Abstract
The modular TRAPP complexes act as nucleotide exchangers to activate the Golgi Ypt/Rab GTPases, Ypt1 and Ypt31/Ypt32. In yeast, TRAPP I acts at the cis-Golgi and its assembly and structure are well characterized. In contrast, TRAPP II acts at the trans-Golgi and is poorly understood. Especially puzzling is the role of Trs20, an essential TRAPP I/II subunit required neither for the assembly of TRAPP I nor for its Ypt1-exchange activity. Mutations in Sedlin, the human functional ortholog of Trs20, cause the cartilage-specific disorder SEDT. Here we show that Trs20 interacts with the TRAPP II-specific subunit Trs120. Furthermore, the Trs20-Trs120 interaction is required for assembly of TRAPP II and for its Ypt32-exchange activity. Finally, Trs20-D46Y, with a single-residue substitution equivalent to a SEDT-causing mutation in Sedlin, interacts with TRAPP I, but the resulting TRAPP complex cannot interact with Trs120 and TRAPP II cannot be assembled. These results indicate that Trs20 is crucial for assembly of TRAPP II, and the defective assembly caused by a SEDT-linked mutation suggests that this role is conserved.
Keywords: TRAPP, Ypt/Rabs, GTPases, Trs20, Trs120, Sedlin, SEDT, SEDL, TRAPPC2, TRAPPC9
Introduction
Transport of secreted proteins begins at the endoplasmic reticulum (ER), progresses through the Golgi apparatus, and proceeds toward the plasma membrane (PM) of the cell. At the yeast Golgi, the Rab GTPases Ypt1 and the functionally redundant pair Ypt31/Ypt32 control two transport steps, ER-to-Golgi and Golgi-to-PM, respectively (1–3). Like all Rabs, these regulators are activated by guanine nucleotide-exchange factors (GEFs) that promote GTP binding by the Rab and, thereby, transport progression. The GEFs for the Golgi Ypts are alternative complexes of TRAPP subunits. One complex, TRAPP I, which contains at least four small subunits, including Bet3, Bet5, Trs23 and Trs31, resides at the cis-Golgi and activates Ypt1 (4–6). TRAPP II, which contains two essential subunits, Trs120 and Trs130, in addition to all those comprising TRAPP I, localizes to the trans-Golgi and can activate Ypt31/Ypt32 in vitro (5, 7). In addition to Golgi-to-PM, Ypt31/32 and TRAPP II also regulate endosome-to-Golgi transport (8, 9). A third complex, TRAPP III, which contains TRAPP I together with another subunit, Trs85, plays a role in autophagy (10–13).
Whereas the crystal structure of recombinant TRAPP I was solved (14, 15), much less is known about the structure of the TRAPP II and TRAPP III complexes (16). We have previously shown that one of the non-essential but similar TRAPP subunits, Trs33 or Trs65, is required for the attachment of Trs130 to TRAPP (17, 18). Currently, it is not clear how Trs120 joins TRAPP.
Trs20, a small TRAPP subunit common to both TRAPP I and TRAPP II, is required for yeast cell viability (5). Sedlin/SEDL/TRAPPC2 is the mammalian homolog of Trs20 and can replace it in yeast (19). The three-dimensional structure of Sedlin was solved both individually and in a sub-complex of TRAPP I that includes Sedlin-mBet3-mTrs31 (15, 20). Mutations in Sedlin cause spondyloepiphyseal dysplasia tarda (SEDT), a recessive X-linked cartilage-specific disorder associated with early onset of osteoarthritis, a degenerative joint disease (21). Most mutations associated with this disease result in truncated proteins. However, four different single substitution mutations were shown to cause SEDT. Three of these mutations result in the substitution of amino acids that lie inside the folded protein, and are hypothesized to disrupt proper folding. The fourth mutation, D47Y, is a substitution of an amino acid that resides on the surface of the protein, but the defect caused by this mutation is unknown (20).
Despite its medical significance, very little is known about the cellular function of Sedlin. While Sedlin is ubiquitously expressed, it is predominant in developing chondrocytes, which secrete large molecules like collagen. Therefore, it was proposed to play a role in transport of large molecules from the ER (22, 23). However, until very recently it was unclear how Sedlin contributes to this transport ((24), see Discussion). In addition, Sedlin was also shown to interact with several transcription factors. However, the role of these interactions is not clear, and, since the Sedlin-D47Y mutant protein is not defective in these interactions, their disruption is probably not the cause of SEDT (25). Using immuno-precipitation (IP) of transiently over-expressed TRAPP subunit pairs, Sedlin was shown to co-IP with TRAPPC8 (mTrs85) and TRAPPC9 (mTrs120), whereas the Sedlin-D47Y mutation results in reduced interactions. Based on these observations, a role for Sedlin as an adaptor between the TRAPP I core and the large subunits was proposed (26). However, neither a direct interaction between Trs20/Sedlin and Trs120/TRAPPC9 nor a clear role for Sedlin in the assembly of any TRAPP complex is currently established.
