exception to these negative results, demonstrating robust and direction-consistent associations with increased blood pressure, higher OR for coronary artery disease and increased retinal venular caliber as opposed to lower levels of eGFR in the general population. Located at 12q24, SH2B3 encodes a member of the SH2B adaptor family of proteins, which play a role in signal, hematopoiesis, and immune response activities by integrin, growth factors and cytokines.24;30;31 The reason for the joint associations between these variants may be due to pleiotropic effects of this gene. The SH2B3 region is recognized for pleiotropy, associating with multiple phenotypes including platelet count,32 susceptibility to insulin-dependent diabetes mellitus33 and celiac disease type 13.34 A recent attempt to measure pleiotropic effects for known kidney and vascular loci demonstrated SH2B3 to have the highest pleiotropic index among all other identified regions.35
Several avenues for future research are suggested by the present analysis, which may improve our understanding of the mechanism of vascular disease in kidney patients, help stratify CKD patients by their risk of developing vascular disease or potentially identify novel pharmacogenomics targets. In particular, studies targeted at achieving a better understanding of the role of SH2B3 in both kidney disease and CVD may prove fruitful. For example, the observation that SH2B3 is strongly associated with cystatin-C-based eGFR but only nominally so with creatinine-based eGFR, suggests that its effects on GFR may be indirect, perhaps mediated by influences on cystatin C production or metabolism, which is a better predictor of CVD than eGFRcr.36 This is supported by the fact that SH2B3 showed no association to urinary ACR due to distinct genetic mechanisms of eGFR and albuminuria, in accordance with a prior interrogation which found a lack of association between CKD-related SNPs and albuminuria.37 Furthermore, as the SH2B3 gene region is large (45,674 nucleotides, based on Ensembl gene annotation release 54) and includes several other genes, fine-mapping studies may identify the causal variant(s) underlying the statistical signals identified in this and other analyses.
The absence of cross-associations does not necessarily imply absence of genetic correlation and there may be other contributing mechanisms to explain these findings. For example, in totality, the previously identified kidney function loci only account for 1.4% of the variation of eGFR,23 with similar values reported by ICBP for the association with blood pressure17 and CARDIoGRAM for coronary artery disease.18 The relatively low explained variance of trait variation may account for the lack of overlap in these analyses, and it remains possible that shared variants may yet be identified as a greater proportion of the underlying genetic variability is explained. However, the ICBP study demonstrated that even SNPs in low linkage disequilibrium can identify overlapping associations with downstream traits,17 supporting our negative findings.
Our findings suggest that non-genetic or environmental factors are primarily responsible for the association between CKD and CVD, either via a direct causal link or by lying in the causal pathway. Examples of such factors include uremic toxins not cleared by the failing kidney, vascular calcification, anemia, inflammation or increased oxidative stress account, which have a higher prevalence in CKD patients as compared to the overall population.12-14 For example, indoxyl sulfate is a uremic toxin that accelerates vascular disease in CKD patients via AHR (aryl hydrocarbon receptor) and other pathways, but which is not relevant to CVD in the overall population.38 Such environmental mechanisms would not be detectable by the present approach. The lack of genetic association between kidney disease and CVD emphasizes the influence of shared risk factors in the CKD-CVD milieu, particularly diabetes and hypertension.
Strengths of this study include the largest to date available GWAS datasets for kidney traits, blood pressure, coronary artery disease and subclinical vascular markers from the CKDGen, ICBP, CARDIoGRAM, CHARGE Eye, CHARGE IMT, NeuroCHARGE and AortaGen consortia, as well as the use of validated SNP variants for these traits. Some limitations also warrant mention. First, as genetic variations for eGFR, albuminuria and CVD explain only about 1% of the variations of these traits, the combined effect size of the SNPs for eGFR and vascular outcomes may be too low to detect shared genetic associations. Second, whereas our study indicates little overlap in the common genetic determinants of CKD and CVD, similar analyses in cohorts targeted at more advanced stages of CKD or containing more cases of both diseases may yield different findings. Third, our study used common genetic variants derived from GWAS and it is therefore possible that low frequency variants could provide more evidence for joint association. Furthermore, future GWAS, exome sequencing or pathway analyses may yet reveal shared genetic underpinnings between these traits. Finally, our study sample was composed of individuals of European ancestry; therefore the generalizability of our findings across other ethnic groups is uncertain.
