Abstract
Graft-versus-host disease (GVHD) induced by donor-derived T cells remains the major limitation of allogeneic bone marrow transplantation (allo-BMT). We previously reported that the pan-Notch inhibitor DNMAML markedly decreased the severity and mortality of acute GVHD mediated by CD4+ T cells in mice. To elucidate the mechanisms of Notch action in GVHD and its role in CD8+ T cells, we studied the effects of Notch inhibition in alloreactive CD4+ and CD8+ T cells using mouse models of allo-BMT. DNMAML blocked GVHD induced by either CD4+ or CD8+ T cells. Both CD4+ and CD8+ Notch-deprived T cells had preserved expansion in lymphoid organs of recipients, but profoundly decreased IFNγ production despite normal T-bet and enhanced Eomesodermin expression. Alloreactive DNMAML T cells exhibited decreased Ras/MAPK and NF-kB activity upon ex vivo restimulation through the TCR. In addition, alloreactive T cells primed in the absence of Notch signaling had increased expression of several negative regulators of T cell activation, including Dgka, Cblb and Pdcd1. DNMAML expression had modest effects on in vivo proliferation but preserved overall alloreactive T cell expansion while enhancing accumulation of preexisting natural regulatory T cells. Overall, DNMAML T cells acquired a hyporesponsive phenotype that blocked cytokine production but maintained their expansion in irradiated allo-BMT recipients, as well as their in vivo and ex vivo cytotoxic potential. Our results reveal parallel roles for Notch signaling in alloreactive CD4+ and CD8+ T cells that differ from past reports of Notch action and highlight the therapeutic potential of Notch inhibition in GVHD.
Introduction
Notch signaling is a highly conserved cell-to-cell communication pathway with multiple functions in health and disease (1). Notch receptors interact with Jagged or Delta-like ligands, leading to proteolytic release of intracellular Notch (ICN). ICN translocates into the nucleus to interact with CSL/RBP-Jk (encoded by Rbpj) and a Mastermind-like (MAML) transcriptional coactivator. ICN, CSL/RBP-Jk and MAML assemble a large complex mediating gene activation. In hematopoiesis, Notch was first identified for its essential role during early T cell development (2). Other functions in B cell and myeloid lineages were subsequently reported (3–5). Moreover, emerging evidence indicates that Notch has important context-dependent effects on mature T cell differentiation and function (6).
Using genetic models of Notch inhibition, we discovered an essential role of Notch signaling in CD4+ T cells mediating graft-versus-host disease (GVHD) after allogeneic bone marrow transplantation (allo-BMT) (7). GVHD is the most significant complication limiting the success of allo-BMT (8). Current strategies to control GVHD rely on global immunosuppression. These strategies are incompletely effective and decrease graft-versus-tumor activity. To improve allo-BMT outcome, it is essential to identify new approaches that limit GVHD without eliminating potent anti-cancer effects. To inhibit Notch signaling downstream of all Notch receptors, we expressed a Cre-inducible dominant negative form of Mastermind-like 1 (DNMAML) under control of the ROSA26 locus. Upon Cd4-Cre expression, pan-Notch inhibition was achieved in mature CD4+ and CD8+ T cells without interference with early stages of T cell development (7, 9, 10). DNMAML blocks the Notch transcriptional activation complex downstream of all Notch receptors, with similar effects to those observed in the absence of CSL/RBP-Jk (the DNA-binding transcription factor that mediates all the effects of canonical Notch signaling). In mouse allo-BMT models, pan-Notch blockade in donor CD4+ T cells led to markedly reduced GVHD severity and improved survival (7). Notch-deprived alloreactive CD4+ T cells had decreased production of inflammatory cytokines, including IFNγ, TNFα, IL-17A, IL-4 and IL-2. Concomitantly, Notch inhibition led to increased accumulation of regulatory T cells (Tregs). However, Notch-deprived CD4+ alloreactive T cells were capable of extensive proliferation, allowing for their enhanced accumulation in lymphoid tissues. Despite reduced cytokine production, Notch-deprived CD4+ T cells retained potent cytotoxic potential in vivo. Thus, Notch signaling in T cells is a new attractive therapeutic target to control GVHD after allo-BMT.
Both CD4+ and CD8+ T cells have pathogenic effects during GVHD. Although we reported an essential function of Notch in CD4+ alloreactive T cells, no information has been available about Notch in CD8+ T cell-driven GVHD. Furthermore, it is unclear if the profound effects of Notch signaling in acute GVHD can be explained by its previously reported effects in T cells, or if new mechanisms are involved. Past work highlighted independent effects of Notch signaling in CD4+ and CD8+ T cells. Notch was shown to regulate Il4 and Gata3 expression during Th2 differentiation (9–12). In Th1 cells, pharmacological inhibitors and a Notch1 antisense strategy suggested that Notch controlled expression of Tbx21, encoding T-bet, a transcription factor regulating Ifng transcription (13). Notch signaling was also shown to influence Th17 and Treg differentiation, as well as CD4+ T cell longevity, at least in vitro (14–17). In CD8+ T cells, Notch was suggested to act directly at the Eomes and Gzmb loci, with an impact on differentiation and function (18–20). However, these findings originate from heterogeneous experimental systems, different immune contexts and variable strategies to manipulate Notch signaling, including gain-of-function approaches and pharmacological inhibitors. These results can be confounded by off-target effects and may not reflect the physiological functions of Notch in T cells.
Here, we investigated the cellular and molecular mechanisms underlying the effects of Notch signaling in alloreactive CD4+ and CD8+ T cells during GVHD. Our strategy relied on in vivo priming of donor T cells in the presence or absence of all canonical CSL/RBP-Jk and MAML-dependent Notch signals specifically in T cells, ensuring that T cells were exposed to relevant Notch ligands in the post-transplantation environment. Notch-deprived alloreactive CD4+ and CD8+ T cells shared a profound defect in IFNγ production, suggesting parallel effects of Notch in both T cell subsets. Decreased IFNγ was observed despite preserved or enhanced expression of the transcription factors T-bet and Eomesodermin, consistent with the absence of a classical Th1 or effector CD8+ T cell differentiation defect. Notch-deprived alloreactive CD4+ and CD8+ T cells acquired a hyporesponsive phenotype with decreased Ras/MAPK and NF-kB signaling. Notch inhibition led to increased expression of selected negative regulators of T cell activation. Some of these characteristics have been observed in anergic T cells, suggesting that Notch-inhibited CD4+ and CD8+ T cells acquire an anergy-like phenotype after allo-BMT, resulting in decreased production of inflammatory cytokines. Despite these changes, Notch inhibition preserved alloreactive T cell expansion in vivo and only had modest effects on their proliferative potential, while increasing expansion of preexisting natural Tregs and preserving high cytotoxic potential. Altogether, our data demonstrate a novel, shared mechanism of Notch action in alloreactive CD4+ and CD8+ T cells during allo-BMT which differs from all previous reports of Notch activity in T cells. Understanding these effects is essential to harness the therapeutic benefits of Notch blockade to control GVHD after allo-BMT.
Methods
Mice
BALB/c (H-2d) and C57BL/6 (B6, H-2b, CD45.2+) mice were from Harlan (Indianapolis, IN); C57BL/6.Ptprca (B6-SJL, H-2b, CD45.1+) from the NCI (Frederick, MD); BALB/b (H-2b) and Foxp3-IRES-RFP (FIR) from Jackson Laboratories (Bar Harbor, ME) (21). NF-kB reporter mice (NGL) were described previously (22). B6.129S6-Tbx21tm1Glm/J mice were provided by Dr. Segal (University of Michigan) (23); Eomesf/fxCd4-Cre mice by Dr. Reiner (Columbia University) (24); Rbpjf/f mice by Dr. Honjo (Kyoto, Japan) (4). ROSA26DNMAMLf mice (DNMAML) contain a Cre-inducible cassette encoding the DNMAML-GFP pan-Notch inhibitor (9, 25). DNMAML and Rbpjf/f mice were crossed to Cd4-Cre mice. All mice were backcrossed to the B6 background (>8 generations). The University of Michigan Committee on Use and Care of Animals approved all experiments.
Induction and assessment of GVHD
Mice underwent allo-BMT as described (7). T cell-depleted bone marrow (TCD BM) was prepared with anti-Thy1.2 antibodies and complement (Cedarlane Labs, Burlington, NC; >95% depletion). BALB/c recipients were irradiated (850–900 rads, 137Cs) 4 hours before allo-BMT. We transplanted donor B6-SJL TCD BM (4–5×106) with/without B6 splenocytes (5–10×106). Where indicated, CD4+ and CD8+ T cells were MACS-purified (Miltenyi, Auburn, CA) and mixed before transplantation into irradiated BALB/b or BALB/c recipients. Clinical GVHD was scored at least weekly.
Flow cytometry
The following antibodies were from BioLegend (San Diego, CA): anti-CD4, CD8α, CD44, CD45.1, CD45.2, CD62L, TCRβ, CD3, H-2Kb, H-2Kd, IFNγ, IL-2, CD28 (37.51), CD152/Ctla-4 (UC10-4B9), and CD279/Pd-1 (RMP1-30). Anti-T-bet (4B10), Eomesodermin (Dan11mag), and CD272/Btla (6F7) antibodies were from eBioscience (San Diego, CA). For T cell restimulation, we used plate-bound anti-CD3 (145-2C11) and anti-CD28 (37.51) (Biolegend, 2.5 µg/ml) or PMA (50 ng/mL) and Ionomycin (50 ng/mL). Intracellular flow cytometry was performed per manufacturer’s instructions after addition of Brefeldin A (>2 hours) (BD). Analysis/sorting were on FACSCanto or FACSAria II/III (BD). Dead cells were excluded with DAPI (Sigma-Aldrich, St. Louis, MO). Files were analyzed in FlowJo (Tree Star, San Carlos, CA).