In yeast, the role that Trs20 plays in either TRAPP I or TRAPP II is not clear. While originally identified as a TRAPP I/II subunit (5), Trs20 is not required for the assembly of the core TRAPP I complex nor for its Ypt1-GEF activity in vitro (15). However, the fact that Trs20 is essential for viability in yeast and is highly conserved indicates a vital role of this subunit, as yet unknown. A deeper knowledge of Trs20 function would enhance our understanding of how TRAPP complexes are involved in regulating transport, and may have implications for disease treatment.
Here, we study the role of Trs20 in the assembly of the TRAPP II complex and the effect of a mutation equivalent to Sedlin-D47Y, Trs20-D46Y, on this role. Using in vivo and in vitro analyses of wild type and mutant Trs20 proteins, we show that Trs20 is required for assembly of the TRAPP II complex. Furthermore, we show that the SEDT-associated Trs20-D46Y mutant protein can join the TRAPP I complex, but this TRAPP is defective in the interaction with Trs120. Together these results provide the first information regarding a specific function of Trs20 in yeast and suggest that this role is conserved.
Results
Trs20 is required for the in vitro association of Trs120 and TRAPP I
Both Trs120 and Trs130 are required for TRAPP II function and GEF activity. We have previously shown that in vivo Trs120 is required for the association of Trs130 with TRAPP I, but not vice versa (7). We also showed that one of the two non-essential TRAPP subunits, Trs33 or Trs65, is required for association of Trs130, but not Trs120, to TRAPP I (17, 18). Therefore, we proposed that the first step in the assembly of TRAPP II is the association of the Trs120 subunit with TRAPP I, and this step is followed by Trs33/Trs65-dependent association of Trs130. However, it is not clear which TRAPP I subunit is required for interaction of Trs120 with this complex. To address this question, we reconstituted the TRAPP I-Trs120 association in vitro using co-precipitation analyses.
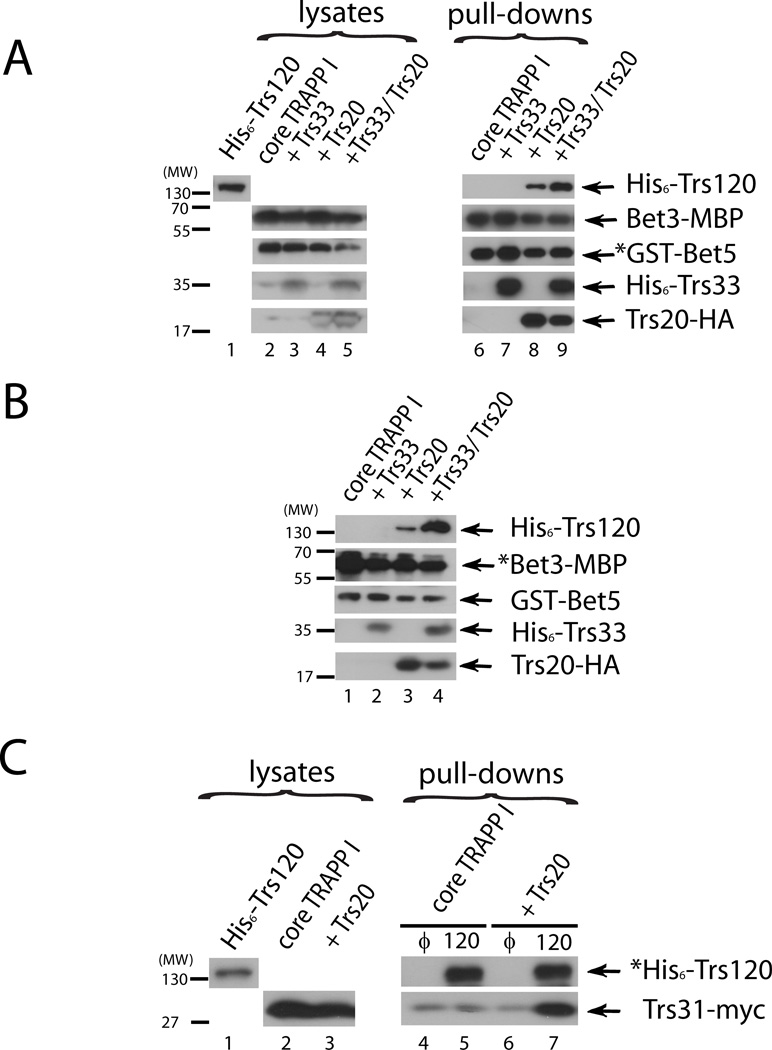
TRAPP I subunits in different combinations were co-expressed from duet vectors in bacteria. His6-tagged Trs120 was expressed separately. When core TRAPP I, which contains the four subunits required for the Ypt1-GEF activity, was pulled down using GST-Bet5 and glutathione beads, Trs120 did not co-precipitate with it. We have previously shown that bacterially expressed Trs33 and Trs120 interact directly (18). However, in the context of TRAPP I without Trs20, Trs33 does not interact with Trs120 (Figure 1, lane 7). Importantly, when Trs20-HA was co-expressed with the core TRAPP I subunits, Trs120 co-precipitated with the GST-Bet5 complex (>5%; Figure 1A, Lane 8). Moreover, in the presence of both Trs20 and Trs33, Trs120 interaction with TRAPP I is further increased by ~2 fold (>10%; Figure 1A, lane 9). The requirement of Trs20 for the association of Trs120 with TRAPP I