In conclusion, we observed overall little genetic overlap between well-validated SNPs for kidney disease and CVD and their related phenotypes in targeted SNP analyses conducted in largest to date available meta-analyses results. Although we confirm one locus, SH2B3, which is associated with both kidney disease and CVD, our primary findings suggest that there is little overlap between kidney disease and CVD risk variants. The interrelationship between these traits may thus be primarily due to clinical factors present in the setting of CKD or CVD.
Acknowledgements
The full information on collaborating scientists of the GWAS Consortia is available in articles previously published by the involved consortia.
Support: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health and Boston University School of Medicine. This work was partially supported by the NHLBI's Framingham Heart Study (Contract No. N01-HC-25195). Full information on funding of the GWAS performed by the involved consortia is available in the consortia's previously published articles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure: Dr Mitchell is owner of Cardiovascular Engineering Inc, a company that designs and manufactures devices that measure vascular stiffness. The company uses these devices in clinical trials that evaluate the effects of diseases and interventions on vascular stiffness. The other authors declare that they have no other relevant financial interests.
References
- 1.Snyder JJ, Foley RN, Collins AJ. Prevalence of CKD in the United States: a sensitivity analysis using the National Health and Nutrition Examination Survey (NHANES) 1999-2004. Am J Kidney Dis. 2009;53(2):218–228. doi: 10.1053/j.ajkd.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298(17):2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 3.Manjunath G, Tighiouart H, Ibrahim H, et al. Level of kidney function as a risk factor for atherosclerotic cardiovascular outcomes in the community. J Am Coll Cardiol. 2003;41(1):47–55. doi: 10.1016/s0735-1097(02)02663-3. [DOI] [PubMed] [Google Scholar]
- 4.Foley RN. Clinical epidemiology of cardiovascular disease in chronic kidney disease. J Ren Care. 2010;36(Suppl 1):4–8. doi: 10.1111/j.1755-6686.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- 5.Menon V, Sarnak MJ. The epidemiology of chronic kidney disease stages 1 to 4 and cardiovascular disease: a high-risk combination. Am J Kidney Dis. 2005;45(1):223–232. doi: 10.1053/j.ajkd.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Levin A, Djurdjev O, Barrett B, et al. Cardiovascular disease in patients with chronic kidney disease: getting to the heart of the matter. Am J Kidney Dis. 2001;38(6):1398–1407. doi: 10.1053/ajkd.2001.29275. [DOI] [PubMed] [Google Scholar]
- 7.Culleton BF, Larson MG, Wilson PW, Evans JC, Parfrey PS, Levy D. Cardiovascular disease and mortality in a community-based cohort with mild renal insufficiency. Kidney Int. 1999;56(6):2214–2219. doi: 10.1046/j.1523-1755.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- 8.Matsushita K, van d V, Astor BC, et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet. 2010;375(9731):2073–2081. doi: 10.1016/S0140-6736(10)60674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351(13):1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 10.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D. Predictors of new-onset kidney disease in a community-based population. JAMA. 2004;291(7):844–850. doi: 10.1001/jama.291.7.844. [DOI] [PubMed] [Google Scholar]
- 11.Weiner DE, Tighiouart H, Elsayed EF, et al. The Framingham predictive instrument in chronic kidney disease. J Am Coll Cardiol. 2007;50(3):217–224. doi: 10.1016/j.jacc.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AK, Sarnak MJ, Yan G, et al. Atherosclerotic cardiovascular disease risks in chronic hemodialysis patients. Kidney Int. 2000;58(1):353–362. doi: 10.1046/j.1523-1755.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- 13.Muntner P, Hamm LL, Kusek JW, Chen J, Whelton PK, He J. The prevalence of nontraditional risk factors for coronary heart disease in patients with chronic kidney disease. Ann Intern Med. 2004;140(1):9–17. doi: 10.7326/0003-4819-140-1-200401060-00006. [DOI] [PubMed] [Google Scholar]