Quantitative reverse-transcription PCR
RNA was isolated using RNEasy MicroKit (Qiagen, Valencia, CA) or TRIzol (Invitrogen, Carlsbad, CA). cDNA was prepared with Superscript II (Invitrogen). qPCR was performed with TaqMan (Applied Biosystems, Carlsbad, CA) or SybrGreen (Fisher, Rockford, IL) on Mastercycler realplex (Eppendorf, Westbury, NY). Relative expression was calculated using the ΔΔCt method. Primers were from PrimerBank (http://pga.mgh.harvard.edu/primerbank/) or Applied Biosystems.
SDS-PAGE and Western blotting
Naïve (CD62LHighCD44Low) and alloreactive CD4+ and CD8+ T cells were negatively selected using an anti-NK1.1/CD19/Gr-1/CD11b/CD11c cocktail. T cells were incubated on ice with anti-CD3/CD28 antibodies (0.2 µg/106 cells) followed by cross-linking with anti-Armenian hamster IgG (0.5 µg/106 cells, Jackson ImmunoResearch, West Grove, PA) in RPMI-1640 at 37°C. Alternatively, T cells were activated with PMA (Sigma-Aldrich, 50 ng/ml) or DMSO at 37°C. Cells were lysed in Laemmli buffer and 2-ME. Samples were run on 4–20% MiniTGX gels (Bio-Rad) and transferred to Immobilon-P membranes (GenHunter, Nashville, TN) (semi-dry transfer, Bio-Rad). Membranes were blocked in 10% FBS+TBS-T (25 mM TrisBase-pH8, 125 mM NaCl, 0.05% Tween). Antibodies were from Cell Signaling (Danvers, MA): anti-MEK1/2 rabbit (47E6), p44/42 MAPK-Erk1/2 mouse (3A7), phospho-MEK1/2 (Ser217/221) rabbit (41G9), phospho-p44/42 MAPK-Erk1/2 (Thr202/Tyr204) (E10). Secondary antibodies were peroxidase-conjugated goat anti-rabbit IgG(H+L) or donkey anti-mouse IgG(H+L) (Jackson ImmunoResearch). Blots were developed with ECL Substrate (Thermo) and HyBlotCL (Denville Scientific, South Plainfield, NJ).
Intracellular cAMP assay
cAMP was measured using ELISA, per manufacturer’s instructions (Enzo LifeSciences, Farmingdale, NY).
Luciferase assay
Sort-purified alloreactive T cells were restimulated with plate-bound anti-CD3/CD28 (Biolegend, 2.5 µg/mL) for 16 hours. Luciferase activity was measured using the Luciferase Assay System (Promega, Madison, WI) and read on PerkinElmer Enspire2300 (Waltham, MA).
In vitro and in vivo proliferation assays
Splenocytes or CD4+ and CD8+ T cells were MACS-purified according to manufacturer’s instructions (Miltenyi) and labeled with 2.5µM CFSE (Sigma-Aldrich). T cells were incubated for 3 days on anti-CD3/CD28+/-hrIL-2 (Peprotech). For BrdU pulse-chase experiments, splenocytes were transplanted into irradiated BALB/c mice. Starting at day 4 post-transplantation, mice received 3 doses of BrdU (1 mg) intraperitoneally, 12 hours apart. Four hours (day 5) or 3 days (day 8) after the last BrdU injection, spleens were harvested for BrdU staining (BD).
Serum cytokine analysis
Serum was collected on day 5 post-transplantation. Serum IFNγ levels were measured by mouse IFNγ duoset (R&D Systems, Minneapolis, MN) per the manufacturer’s instructions (Immunology Core, University of Michigan Cancer Center).
In vivo cytotoxicity
Experiments were performed as previously described (7). Briefly, lethally irradiated (900 rads) BALB/c mice received T cell depleted bone marrow (5×106 cells) alone or with WT or DNMAML splenocytes (8×106). On day 13, BALB/c recipients were challenged with a 1:1 mixture of CFSEHigh BALB/c allogeneic targets and CFSELow B6 syngeneic control cells (107 each). After 18 hours, elimination of BALB/c allogeneic targets was measured by flow cytometry in the spleen.
51Chromium release assay
Spleen and lymph nodes were harvested on day 8 post-transplantation. WT and DNMAML CD8+ T cells were purified by MACS according to the manufacturer’s instructions (Miltenyi). A20 (H-2d) and P815 (H-2d) cells were used as allogeneic targets, with EL4 (H-2b) as syngeneic control targets. Tumor targets were labeled with 2MBq of Na251CrO4 (PerkinElmer Life, Boston, MA, USA) for 2 hours. After washing 3 times, labeled targets were plated at 5 × 103 cells per well in U-bottom 96-well plates (Corning-Costar Corp., Cambridge, MA). Splenocytes were added in triplicate wells at varying E:T ratios, and incubated for 5 hours. 51Cr activity in supernatants was read in a LumaPlate (PerkinElmer, Waltham, MA) in an auto-gamma counter (Packard Instrument Company, Meriden, CT). Maximal and background release were determined by the addition of 2% Triton X-100 (Sigma) or media alone to targets, respectively. The percentage of specific lysis was calculated as 100 × (sample count – background count)/(maximal count – background count).
Statistical analysis
Comparison of two means was performed with 2-tailed unpaired Student t test. When less than five data points were available per group, we used the unpaired Mann-Whitney U test. Linear regression analysis was used for comparison of cytotoxicity curves. Survival was compared using log-rank statistics (GraphPad-Prism, La Jolla, CA).
Results
Notch inhibition blocks acute graft-versus-host disease mediated by CD4+ or CD8+ T cells
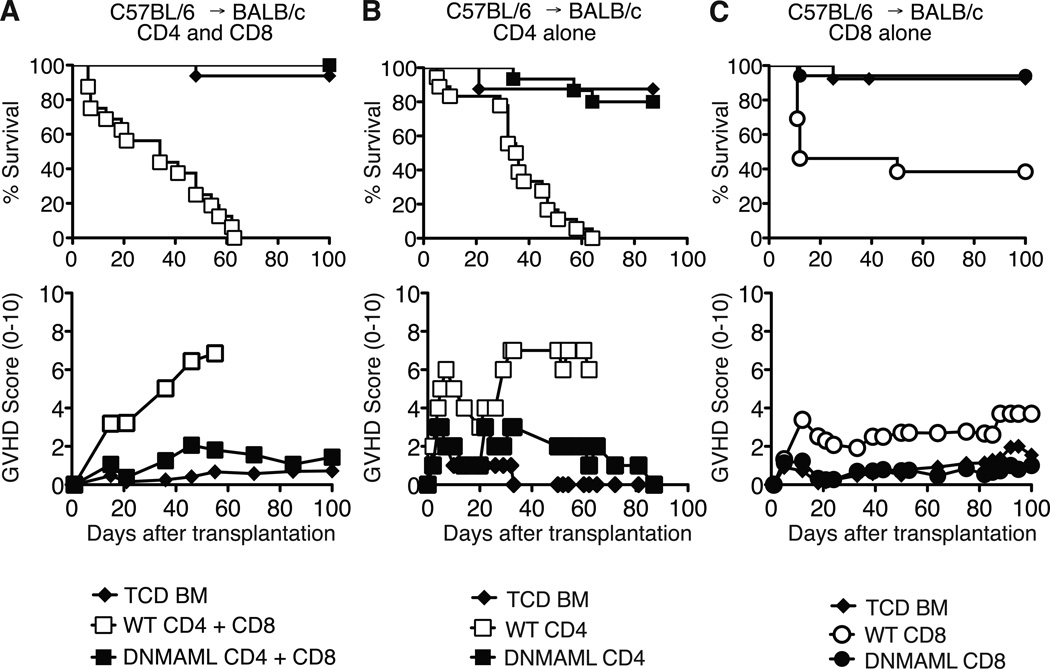
We previously reported an essential role for Notch in CD4+ T cells during acute GVHD in a major histocompatibility complex-mismatched allo-BMT model (B6 anti-BALB/c) (7). To assess if alloreactive CD8+ T cells were also sensitive to Notch signaling, we used ROSA26DNMAMLf x Cd4-Cre (DNMAML) mice as source of Notch-deprived CD4+ and CD8+ T cells. These mice express the DNMAML pan-Notch inhibitor in all mature CD4+ and CD8+ T cells. Wild-type (WT) or DNMAML B6 splenocytes were transplanted into irradiated BALB/c mice. Recipients were monitored for survival and GVHD severity. Allo-BMT recipients of B6 T cells died rapidly with severe GVHD (Fig. 1A). In contrast, recipients of DNMAML CD4+ and CD8+ T cells survived as well as mice infused only with T cell-depleted bone marrow (Fig. 1A). We then performed allo-BMT with purified CD4+ or CD8+ T cells. DNMAML expression blocked severe GVHD induced by CD4+ T cells (Fig. 1B). Purified CD8+ T cells also induced significant lethality post-transplantation with <40% long-term survival (Fig. 1C). In contrast, DNMAML CD8+ T cell recipients had >94% survival by day 100, similar to mice receiving no T cells (Fig. 1C). These data demonstrate that Notch is an essential regulator of GVHD induced by either or both CD4+ and CD8+ T cells after MHC-mismatched allo-BMT.
Figure 1.
DNMAML inhibits graft-versus-host disease mediated by CD4+ and/or CD8+ T cells after MHC-mismatched bone marrow transplantation. Lethally irradiated BALB/c mice (H-2d) were transplanted with B6 T cell-depleted bone marrow (TCD BM, 5×106 cells) alone or with (A) B6 (H-2b) splenocytes containing CD4+ and CD8+ T cells from wild-type (WT) or DNMAML mice (10×106 cells; 16 mice/group) (p<0.001 for WT vs. TCD and WT vs. DNMAML survival); (B) purified B6 WT or DNMAML CD4+ T cells (2×106 cells; 8–17 mice/group) (pp<0.001 for WT vs. TCD and WT vs. DNMAML survival); (C) purified B6 WT or DNMAML CD8+ T cells (5×106 cells; 13–17 mice/group) (p<0.01 for WT vs. TCD; p<0.001 for WT vs. DNMAML survival). Recipients were monitored over time for survival and GVHD severity after transplantation (clinical GVHD score, 0–10).