was verified using another tagged TRAPP I subunit, MBP-tagged Bet3, and amylose resin precipitation. In this case too, Trs120 co-precipitated with Bet3-MBP only when Trs20 was co-expressed with the TRAPP I core, and inclusion of Trs33 resulted in an increased co-precipitation (>5% and >10%, Figure 1B, lanes 3 and 4, respectively). Finally, in a reciprocal experiment, co-precipitation of TRAPP I with His6-Trs120 was determined using Ni2+ resin. The TRAPP I subunit Trs31-myc co-precipitated with Trs120 only from bacterial lysates co-expressing the TRAPP I core with Trs20, but not without Trs20 (Figure 1C, lanes 5 and 7). Together, these results strongly suggest that Trs120 interacts directly with the TRAPP I complex, and that this interaction requires Trs20 and can be further enhanced by Trs33.
Figure 1. Trs20 is required for interaction of Trs120 with recombinant TRAPP I.
A. Pull down of His6-Trs120 with GST-Bet5 purified complexes. GST was pulled down, using glutathione sepharose resin, from lysates of bacteria expressing core TRAPP I (GST-Bet5, Trs23-S, Trs31-myc, and Bet3-MBP), core TRAPP I plus Trs33 (His6-Trs33), core TRAPP I plus Trs20 (Trs20-HA), or core TRAPP I plus Trs20 and Trs33. Cleared lysate (S100) from different bacterial cells expressing His6-Trs120 was then incubated with the resin and the level of proteins associated with the resin after precipitation was determined using immuno-blot analysis and antibodies against the tags (Trs120, Bet5, Trs33, Trs20) or the protein (Bet3). Trs120 co-purifies with GST-Bet5 complex (>5%), but not with GST, and only in the presence of Trs20. More Trs120 co-purifies with TRAPP when Trs33 is present (>10%). The expression levels of the different proteins in lysates are shown on the left (10% input for the Trs120 lysate). The full anti-His6 immuno-blot is shown in Figure S1A. B. Pull down of His6-Trs120 with Bet3-MBP purified complexes. Binding of His6-Trs120 TRAPP I was determined as in part A (using the same lysates), except that amylose resin was used to pull down Bet3-MBP within the core TRAPP I complex. Trs120 co-purifies with TRAPP I only in the presence of Trs20 (>5%), and this level is higher in the presence of Trs33 (>10%). C. Pull down of core TRAPP I with His6-Trs120. Lysates from bacteria expressing His6-Trs120, or empty plasmid (θ) as a negative control, were purified on Ni2+ resin. The resin was then incubated with lysates from cells expressing either core TRAPP I, or core TRAPP I plus Trs20-HA. The level of proteins associated with the resin after precipitation was determined using immuno-blot analysis and antibodies against the tags. TRAPP I, scored by the level of Trs31-myc, co-purified with Trs120, but not with the empty plasmid control, and only in the presence of Trs20. In A–C, asterisks indicate the tagged protein being bound directly to the resin. Results in this figure are representative of at least two independent experiments.