- 14.Shlipak MG, Fried LF, Cushman M, et al. Cardiovascular mortality risk in chronic kidney disease: comparison of traditional and novel risk factors. JAMA. 2005;293(14):1737–1745. doi: 10.1001/jama.293.14.1737. [DOI] [PubMed] [Google Scholar]
- 15.Clase CM, Gao P, Tobe SW, et al. Estimated glomerular filtration rate and albuminuria as predictors of outcomes in patients with high cardiovascular risk: a cohort study. Ann Intern Med. 2011;154(5):310–318. doi: 10.7326/0003-4819-154-5-201103010-00005. [DOI] [PubMed] [Google Scholar]
- 16.Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012;379(9811):165–180. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 17.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bis JC, Kavousi M, Franceschini N, et al. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet. 2011;43(10):940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell GF, Verwoert GC, Tarasov KV, et al. Common Genetic Variation in the 3-BCL11B Gene Desert Is Associated with Carotid-Femoral Pulse Wave Velocity and Excess Cardiovascular Disease Risk: The AortaGen Consortium. Circ Cardiovasc Genet. 2012;5(1):81–90. doi: 10.1161/CIRCGENETICS.111.959817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikram MK, Sim X, Jensen RA, et al. Four novel Loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet. 2010;6(10):e1001184. doi: 10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol. 2011;69(6):928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boger CA, Chen MH, Tin A, et al. CUBN is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22(3):555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu CT, Garnaas MK, Tin A, et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7(9):e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 27.Stevens LA, Coresh J, Schmid CH, et al. Estimating GFR using serum cystatin C alone and in combination with serum creatinine: a pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis. 2008;51(3):395–406. doi: 10.1053/j.ajkd.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 29.Mattix HJ, Hsu CY, Shaykevich S, Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J Am Soc Nephrol. 2002;13(4):1034–1039. doi: 10.1681/ASN.V1341034. [DOI] [PubMed] [Google Scholar]
- 30.Devalliere J, Chatelais M, Fitau J, et al. LNK (SH2B3) is a key regulator of integrin signaling in endothelial cells and targets alpha-parvin to control cell adhesion and migration. FASEB J. 2012;26(6):2592–2606. doi: 10.1096/fj.11-193383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takaki S, Morita H, Tezuka Y, Takatsu K. Enhanced hematopoiesis by hematopoietic progenitor cells lacking intracellular adaptor protein, Lnk. J Exp Med. 2002;195(2):151–160. doi: 10.1084/jem.20011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soranzo N, Spector TD, Mangino M, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt KA, Zhernakova A, Turner G, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Johnson AD, O'Donnell CJ. PRIMe: a method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27(9):1201–1206. doi: 10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ix JH, Shlipak MG, Chertow GM, Whooley MA. Association of cystatin C with mortality, cardiovascular events, and incident heart failure among persons with coronary heart disease: data from the Heart and Soul Study. Circulation. 2007;115(2):173–179. doi: 10.1161/CIRCULATIONAHA.106.644286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis JW, Chen MH, Foster MC, et al. Validated SNPs for eGFR and their associations with albuminuria. Hum Mol Genet. 2012;21(14):3293–3298. doi: 10.1093/hmg/dds138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CJ, Liu HL, Pan CF, et al. Indoxyl sulfate predicts cardiovascular disease and renal function deterioration in advanced chronic kidney disease. Arch Med Res. 2012;43(6):451–456. doi: 10.1016/j.arcmed.2012.08.002. [DOI] [PubMed] [Google Scholar]