To further investigate the role of Notch in CD4+ and CD8+ alloreactive T cells, we used the B6 anti-BALB/c model as well as a minor histocompatibility antigen (miHA)-mismatched model (B6 anti-BALB/b) in which lethal GVHD is mediated by CD4-dependent CD8+ T cells (27). Together, WT CD4+ and CD8+ T cells induced significant lethality with <20% long-term survival in both models (Fig. S1A, B). DNMAML CD4+ and CD8+ T cell recipients achieved ~80% survival (Fig. S1A, B). When either CD4+ or CD8+ T cells expressed DNMAML, recipients were protected from severe GVHD with 80–90% of the mice surviving long-term (Fig. S1A, B). These data indicate that Notch is required in both CD4+ and CD8+ T cells to fully induce GVHD in MHC- and miHA-mismatched allo-BMT models.
Alloreactive DNMAML CD8+ T cells display intrinsic and CD4+ T cell-dependent defects in IFNγ production
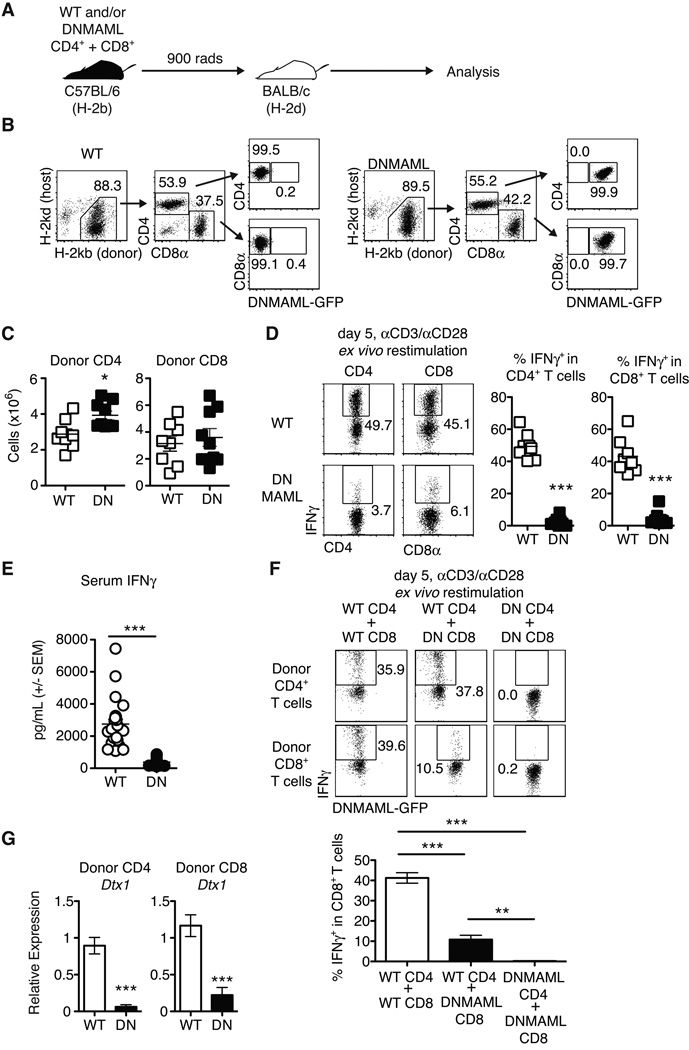
Notch inhibition preserves CD4+ T cell expansion after allo-BMT, but profoundly decreases IFNγ production (7). Since CD8+ T cells are a major source of IFNγ during GVHD, we assessed DNMAML CD8+ T cell expansion and IFNγ production. After transplantation in the B6 anti-BALB/c model (Fig. 2A), we recovered similar numbers of donor-derived WT and DNMAML CD4+ and CD8+ T cells from lymphoid organs of allo-BMT recipients, indicating that Notch blockade did not prevent in vivo expansion of these cells (Fig. 2B-C). In contrast, DNMAML expression markedly decreased IFNγ production by both alloreactive CD4+ and CD8+ T cells (Fig. 2D), suggesting parallel effects of Notch in these two subsets. These differences in IFNγ production could not be explained by increased apoptosis of Notch-deprived T cells during the restimulation period (data not shown). To determine if changes in IFNγ production by Notch-deprived alloreactive T cells were biologically relevant, serum IFNγ levels were measured on day 5 after transplantation (Fig. 2E). Serum IFNγ levels were markedly reduced in recipients of DNMAML T cells. Since IFNγ production by CD8+ T cells is influenced by cell-intrinsic signals and CD4+ T cell help, we studied DNMAML CD8+ T cells in the presence of WT or DNMAML alloreactive CD4+ T cells (Fig. 2F). WT CD4+ and CD8+ T cells transplanted together produced abundant IFNγ. In the presence of WT CD4+ T cells, DNMAML expression in CD8+ T cells markedly decreased but did not abolish IFNγ production. When both DNMAML CD4+ and CD8+ T cells were infused, CD8+ T cells had little to no IFNγ production. Thus, Notch promotes maximal IFNγ production by alloreactive CD8+ T cells via cell-intrinsic changes in the CD8+ compartment and effects on CD4+ T cell help.
Figure 2.
Alloreactive DNMAML CD4+ and CD8+ T cells have preserved in vivo expansion but markedly decreased IFNγ production. (A) Experimental design: lethally irradiated BALB/c mice were transplanted with combinations of WT or DNMAML CD4+ and CD8+ B6 T cells; (B) Donor-derived T cells were tracked by flow cytometry based on expression of donor/host MHC class I molecules and DNMAML-GFP; (C) Preserved expansion of donor-derived H-2Kb+H-2Kd– DNMAML CD4+ and CD8+ T cells as compared to WT T cells on day 5 post-transplantation (n=3–5 mice/group, 2 independent experiments); (D) Intracellular staining for IFNγ in donor-derived H-2Kb+H-2Kd– CD4+ and CD8+ spleen T cells after ex vivo anti-CD3/CD28 restimulation (n=3–5 mice/group, representative of >3 experiments). DNMAML inhibited IFNγ production by CD4+ and CD8+ T cells; (E) Serum was collected on day 5 after transplantation and IFNγ levels were measured by ELISA (n=5 mice/group, >2 independent experiments);(F) Mixed populations of WT or DNMAML CD4+ and CD8+ T cells (2×106 each) were transplanted into lethally irradiated BALB/c recipients. IFNγ production was measured by intracellular flow cytometry on day 5 post-transplantation. DNMAML blocked IFNγ production in CD8+ T cells both through cell-intrinsic and CD4-dependent effects (n=3/group, representative of two experiments); (G) Abundance of Dtx1 Notch target gene mRNA in sort-purified alloreactive CD4+ and CD8+ T cells on day 5 post-transplantation (n=3 mice/group, representative of >3 experiments). **p<0.01; ***p<0.001.
To verify that Notch signaling was potently inhibited in both alloreactive DNMAML CD4+ and CD8+ T cells, we assessed expression of the Notch target gene Dtx1 (Fig. 2G). Dtx1 transcripts were significantly reduced in both subsets of alloreactive DNMAML T cells. Collectively, inhibition of Notch signaling in alloreactive T cells preserved their expansion but profoundly reduced T cell-dependent IFNγ production and systemic IFNγ levels.
Notch inhibition blocks IFNγ production in alloreactive CD4+ and CD8+ T cells despite preserved T-bet and Eomesodermin expression
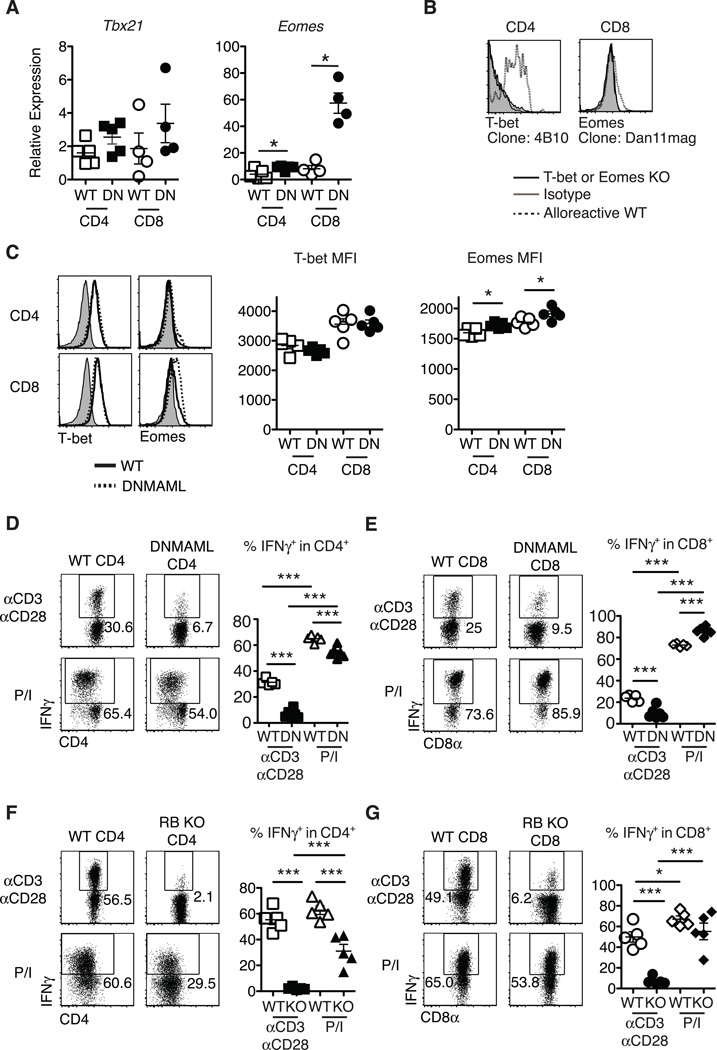
Notch was suggested to control the expression of Tbx21 (encoding T-bet) in CD4+ T cells and Eomes (encoding Eomesodermin) in CD8+ T cells (13, 18, 19). To assess if these mechanisms accounted for decreased IFNγ production by Notch-deprived CD4+ and CD8+ T cells after allo-BMT, we measured T-bet and Eomesodermin levels at the peak of the effector response. Alloreactive DNMAML CD4+ and CD8+ T cells had preserved Tbx21 and increased Eomes transcripts as compared to WT T cells (Fig. 3A). We used flow cytometry to assess intracellular T-bet and Eomesodermin and established antibody specificity by staining alloreactive T cells from WT mice compared to Tbx21−/− and Eomesf/f×Cd4-Cre mice (Fig. 3B). Consistent with mRNA findings, alloreactive DNMAML CD4+ and CD8+ T cells had preserved intracellular T-bet and increased Eomesodermin (Fig. 3C). As expected, we observed more Eomesodermin protein in CD8+ T cells than CD4+ T cells, consistent with accurate detection of Eomesodermin (28). These data indicate that decreased IFNγ production by DNMAML CD4+ and CD8+ T cells was not caused by decreased Th1 and effector CD8+ T cell differentiation resulting from reduced expression of these master transcription factors.