A second Sedlin yeast homolog, Tca17/TRAPPC2L, was shown to interact with Trs130 using the yeast two hybrid assay and to co-precipitate with TRAPP from yeast lysates in the presence of Trs33 or Trs65 (27–29). Although TCA17 is not essential for cell viability, we wanted to test whether, like Trs20, Tca17 can mediate the Trs120 interaction with TRAPP. Bacterially expressed Tca17 was shown to co-purify with the His6-tagged Bet3 and Trs31 heterodimer (29), however, co-purification of recombinant Tca17 with the core TRAPP I complex has not been reported. S-tagged Tca17 was expressed in bacteria together with the core TRAPP I subunits and Trs33 (see Figure 1). Whereas Trs23 was found in the GST-Bet5 pull down, Tca17 did not co-purify with the TRAPP complex and did not affect the co-precipitation of Trs120 with the complex (Trs20 serves as a positive control in this experiment; Figure S1B). Therefore, under our experimental conditions, recombinant Tca17 does not associate with the TRAPP complex and its relevance to the association of Trs120 to TRAPP cannot be assessed in this assay.
Trs20 mutations cause defects in the cellular localization of TRAPP II-specific subunits
To explore the role of the Trs20 subunit of TRAPP in vivo, a temperature-sensitive trs20 mutant strain, hereafter called trs20ts, was used (29). The trs20ts mutant allele was sequenced and it contains two amino acid alterations: V92A and F133S. This strain, along with a wild-type control, was used in experiments to test the effect of this double mutation on TRAPP complexes in yeast.
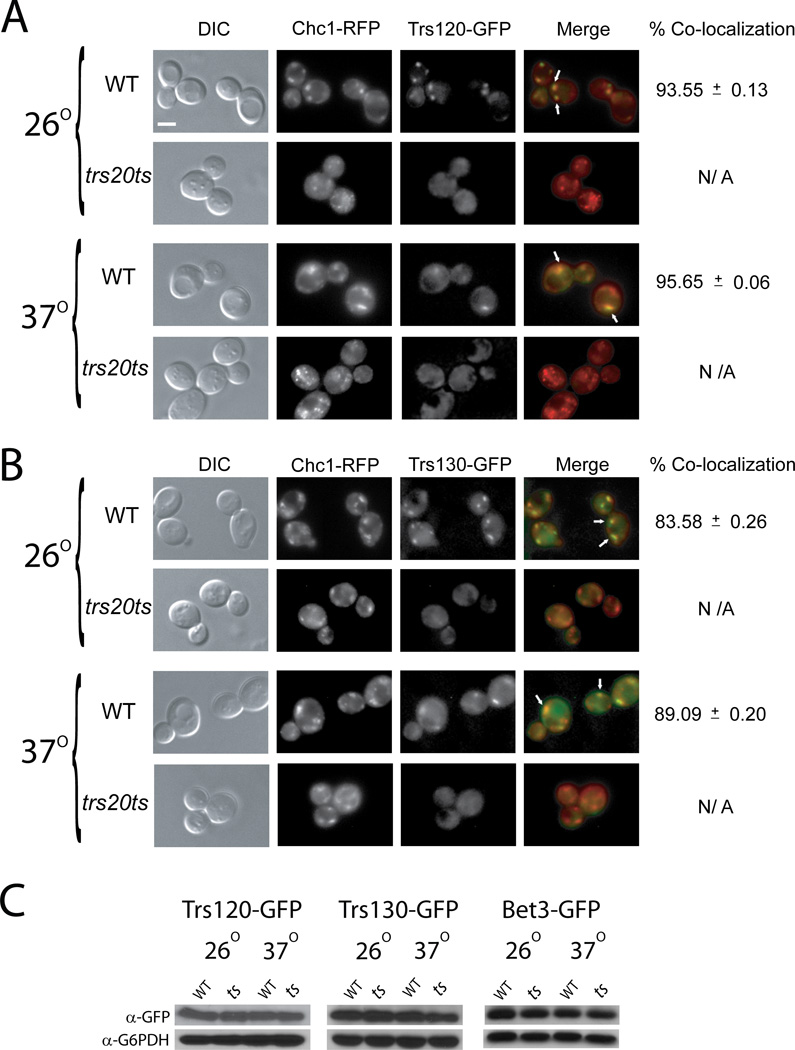
First, the effect of the trs20ts mutation on the cellular localization of TRAPP subunits was determined. The TRAPP I/II subunit Bet3, and the two essential TRAPP II-specific subunits Trs120 and Trs130 were tagged with GFP on the chromosome in wild type and trs20ts mutant strains. The localization patterns of these TRAPP subunits were determined using live-cell microscopy in strains that also express RFP-tagged Golgi subunits. In wild type cells, all three TRAPP subunits appear as puncta. When expressed in cells that also express red Golgi markers, ~95% and 85% of the TRAPP II-specific subunits Trs120 and Trs130, respectively, co-localized with the trans Golgi marker Chc1 (Figure 2, A–B). A lower percentage of the TRAPP I/II subunit Bet3 co-localized with the trans Golgi marker Chc1 (~66%), and ~20% of the Bet3 co-localized with the cis Golgi marker COPI (Figure 3).