Figure 3.
Preserved T-bet and enhanced Eomesodermin expression in alloreactive Notch-deprived CD4+ and CD8+ T cells, allowing restoration of IFNγ production after treatment with PMA and ionomycin. Lethally irradiated BALB/c mice (900 rads) were transplanted with B6-CD45.1 TCD BM (5×106 cells) and splenocytes (10×106 cells) from WT or DNMAML B6 mice. (A) Preserved Tbx21 mRNA (encoding T-bet) and enhanced Eomes transcripts in DNMAML CD4+ and CD8+ T cells. Donor-derived H-2Kb+H-2Kd–CD45.2+ CD4+ and CD8+ T cells were sort-purified and subjected to qRT-PCR (day 14, n=4–5 mice, representative of 3 experiments); (B) Specific detection of T-bet and Eomesodermin in alloreactive T cells with anti-T-bet (4B10) and anti-Eomesodermin (Dan11mag) antibodies. Splenocytes from WT, B6.129S6-Tbx21tm1Glm/J or Eomesf/fxCd4-Cre mice were transplanted into irradiated BALB/c recipients. Histograms show intracellular staining with isotype control or specific antibodies in donor-derived H-2Kb+H-2Kd–CD45.2+ CD4+ or CD8+ T cells (day 5) (n=2); (C) Representative intracellular flow cytometry plots and mean fluorescence intensity (MFI) for T-bet and Eomesodermin expression in alloreactive WT and DNMAML CD4+ and CD8+ T cells (day 14, n=4–5 mice, representative of 4 experiments); (D-G) At day 5 after transplantation, spleen and lymph node cells were incubated for 6 hours with either anti-CD3/anti-CD28 (2.5 µg/ml each), or PMA and ionomycin (50 ng/ml and 500 ng/ml, respectively). Percent IFNγ+ cells as measured by intracellular flow cytometry in WT and DNMAML donor-derived (D) CD4+ and (E) CD8+ T cells, or WT and CSL/RBP-Jk-deficient donor-derived (F) CD4+ and (G) CD8+ T cells (n=5 mice/group, representative of >2 experiments). Representative flow cytometry plots are shown. Numbers indicate the percentage of cells in each quadrant. * p<0.05; *** p<0.001.
PMA (a diacylglycerol analog) and ionomycin (a calcium ionophore) are often used to elicit cytokine production to study T cell differentiation. This strategy can reveal defective IFNγ production by Tbx21−/− T cells (29). In contrast, restimulation of alloreactive DNMAML T cells with PMA/ionomycin restored IFNγ production by both CD4+ (Fig. 3D) and CD8+ T cells (Fig. 3E) close to levels observed in WT T cells. Partial rescue of IL-2 production was also apparent (Fig. S2). We confirmed the effects of DNMAML-mediated Notch blockade on IFNγ production using Rbpj−/− T cells lacking CSL/RBP-Jk, a central component of the Notch transcriptional activation complex (Fig. 3F-G). Altogether, our observations were consistent with the presence of functional T-bet and Eomesodermin capable of activating Ifng transcription in DNMAML or CSL/RBP-Jk-deficient alloreactive T cells, suggesting that PMA/ionomycin restored activation of other pathways regulating cytokine production in these cells.
Notch-deprived alloreactive CD4+ and CD8+ T cells develop blunted Ras/MAPK and NF-kB activation
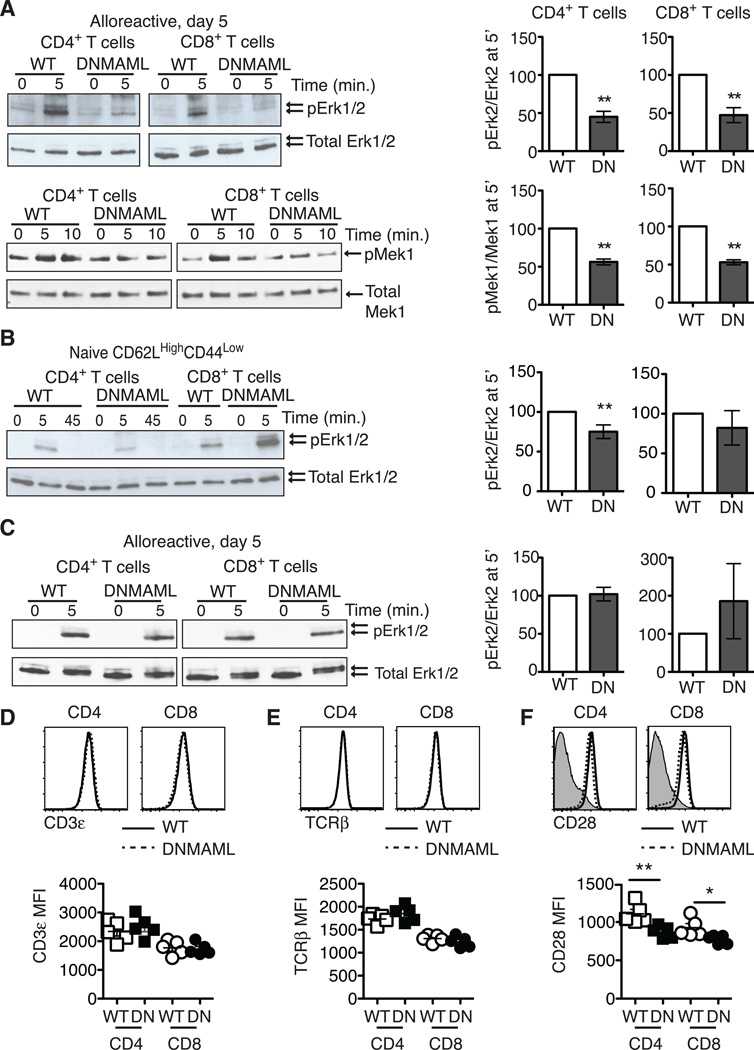
One of the major pathways activated by PMA is Ras/MAPK signaling, a key contributor to T cell cytokine gene transcription (30). To assess if alloreactive DNMAML T cells had decreased Ras/MAPK activation, we measured phosphorylation of Erk1/2 and its upstream kinase Mek1. Notch-deprived CD4+ and CD8+ T cells were primed in vivo before sort-purification and ex vivo restimulation through the TCR and CD28 co-receptor. Both alloreactive DNMAML CD4+ and CD8+ T cells had a significant reduction in Mek1 and Erk1/2 phosphorylation (Fig. 4A). Interestingly, naïve CD62LHighCD44Low DNMAML CD4+ T cells only had slightly decreased Erk1/2 phosphorylation, while naïve DNMAML CD8+ T cells activated Erk1/2 normally, suggesting that the majority of the Ras/MAPK defect was acquired in vivo in the absence of Notch signaling (Fig. 4B). Next, we assessed if PMA rescued Ras/MAPK activation in DNMAML alloreactive T cells. PMA induced similar Erk2 phosphorylation in WT and DNMAML T cells, indicating restoration of Ras/MAPK signaling in DNMAML T cells (Fig. 4C).
Figure 4.
Alloreactive Notch-deprived CD4+ and CD8+ T cells have an acquired defect in Ras/MAPK pathway activation that is rescued by PMA. (A) WT or DNMAML B6 splenocytes were transplanted into lethally irradiated BALB/c recipients. On day 5, H-2Kb+H-2Kd– alloreactive WT and DNMAML CD4+ and CD8+ T cells were sort-purified and restimulated in vitro for 5–10’ at 37°C with anti-CD3/anti-CD28 and IgG crosslinking. Baseline activation was assessed by incubating cells at 37°C with IgG crosslinker alone (0’ time point). Phosphorylated Erk1/2 and Mek1 were detected by Western blotting as compared to total Erk1/2 and Mek1; (B) Naïve CD62LHighCD44Low CD4+ and CD8+ T cells were sort-purified from WT and DNMAML mice. Mek1 phosphorylation was assessed after anti-CD3/CD28 restimulation; (C) Sort-purified, day 5 alloreactive WT and DNMAML CD4+ or CD8+ T cells were restimulated ex vivo with PMA for 5’ (or DMSO as negative control). In all experiments, the abundance of phosphorylated proteins was measured by densitometry relative to total protein levels. WT T cells were set to 100% (n=2–4 individual experiments, 6 mice/group in each experiment); (D) cell surface CD3, (E) TCRβ, and (F) CD28 levels were assessed in alloreactive WT and DNMAML CD4+ and CD8+ T cells on day 5 post-transplantation. Representative flow cytometry plots and mean fluorescence intensity are shown. **p<0.01.
NF-kB is another major pathway activated downstream of diacylglycerol that can regulate Ifng transcription in T cells (31). To capture the overall NF-kB activity in Notch-deprived alloreactive T cells, we bred DNMAML mice to transgenic NFkB-GFP-Luciferase (NGL) reporter mice and used F1 progeny as donors for allo-BMT (Fig. S3) (22). On day 5 post-transplantation, alloreactive NGL and NGL/DNMAML CD4+ and CD8+ T cells were sort-purified and restimulated with anti-CD3/CD28 antibodies. DNMAML CD4+ and CD8+ T cells had significantly reduced NF-kB/luciferase activity. Blunted signal transduction downstream of the TCR occured in the absence of any changes in TCRβ (Fig. 4D) or CD3ε (Fig. 4E) surface expression, although we observed a slight but significant decrease in CD28 expression (Fig. 4F). Altogether, both DNMAML CD4+ and CD8+ alloreactive T cells acquired a blunted capacity for Ras/MAPK and NF-kB activation downstream of TCR/CD28 signals after allo-BMT.
Alloreactive DNMAML CD4+ and CD8+ T cells have increased expression of multiple negative regulators of T cell activation
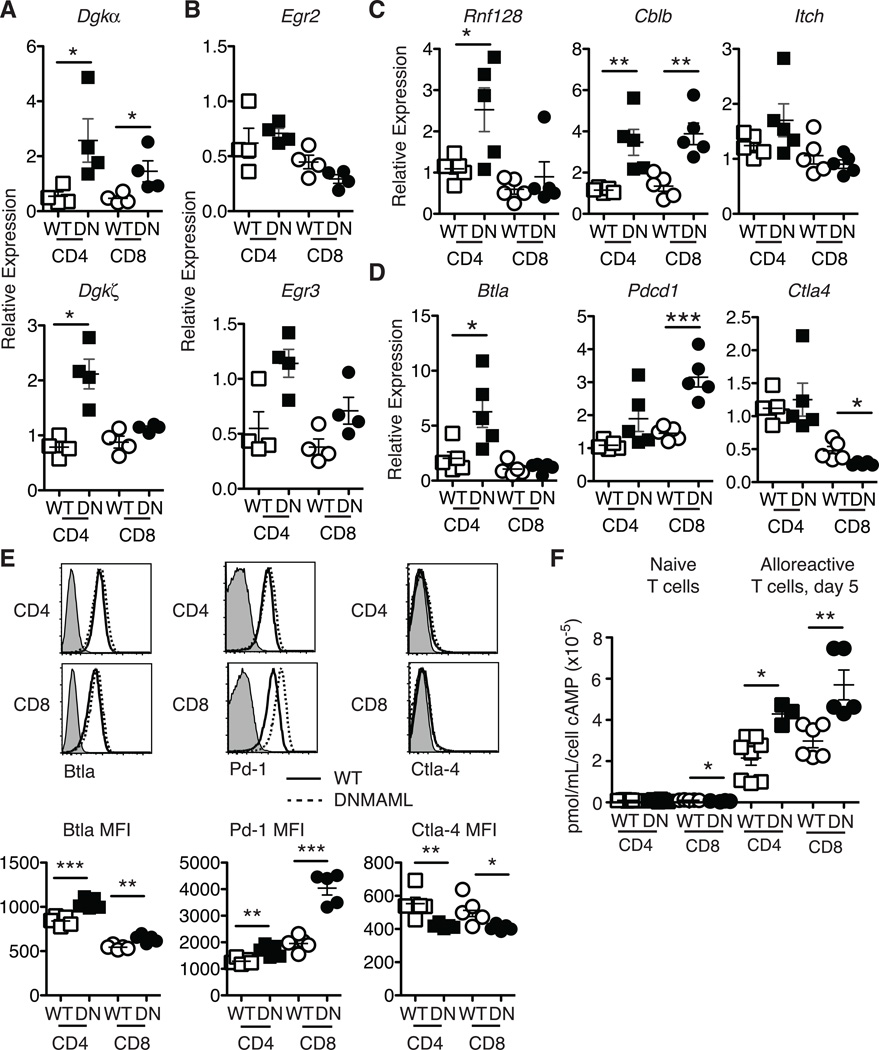
Decreased Ras/MAPK signaling leading to reduced cytokine production has been described in certain forms of T cell hyporesponsiveness or anergy (32, 33). Mechanistically, increased expression of diacylglycerol kinases (Dgk) such as Dgkα and Dgkζ was linked to increased degradation of diacylglycerol into phosphatidic acid, resulting in blunted Erk1/Erk2 activation and cytokine production (34, 35). Interestingly, restimulated alloreactive DNMAML CD4+ T cells had elevated levels of Dgka and Dgkz mRNA, while DNMAML CD8+ T cells had increased Dgka and a trend for more Dgkz transcripts (Fig. 5A). This constellation of effects was reminiscent of T cell anergy. Therefore, we studied a panel of anergy-associated genes, starting with the NFAT-dependent genes, Egr2/3 (36, 37). Egr3 but not Egr2 transcripts were increased in alloreactive DNMAML CD4+ and CD8+ T cells (Fig. 5B). We also observed increased expression of Rnf128 and Cblb, encoding Grail and Cbl-b, two E3 ubiquitin ligases that function as negative regulators of T cell activation (38, 39). In contrast, Itch expression was not significantly changed (Fig. 5C). As co-inhibitory receptors can also regulate alloreactive T cell activation and function, we investigated expression of BtlaCtla4 and Pdcd1 (encoding Pd-1) in DNMAML T cells. Although Ctla4 expression was slightly decreased, alloreactive DNMAML CD4+ T cells had increased Btla and DNMAML CD8+ T cells elevated Pdcd1 mRNA (Fig. 5D). These mRNA changes correlated well to protein expression with decreased Ctla-4 and increased Blta and Pd-1 in alloreactive DNMAML CD4+ and CD8+ T cells (Fig. 5E). Finally, DNMAML alloreactive T cells had increased intracellular cAMP, a second messenger that provides negative feedback regulation of T cell activation (Fig. 5F) (40).
Figure 5.
Increased levels of Dgka mRNA and other negative regulators of T cell activation in alloreactive DNMAML CD4+ and CD8+ T cells. H-2Kb+H-2Kd–CD45.2+alloreactive WT or DNMAML T cells were sort-purified at day 5 after transplantation. (A) Abundance of Dgka/Dgkz and (B) Egr2/Egr3 transcripts in anti-CD3/anti-CD28 restimulated alloreactive WT or DNMAML CD4+ and CD8+ T cells (n=4 mice, representative of >2 experiments); (C) Expression of anergy-associated E3 ubiquitin ligases Cblb, Rnf128, and Itch and (D) expression of T cell co-inhibitory receptors Btla, Ctla4, Pd-1 in sort-purified WT or DNMAML CD4+ and CD8+ T cells. Each symbol represents triplicate qRT-PCR data for cells purified from one individual recipient (n=5 mice, representative of 2 experiments); (E) Cell surface levels of T cell co-inhibitory receptors in WT and DNMAML CD4+ and CD8+ T cells (n=5 mice/group, representative of 2 experiments). Representative flow cytometry plots and mean fluorescence intensity are shown; (F) Increased intracellular cAMP in alloreactive DNMAML CD4+ and CD8+ T cells relative to WT T cells on day 5 after transplantation. cAMP levels were very low in naïve cells irrespectively of DNMAML expression (n=3 experiments, 6 mice/group in each experiment). *p<0.05; **p<0.01; ***p<0.001.
Altogether, Notch-deprived alloreactive CD4+ and CD8+ T cells acquired features of hyporesponsive T cells, including increased expression of several negative regulators of T cell activation, some of which are NFAT-dependent. Importantly, naïve DNMAML CD4+ and CD8+ T cells expressed normal levels of these negative regulators (except for Cblb and Itch which were mildly elevated in naïve DNMAML CD4+ T cells) (Fig. S4). These findings suggest that Notch deprivation results in changes in naïve T cells that only become apparent after allo-BMT or that Notch inhibition has minimal influence on naïve T cells, but profound effects on T cells in vivo during allo-BMT.