Figure 2. The trs20ts mutation affects the cellular localization of TRAPP II-specific subunits.
(A) The TRAPP II-specific subunit Trs120, and the trans Golgi marker Chc1, were tagged on the chromosome at their C-termini with GFP and RFP, respectively, in wild type (NSY1471) and trs20ts (NSY1472) yeast strains. Cells were grown to mid-log phase, and were either left at 26° or shifted to 37° for 90 minutes. GFP and RFP were visualized by live-cell deconvolution microscopy. From left to right: DIC, RFP, GFP, and merge (arrows indicate co-localization). Percent co-localization of GFP with RFP was determined (a total of 64 cells were visualized in two independent experiment for each set; NA: not applicable). In wild-type cells, ~95% of Trs120 co-localizes with Chc1. In trs20ts mutant cells, Trs120, but not Chc1, loses its punctate localization pattern even at the permissive temperature. B. The TRAPP II-specific subunit, Trs130, and the trans Golgi marker, Chc1, were tagged on the chromosome at their C-termini with GFP and RFP, respectively, in wild type and trs20ts yeast strains. The experiment was done as described above (panel A). In wild-type cells, ~85% of Trs130 co-localizes with Chc1. In trs20ts mutant cells, Trs130, but not Chc1, loses its punctate localization pattern even at the permissive temperature. C. The protein levels of Trs120-GFP, Trs130-GFP, and Bet3-GFP were compared in lysates from wild type and trs20ts mutant cells expressing GFP-tagged endogenous TRAPP subunits and probing the samples with α-GFP antibodies, or α-G6PDH as a loading control, using immuno-blot analysis. The protein levels of Trs120, Trs130 and Bet3 in trs20ts mutant cells are similar to those seen in wild type cells. Results in this figure are representative of at least two independent experiments. Scale bar, 5 µm.
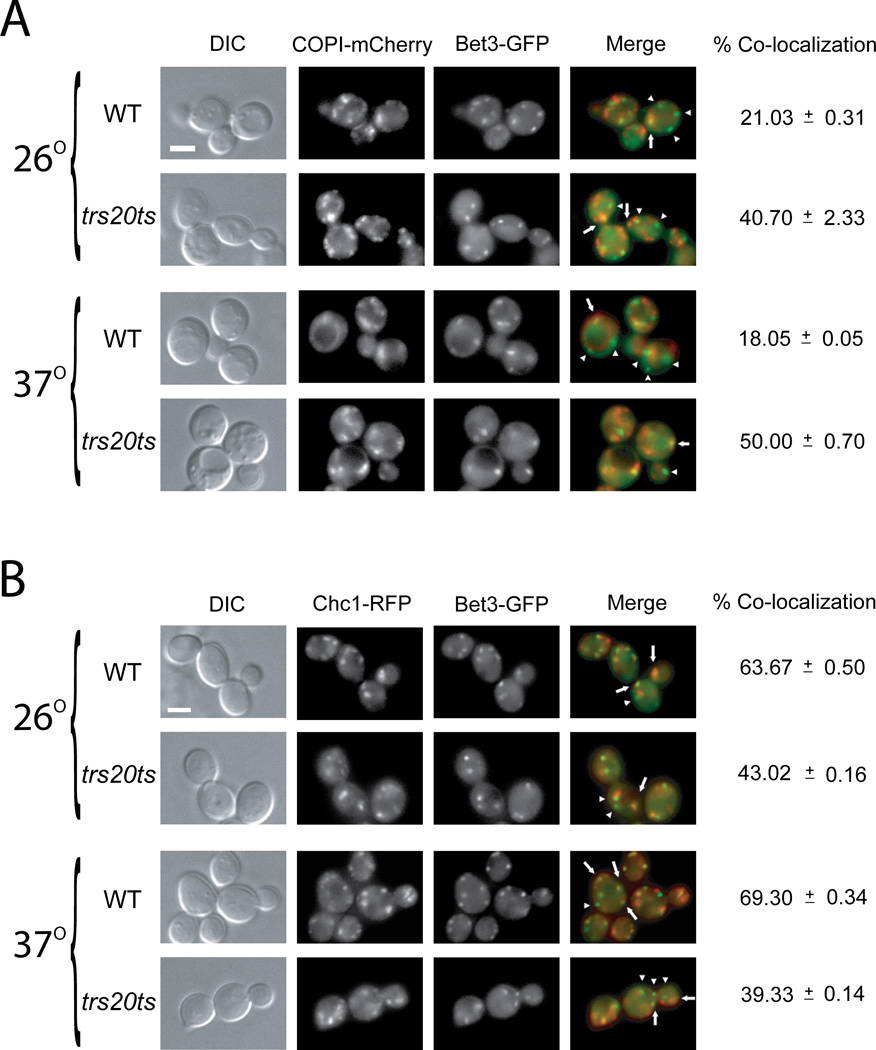
Figure 3. The trs20ts mutation affects the distribution of the TRAPP I/II subunit Bet3 between the cis and trans Golgi.