Notch inhibition modestly reduces in vivo proliferation of alloreactive CD4+ and CD8+ T cells while enhancing expansion of natural Tregs
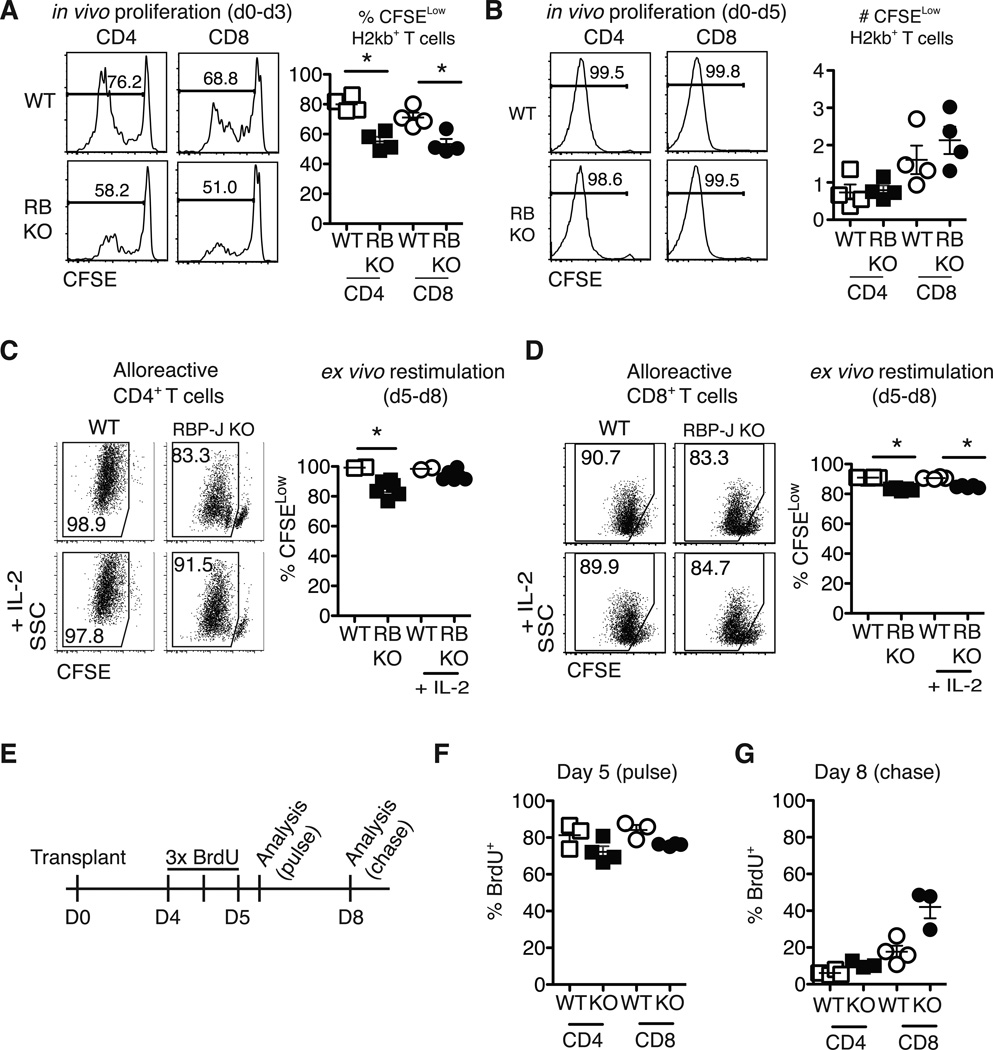
DNMAML alloreactive T cells acquire increased levels of negative regulators that have previously been associated with decreased proliferation, at least using in vitro models of T cell anergy (41). However, in vitro and in vivo T cell proliferation are regulated by different stimuli. To evaluate in detail the impact of Notch blockade on T cell proliferation in vivo after allo-BMT, we used Rbpjf/f×Cd4-Cre mice (Fig. 6). As DNMAML T cells, CSL/RBP-Jk-deficient T cells fail to respond to Notch signals, but lack DNMAML-GFP fluorescence, allowing use of CFSE to track proliferation. At day 3 after allo-BMT, Notch-deficient T cells had proliferated slightly less than WT T cells (Fig. 6A). However, by day 5, >99% of donor WT and CSL/RBP-Jk-deficient T cells had proliferated for >6–8 divisions (Fig. 6B). Thus, despite modestly reduced initial proliferation in the absence of Notch signaling, Notch-deprived T cells accumulated to levels similar to WT T cells by day 5 after allo-BMT. After this initial burst, ex vivo restimulation of Notch-deprived alloreactive T cells revealed only modest cycling defects (Fig. 6C-D). Similar in vivo and in vitro results were observed when DNMAML T cells were labeled with eFluor670 (data not shown). To assess ongoing in vivo proliferation, we performed a BrdU pulse-chase experiment 5–8 days after allo-BMT (Fig. 6E-G). CSL/RBP-Jk-deficient T cells demonstrated slightly reduced initial BrdU incorporation during the pulse phase (Fig. 6F). During chase, decreased loss of BrdU was apparent in Notch-deprived T cells (Fig. 6G). Thus, Notch inhibition modestly reduced in vivo proliferation of alloreactive CD4+ and CD8+ T cells, while preserving their massive initial expansion.
Figure 6.
Notch-deprived alloreactive CD4+ and CD8+ T cells have a preserved initial proliferative burst but subsequent reduced proliferation in vitro and in vivo. CFSE-labeled T cells from WT or Rbpjf/f x Cd4-Cre mice (RB KO, lacking all CSL/RBP-Jk-mediated Notch signals) were transplanted into lethally irradiated BALB/c recipients (900 rads). Flow cytometry plots show CFSE dilution at (A) day 3 and (B) day 5 in donor-derived H-2Kb+H-2Kd– CD4+ and CD8+ T cells. WT or RBP-Jk KO T cells were transplanted into lethally irradiated BALB/c recipients. On day 5, purified WT and RBP-Jk KO (C) CD4+ or (D) CD8+ T cells were CFSE-labeled and restimulated in vitro for 3 days with anti-CD3/CD28 +/− IL-2. Division history of donor-derived H-2Kb+H-2Kd– B6 T cells was measured by flow cytometry (n=6–7 mice/group in each experiment, representative of >2 experiments); (E) Design of BrdU pulse-chase experiment. Lethally irradiated BALB/c recipients were transplanted with splenocytes from WT or RBP-Jk KO mice. Between days 4 and 5, transplant recipients received 3 doses of BrdU 12 hours apart; (F) Four hours after the last BrdU injection, mice were euthanized for day 5 BrdU incorporation analysis (pulse); (G) At day 8 after transplantation, residual BrdU content was assessed (chase). n=5 mice/group in each experiment, representative of 2 experiments. *p<0.05
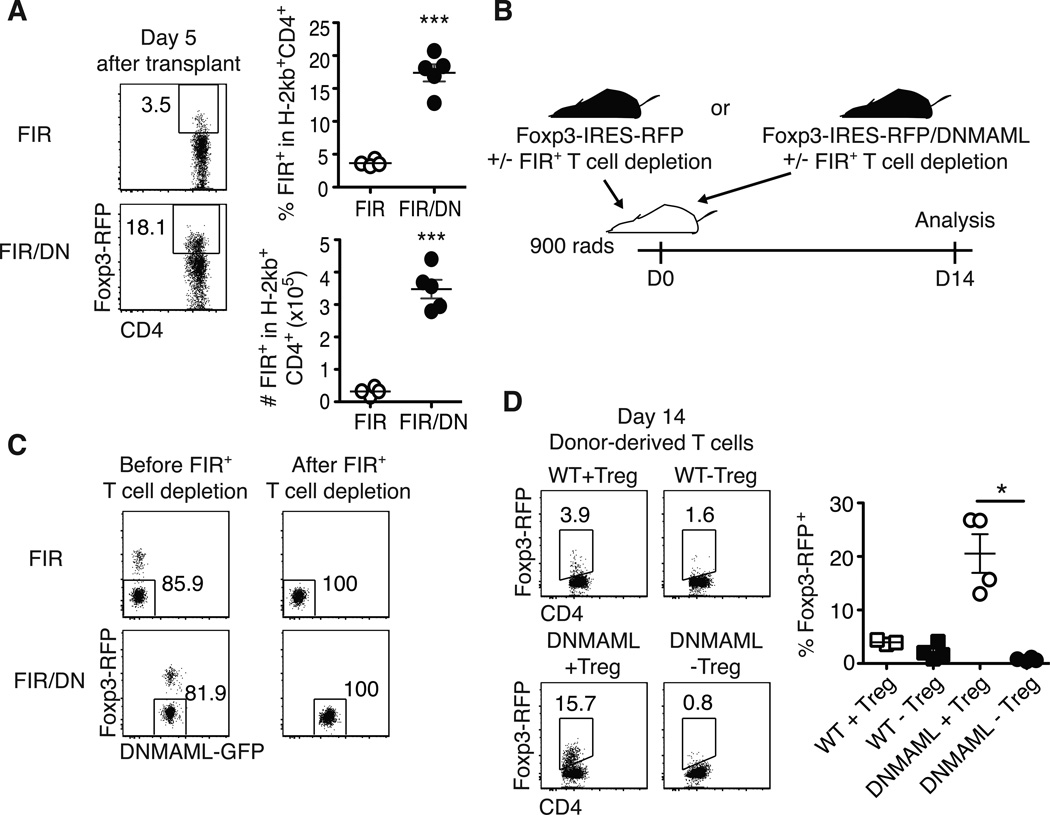
Traditional immunosuppressants often prevent Treg expansion, which can increase GVHD severity (42). If inhibition of Notch signaling is to be used to prevent or treat GVHD, we needed to assess the impact of Notch deprivation on Treg expansion. Using Foxp3-IRES-RFP+ DNMAML mice, we observed increased accumulation of DNMAML as compared to WT Foxp3+ donor T cells (Fig. 7A) (21). Furthermore, depletion of Tregs from the donor DNMAML inoculum completely prevented Treg expansion (Fig. 7B-D). These data indicate that rather than increasing conversion to induced Tregs, Notch blockade enhanced expansion of preexisting Tregs after allo-BMT.
Figure 7.
Notch inhibition enhances the accumulation of natural Foxp3+ regulatory T cells after allo-BMT. (A Detection of live regulatory T cell (Tregs) on day 5 post-transplantation using Foxp3-IRES-RFP (FIR) mice crossed to ROSA26DNMAMLf×Cd4-Cre mice, showing expanded Tregs among Notch-deprived alloreactive T cells; (B) To determine the origin of the expanded Tregs, lethally irradiated BALB/c recipients were transplanted with WT or DNMAML CD4+ T cells with or without FIR+ Tregs and analyzed on day 14 post-transplantation; (C) Post-sort purity of WT and DNMAML CD4+ T cell fractions after depletion of FIR+ cells; (D) Frequency of donor-derived FoxP3-IRES-RFP+ T cells as assessed by flow cytometry after transplantation of WT or DNMAML CD4+ T cells, including Tregs (+Treg), or depleted of Tregs (-Treg). Day 14 alloreactive DNMAML Tregs were derived from preexisting Tregs. *p<0.05; *** p<0.001.