(A) The TRAPP I/II subunit Bet3, and the cis Golgi marker COPI, were tagged on the chromosome at their C-termini with GFP and m-Cherry, respectively, in wild type (NSY1471) and trs20ts (NSY1472) yeast strains. Cells were grown to mid-log phase, and were either left at 26° or shifted to 37° for 90 minutes. GFP and mCherry were visualized by live-cell deconvolution microscopy. From left to right: DIC, mCherry, GFP, and merge (arrows point to co-localized puncta; arrowheads point to Bet3-GFP puncta that do not co-localize with the red cis Golgi marker). Percent co-localization of GFP with m-Cherry was determined (a total of 64 cells were visualized in two independent experiment for each set; NA: not applicable). In wild-type cells, ~20% of Bet3 co-localizes with COPI. In trs20ts mutant cells, the co-localization of Bet3 with COPI is higher even at the permissive temperature. B. The TRAPP I/II subunit, Bet3, and the late Golgi marker, Chc1, were tagged on the chromosome at their C-termini with GFP and RFP, respectively, in wild type and trs20ts yeast strains. The experiment was done as described above (panel A). From left to right: DIC, RFP, GFP, and merge (arrows point to co-localized puncta; arrowheads point to Bet3-GFP puncta that do not co-localize with the red trans Golgi marker). In wild-type cells, ~65% of Bet3 co-localizes with Chc1. In trs20ts mutant cells, the co-localization of Bet3 with Chc1 is decreased even at the permissive temperature. Results in this figure are representative of at least two independent experiments. Scale bar, 5 µm.
In trs20ts mutant cells, both the cis and trans Golgi markers, COPI and Chc1, respectively, appeared normal at permissive and restrictive temperatures (Figure 2A–B; and Figure 3). In contrast, the GFP-tagged TRAPP II-specific subunits Trs120 and Trs130 appeared diffuse in trs20ts mutant cells already at permissive temperature (Figure 2A and B, respectively). The GFP-tagged TRAPP I/II subunit Bet3 appears as puncta in trs20ts mutant cells even at their restrictive temperature, with ~30% less Bet3 puncta in mutant cells when compared to wild type cells. Importantly, in these mutant cells, the co-localization of GFP-Bet3 with the cis and trans Golgi markers, COPI and Chc1, respectively, is changed even at permissive temperature, with a higher proportion of Bet3 co-localizing with COPI and a lower proportion with Chc1 (Figure 3 A and B, respectively). The diffuse pattern of GFP-tagged Trs120 and Trs130 was not due to lower protein levels because, like Bet3-GFP, their level is not reduced in trs20ts mutant cells (Figure 2C). Cell fractionation analyses of GFP-tagged Trs120 and Trs130 from wild type and trs20ts mutant cell lysates showed reduction in the levels of both subunits in the 100,000 × g pellet (P100) of mutant cell lysates (Figure S2), which contains Golgi membranes (30). This result is in agreement with the microscopy analysis. Together, the localization analysis of TRAPP subunits suggests that in trs20ts mutant cells TRAPP II is not efficiently formed, whereas up to 50% of Bet3-containing TRAPP accumulates in the cis Golgi (an increase from ~20%).