Preserved anti-host and anti-tumor cytotoxicity of alloreactive Notch-deprived CD8+ T cells
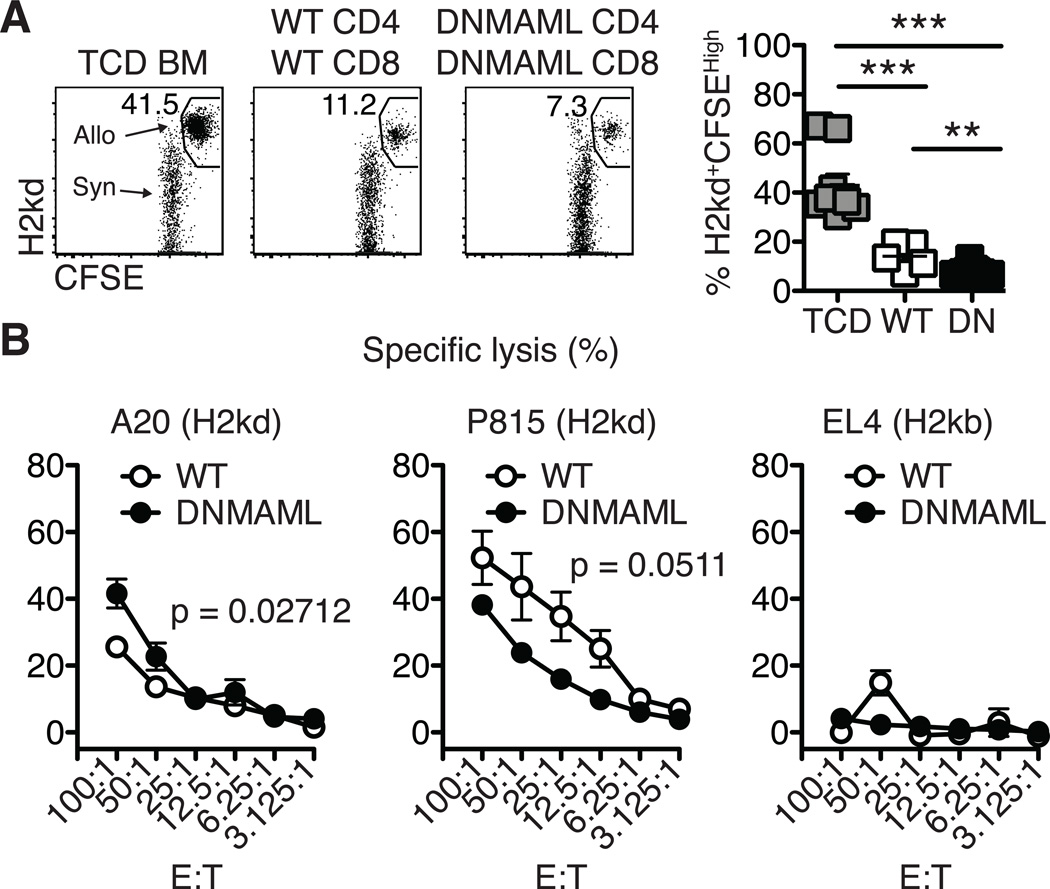
Our previous work showed that alloreactive Notch-deprived CD4+ T cells had preserved cytotoxicity against allogeneic host and tumor cells after transplantation (7). To test the overall in vivo cytotoxic potential of DNMAML B6 CD4+ and CD8+ T cells, BALB/c transplant recipients were challenged with a 1:1 mix of CFSEHigh BALB/c and CFSELow B6-CD45.1 splenocytes. Cytotoxicity against BALB/c targets was measured by flow cytometry (Fig. 8A). While recipients receiving TCD BM could not elicit cytotoxicity against the BALB/c targets, mice receiving WT or DNMAML splenocytes showed high and similar cytotoxicity against CFSEHigh BALB/c targets (Fig. 8A).
Figure 8.
Alloreactive Notch-deprived CD8+ T cells maintain high cytotoxic potential. (A) In vivo cytotoxicity assay. Transplanted BALB/c recipient mice were challenged on day 14 with a 1:1 mixture of CFSE-labeled allogeneic targets and control cells (CFSEHigh H-2Kd+ BALB/c and CFSElow control H-2Kb+ B6-CD45.1 splenocytes, respectively). After 18 hours, elimination of the BALB/c targets was assessed in the spleen by flow cytometry; (B) BALB/c recipients were transplanted with TCD BM and WT or DNMAML splenocytes (5×106 each). On day 8, alloreactive WT and DNMAML and naïve CD8+ T cells were MACS-purified and incubated with 51Chromium-labeled tumor cells at various E:T ratios for 5 hours (representative of n=3 independent experiments). A20 and P815 cells were allogeneic targets (H2kd). EL4 cells were syngeneic controls (H2kb). **p<0.01; ***p<0.001.
To determine if alloreactive purified DNMAML CD8+ T cells could lyse allogeneic tumor cells, an in vitro cytotoxicity assay was used. T cells were primed in vivo in lethally irradiated BALB/c recipients before assessing cytotoxicity ex vivo. Incubation of alloreactive WT or DNMAML CD8+ T cells with 51Chromium-labeled allogeneic A20 and P815 (H2kd) tumor cells showed efficient cytotoxicity, with preserved cytotoxicity against A20 cells and preserved or only slightly reduced cytotoxicity against P815 cells (Fig. 8B). As expected, syngeneic EL4 (H2kb) tumor cells were not killed (Fig. 8B). Collectively, these data demonstrate that Notch inhibition in alloreactive CD8+ T cells preserved a high degree of cytotoxic potential after transplantation.
Discussion
Our findings highlight a new, shared mechanism of Notch action in alloreactive CD4+ and CD8+ T cells that differs from all previously reported Notch functions in T cells. We used pan-Notch inhibition specifically in T cells to study the effects of Notch signaling in CD4+ and CD8+ T cell differentiation and function in several models of acute GVHD. Notch was absolutely required in both CD4+ and CD8+ T cells to mediate lethal acute GVHD. Notch inhibition preserved in vivo T cell expansion, but led to profoundly decreased IFNγ production by both T cell subsets. Decreased IFNγ production was not explained by an overall defect in Th1 CD4+ or effector CD8+ T cell differentiation, as expression of the master transcription factors T-bet and Eomesodermin was preserved or even enhanced in the absence of Notch signaling. In contrast, analysis of signal transduction pathways downstream of the TCR revealed defects in Ras/MAPK and NF-kB activation in Notch-deprived T cells, in addition to increased expression of multiple negative regulators of T cell activation. These features of hyporesponsiveness were observed in vivo upon T cell activation without Notch signaling. Importantly, Notch inhibition preserved the overall expansion of alloreactive T cells and efficient cytotoxic potential, while leading to increased accumulation of preexisting natural Foxp3+ Tregs. This constellation of effects led to beneficial immunomodulation, indicating that Notch is an attractive therapeutic target to control GVHD after allo-BMT.
Past work in other immune contexts suggested that Notch could directly regulate Tbx21 transcription in CD4+ T cells and Eomes expression in CD8+ T cells (13, 18, 19). In contrast, Notch appeared dispensable for transcriptional activation of these genes in alloreactive T cells during GVHD (Fig. 3A-C) (7). It is possible that use of different experimental systems to manipulate Notch signaling could underlie these discrepant observations. Others reported that Notch receptors could have “non-canonical” effects independent of CSL/RBP-Jk and MAML, although the nature of these effects is not well defined (43). However, in alloreactive T cells at least, the dominant effects of Notch are mediated by “canonical” signaling and do not involve decreased Tbx21 and Eomes expression. Alternatively, different signaling pathways may be required to activate Tbx21 and Eomes transcription in distinct immune responses. After allo-BMT, T cell exposure to abundant alloantigens in a highly inflammatory environment may bypass the requirement for Notch signaling to activate these genes. Moreover, an interesting feature of Notch-deprived alloreactive T cells was increased Eomes expression, a finding that was particularly apparent in CD8+ T cells but also detected in the CD4+ compartment. Although the regulation of Eomes expression during T cell differentiation is incompletely understood, decreased IL-12 signaling and increased Foxo1 activity were reported to enhance Eomes at the expense of Tbx21 expression (44, 45). The effects of Notch deprivation on Eomes could be mediated via interference with these pathways. Interestingly, increased Eomes expression was reported in memory CD8+ T cells that acquire an enhanced ability for long-term persistence, as opposed to terminal differentiation (44, 46). This mechanism could contribute to the enhanced expansion and survival of Notch-deprived alloreactive T cells after allo-BMT.
In contrast to past findings suggesting that Notch regulates independent aspects of CD4+ and CD8+ T cell biology, our results revealed similar effects of Notch in the CD4+ and CD8+ T cell compartments during GVHD. Notch-deprived CD4+ and CD8+ T cells shared key functional properties and gene expression changes after allo-BMT. During in vivo activation, DNMAML alloreactive T cells acquired elevated levels of several negative regulators of T cell activation, including genes whose activation is NFAT-dependent. A prominent feature of Notch-deprived T cells was decreased activation of Ras/MAPK and NF-kB pathways. Ras/MAPK and NF-kB activation are controlled by diacylglycerol, whose levels are negatively regulated by the lipid kinases Dgkα/ζ. Elevated Dgka and Dgkz expression in DNMAML T cells is reminiscent of past observations in models of T cell hyporesponsiveness with decreased Ras/MAPK activation (34, 35). However, DNMAML T cells also acquired elevated expression of other negative regulators of T cell activation, including E3 ubiquitin ligases and co-inhibitory receptors, in addition to increased intracellular cAMP. Thus, the functional properties of Notch-deprived alloreactive T cells may not be explained entirely by decreased Ras-MAPK and NF-kB activation.
Despite the profound effects of Notch inhibition on cytokine production and a slight reduction in their initial proliferative burst, the overall accumulation of DNMAML T cells was preserved in vivo. These findings contrast with many other interventions that decrease expansion of alloreactive T cells in vivo. Interestingly, past work suggested that in vivo proliferation of alloreactive T cells was independent of IL-2 and may be controlled by IL-15, at least in MHC-mismatched allo-BMT models (47). Furthermore, classical models of T cell hyporesponsiveness and anergy have often been examined in vitro, a situation in which IL-2 production may play a critical role to support proliferation that does not reflect its effects in vivo (41). Other possible mechanisms accounting for the preserved or even enhanced in vivo expansion of DNMAML alloreactive T cells include decreased activation-induced cell death, for example as a result of reduced exposure to IFNγ (48). In parallel to these effects on conventional alloreactive T cells, Notch inhibition markedly enhanced in vivo expansion of Foxp3+ Tregs without inhibiting effector T cell expansion, which was explained by increased expansion of preexisting natural Tregs present in the transplant inoculum. These findings may contribute to the protective effects of Notch inhibition in GVHD and contrast with a shortcoming of many immunosuppressive strategies, including calcineurin inhibitors, which limit Treg proliferation (42).