Trs20 mutations cause defects in TRAPP II assembly in vivo
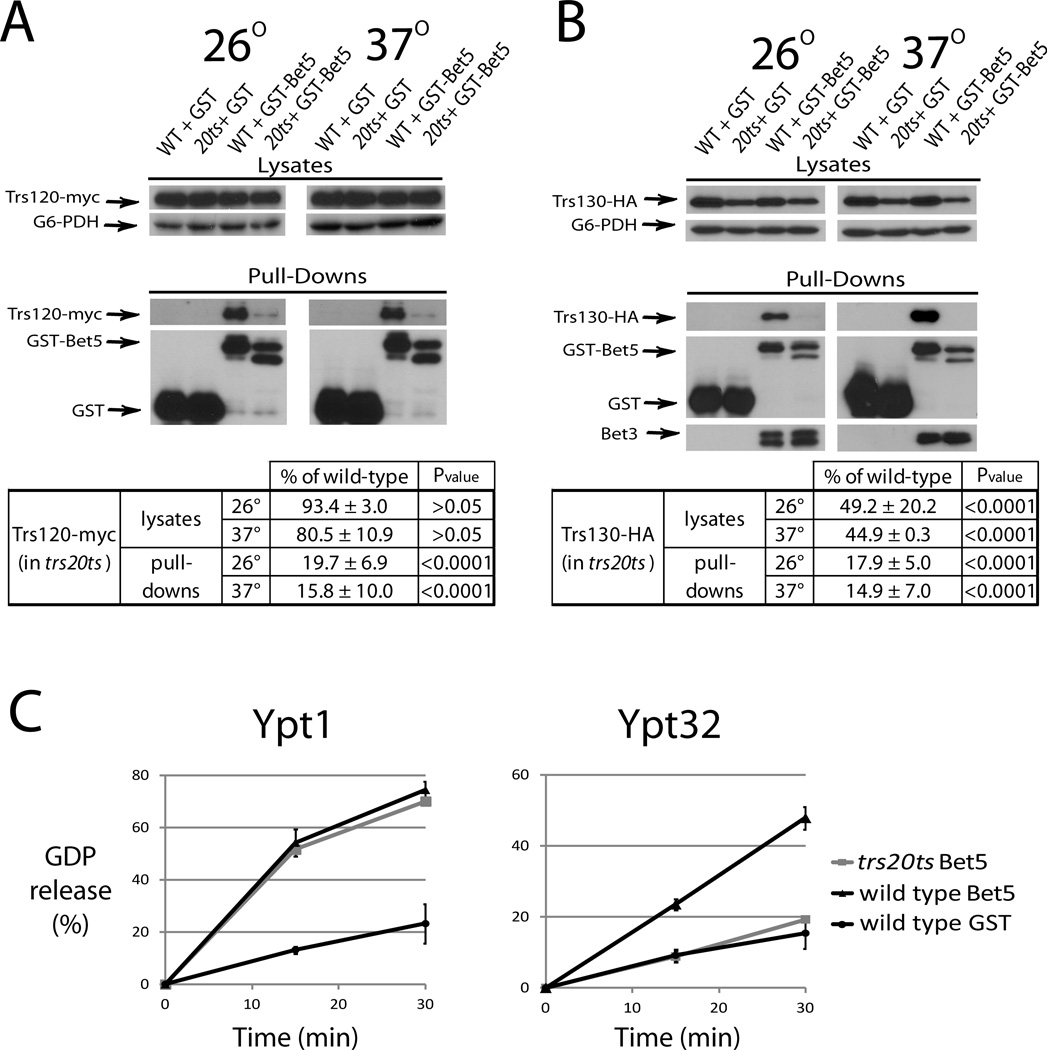
The Trs120 and Trs130 localization defects in trs20ts mutant cells are highly reminiscent of those previously published for the trs33ts and trs65ts strains, which are defective in TRAPP II assembly (17, 18). To determine whether Trs20, like Trs33 and Trs65, is required for TRAPP II assembly, the effect of the trs20ts double mutation on purified TRAPP II composition and GEF activity was tested. TRAPP complexes were purified from lysates of wild type and trs20ts mutant cells over-expressing GST-Bet5 in which Trs120 was tagged on the chromosome with myc, or Trs130 with HA. We have previously shown that whereas Trs120 can join TRAPP in the absence of Trs130, Trs130 requires Trs120 to join TRAPP (7). The level of Trs120-myc is similar in lysates of wild type and trs20ts mutant cells grown at 26 or 37°C (Figure 4A, top). TRAPP complexes were purified from these lysates using GST pull down and the level of Trs120-myc that co-precipitated with these complexes was determined by immuno-blot analysis. The level of Trs120 was significantly lower in GST-Bet5 complexes purified from trs20ts lysates when compared to wild type lysates (~20 and 16% at 26 and 37°C, respectively, Figure 4A, bottom). The level of Trs130-HA was lower in lysates of trs20ts mutant cells when compared to those of wild type cells (~50 and 45% at 26 and 37°C, respectively, Figure 4B, top). The fact that, unlike Trs130-HA, the level of Trs130-GFP is not lower in trs20ts mutant cells (Figure 2C) might reflect the fact that the GFP tag can stabilize proteins, and/or the effect of over-expressing GST-Bet5 in these mutant cells. Whereas the level of Bet3 co-purified with GST-Bet5 from wild type and trs20ts mutant cells was similar, the level of Trs130-HA co-purified with GST-Bet5 from trs20ts cell lysates was significantly lower than that purified from wild type cell lysates (~18 and 14% at 26 and 37°C, respectively, Figure 4B, bottom). Together with the microscopy data, these results indicate that assembly of the TRAPP II-specific subunits, Trs130 and Trs120, with TRAPP is defective when Trs20 functionality is compromised. The observation that the level of Trs130-HA, but not Trs120, is lower in trs20ts mutant cells than in wild type cells agrees with the idea that when Trs120 does not join TRAPP, Trs130 is sensitive to degradation (7).
Figure 4. TRAPP purified from trs20ts mutant cells does not contain TRAPP II and does not act as a Ypt32 GEF.