Alloreactive Notch-deprived CD8+ T cells had largely preserved cytotoxicity after allo-BMT similar to our previously reported effects of Notch deprivation in CD4+ T cells (7). An interesting feature of alloreactive Notch-deprived T cells is their decreased cytokine production but preserved cytotoxic potential. Dissociation of cytokine production from cytotoxicity responses could reflect differential sensitivity of these pathways to Notch inhibition. Preserved or even enhanced T-bet and Eomesodermin expression in Notch-deprived T cells could be responsible for maintaining their cytotoxic potential after allo-BMT. Prior work has shown that T-bet and Eomesodermin are important for transcription of cytotoxic molecules (49). Collectively, Notch inhibition in alloreactive CD4+ and CD8+ T cells preserved efficient cytotoxicity while minimizing GVHD.
Altogether, our findings reveal a broad effect of Notch signaling in CD4+ and CD8+ alloreactive T cells during GVHD. Notch-deprived alloreactive T cells acquired features previously associated with hyporesponsiveness or anergy. However, this had differential effects on T cell effector functions in vivo in the post-transplantation environment, with profound inhibition of cytokine production but preserved T cell expansion, cytotoxic potential, and natural Treg accumulation. Notch-deprived T cells maintained potent cytotoxic activity in vivo, suggesting a “split anergy” phenotype (7, 50). Overall, Notch inhibition in T cells induced a unique combination of effects that potently inhibited GVHD, highlighting the promise of this new therapeutic strategy after allo-BMT.
Supplementary Material
Acknowledgements
We thank Dr. Tasuku Honjo (Kyoto University) for Rbpjf/f mice, Dr. Steve Reiner (Columbia University) for Eomesf/f×Cd4-Cre mice and Dr. Benjamin Segal (University of Michigan) for Tbx21−/− mice.
This work was supported by a Damon Runyon-Rachleff Innovation award (DRR-05A-09), a Scholar Award of the American Society of Hematology and the National Institutes of Health (RO1-AI091627) (IM). Individual support included T32 training grants (AI007413-17 to AS, GM007315 to JC), a Trainee Research Award from the American Society of Hematology (GTS) and a Miller Award for innovative immunology research (AS). Flow cytometry was partially supported by a University of Michigan Cancer Center Grant (P30-CA46592).
References
- 1.Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Seminars in cell & developmental biology. 2012;23:473–480. doi: 10.1016/j.semcdb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 3.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, Liu C, Ibrahim S, Beran M, Zavadil J, Efstratiadis A, Taghon T, Michor F, Levine RL, Aifantis I. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 5.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Sandy AR, Wang J, Radojcic V, Shan GT, Tran IT, Friedman A, Kato K, He S, Cui S, Hexner E, Frank DM, Emerson SG, Pear WS, Maillard I. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood. 2011;117:299–308. doi: 10.1182/blood-2010-03-271940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nature reviews. Immunology. 2012;12:443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 13.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, Kostura L, Fauq AH, Simpson K, Such KA, Miele L, Golde TE, Miller SD, Osborne BA. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 14.Helbig C, Gentek R, Backer RA, de Souza Y, Derks IA, Eldering E, Wagner K, Jankovic D, Gridley T, Moerland PD, Flavell RA, Amsen D. Notch controls the magnitude of T helper cell responses by promoting cellular longevity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9041–9046. doi: 10.1073/pnas.1206044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-7 production and Rorc activation. J Immunol. 2009;182:7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, Das P, Golde TE, Osborne BA. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 2008;112:1813–1821. doi: 10.1182/blood-2008-03-144980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou-Yang HF, Zhang HW, Wu CG, Zhang P, Zhang J, Li JC, Hou LH, He F, Ti XY, Song LQ, Zhang SZ, Feng L, Qi HW, Han H. Notch signaling regulates the FOXP3 promoter through RBP-J- and Hes1-dependent mechanisms. Mol Cell Biochem. 2009;320:109–114. doi: 10.1007/s11010-008-9912-4. [DOI] [PubMed] [Google Scholar]
- 18.Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I, Chiba S, Sone S, Yasutomo K. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9:1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- 19.Cho OH, Shin HM, Miele L, Golde TE, Fauq A, Minter LM, Osborne BA. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182:3380–3389. doi: 10.4049/jimmunol.0802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto K, Maekawa Y, Kitamura A, Nishida J, Koyanagi A, Yagita H, Kojima H, Chiba S, Shimada M, Yasutomo K. Notch2 signaling is required for potent antitumor immunity in vivo. Journal of immunology. 2010;184:4673–4678. doi: 10.4049/jimmunol.0903661. [DOI] [PubMed] [Google Scholar]
- 21.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everhart MB, Han W, Sherrill TP, Arutiunov M, Polosukhin VV, Burke JR, Sadikot RT, Christman JW, Yull FE, Blackwell TS. Duration and intensity of NF-kappaB activity determine the severity of endotoxin-induced acute lung injury. J Immunol. 2006;176:4995–5005. doi: 10.4049/jimmunol.176.8.4995. [DOI] [PubMed] [Google Scholar]
- 23.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 24.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- 26.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood. 2004;103:2677–2682. doi: 10.1182/blood-2003-06-2070. [DOI] [PubMed] [Google Scholar]
- 27.Berger M, Wettstein PJ, Korngold R. T cell subsets involved in lethal Graft-versus-Host-disease directed to immunodominant minor histocompatibility antigens. Transplantation. 1994;57:1095–1102. [PubMed] [Google Scholar]
- 28.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 29.Usui T, Preiss JC, Kanno Y, Yao ZJ, Bream JH, O'Shea JJ, Strober W. T-bet regulates Th1 responses through essential effects on GATA- function rather than on IFNG gene acetylation and transcription. The Journal of experimental medicine. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egerton M, Fitzpatrick DR, Kelso A. Activation of the extracellular signal-regulated kinase pathway is differentially required for TCR-stimulated production of six cytokines in primary T lymphocytes. International immunology. 1998;10:223–229. doi: 10.1093/intimm/10.2.223. [DOI] [PubMed] [Google Scholar]
- 31.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. The Journal of biological chemistry. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 32.Fields PE, Gajewski TF, Fitch FW. Blocked Ras activation in anergic CD4+ T cells. Science. 1996;271:1276–1278. doi: 10.1126/science.271.5253.1276. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Whaley CD, Mondino A, Mueller DL. Blocked signal transduction to the ERK and JNK protein kinases in anergic CD4+ T cells. Science. 1996;271:1272–1276. doi: 10.1126/science.271.5253.1272. [DOI] [PubMed] [Google Scholar]
- 34.Olenchock BA, Guo R, Carpenter JH, Jordan M, Topham MK, Koretzky GA, Zhong XP. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nature immunology. 2006;7:1174–1181. doi: 10.1038/ni1400. [DOI] [PubMed] [Google Scholar]
- 35.Zha Y, Marks R, Ho AW, Peterson AC, Janardhan S, Brown I, Praveen K, Stang S, Stone JC, Gajewski TF. T cell anergy is reversed by active Ras and is regulated by diacylglycerol kinase-alpha. Nature immunology. 2006;7:1166–1173. doi: 10.1038/ni1394. [DOI] [PubMed] [Google Scholar]
- 36.Collins S, Lutz MA, Zarek PE, Anders RA, Kersh GJ, Powell JD. Opposing regulation of T cell function by Egr-/NAB2 and Egr-/Egr- European journal of immunology. 2008;38:528–536. doi: 10.1002/eji.200737157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, Blackford A, Horton MR, Drake C, Schwartz RH, Powell JD. Egr- and Egr- are negative regulators of T cell activation. Nature immunology. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 38.Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity. 2003;18:535–547. doi: 10.1016/s1074-7613(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 39.Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, Bouchard D, Jones R, Gronski M, Ohashi P, Wada T, Bloom D, Fathman CG, Liu YC, Penninger JM. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–177. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Yee C, Beavo JA. CD3- and CD28-dependent induction of PDE7 required for T cell activation. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz RH. T cell anergy. Annual review of immunology. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 42.Lim DG, Joe IY, Park YH, Chang SH, Wee YM, Han DJ, Kim SC. Effect of immunosuppressants on the expansion and function of naturally occurring regulatory T cells. Transplant immunology. 2007;18:94–100. doi: 10.1016/j.trim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Minter LM, Osborne BA. Canonical and Non-Canonical Notch Signaling in CD4(+) T Cells. Current topics in microbiology and immunology. 2012;360:99–114. doi: 10.1007/82_2012_233. [DOI] [PubMed] [Google Scholar]
- 44.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-2 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. Journal of immunology. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 45.Rao RR, Li Q, Gubbels Bupp MR, Shrikant PA. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. Journal of immunology. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XC, Demirci G, Ferrari-Lacraz S, Groves C, Coyle A, Malek TR, Strom TB. IL-5 and IL-: a matter of life and death for T cells in vivo. Nat Med. 2001;7:114–118. doi: 10.1038/83253. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Janeway CA., Jr Interferon gamma plays a critical role in induced cell death of effector T cell: a possible third mechanism of self-tolerance. The Journal of experimental medicine. 1990;172:1735–1739. doi: 10.1084/jem.172.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature immunology. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 50.Otten GR, Germain RN. Split anergy in a CD8+ T cell: receptor-dependent cytolysis in the absence of interleukin-2 production. Science. 1991;251:1228–1231. doi: 10.1126/science.1900952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.