A. The protein level of Trs120-myc is significantly lower in purified TRAPP complexes, but not in lysates, from trs20ts when compared to wild type cells. GST-Bet5 and GST, as a negative control, were over-expressed in wild type (NSY1471) and trs20ts (NSY1472) mutant cells also expressing endogenously tagged Trs120-myc. Cells grown to mid-log phase were either left at 26° or shifted to 37° for 70 minutes and then harvested. Cell lysates were prepared and GST-Bet5 complexes were purified on glutathione sepharose resin. The level of Trs120-myc was determined in lysates (top) and pull-downs (bottom) using immuno-blot analysis; G6PDH level was used as a loading control for lysates; GST-Bet5 and GST levels are used for the pull down yield. B. The protein level of Trs130-HA is significantly lower in lysates and TRAPP complexes purified from trs20ts when compared to wild type cells. Same as in panel A, except that cells were expressing endogenously tagged Trs130-HA, and the pull-down of the TRAPP I/II subunit Bet3 was verified using anti-Bet3 antibodies. The partial degradation of over-expressed GST-Bet5 in trs20ts mutant cells in likely due to the instability of TRAPP complexes in these cells. For panels A and B, the level of Trs120-myc or Trs130-HA was quantified and shown under the immuno-blots as percent of wild type cells. Protein level in lysates was corrected for the loading control, while in pull downs it was corrected for the full-length GST-Bet5; +/− represents SEM; P values are shown on the right (values <0.05 represent significant difference). C. TRAPP complexes purified from trs20ts mutant cells can act as GEF for Ypt1, but not Ypt32. TRAPP complexes were purified from the strains shown in panel B, except that GST-purified complexes were eluted from the glutathione resin with glutathione. Eluates were used in a GDP-release assay with Ypt1 or Ypt32 as substrates to determine GEF functionality. Error bars represent std. dev. Results in this figure are representative of at least two independent experiments.
We have previously shown that TRAPP complexes purified from cells defective in TRAPP II assembly can act as Ypt1 GEF, but not Ypt31/Ypt32 GEF (7). To determine whether this is true also for TRAPP complexes purified from trs20ts mutant cells, the GEF activity of GST-Bet5 purified complexes was assessed using a GDP-release assay. The Ypt1 GEF activity of TRAPP complexes purified from trs20ts mutant cells is similar to that of complexes purified from wild type cells. In contrast, the same complexes purified from trs20ts cannot act as Ypt32 GEF under conditions in which wild type complexes can (Figure 4C). Collectively, these data strongly indicate that TRAPP II complex assembly, localization and GEF activity are defective in trs20ts mutant cells, while the assembly and Ypt1 GEF activity of TRAPP I complex is not affected.
SEDT-associated mutation in Trs20 results in a Trs120-TRAPP I interaction defect
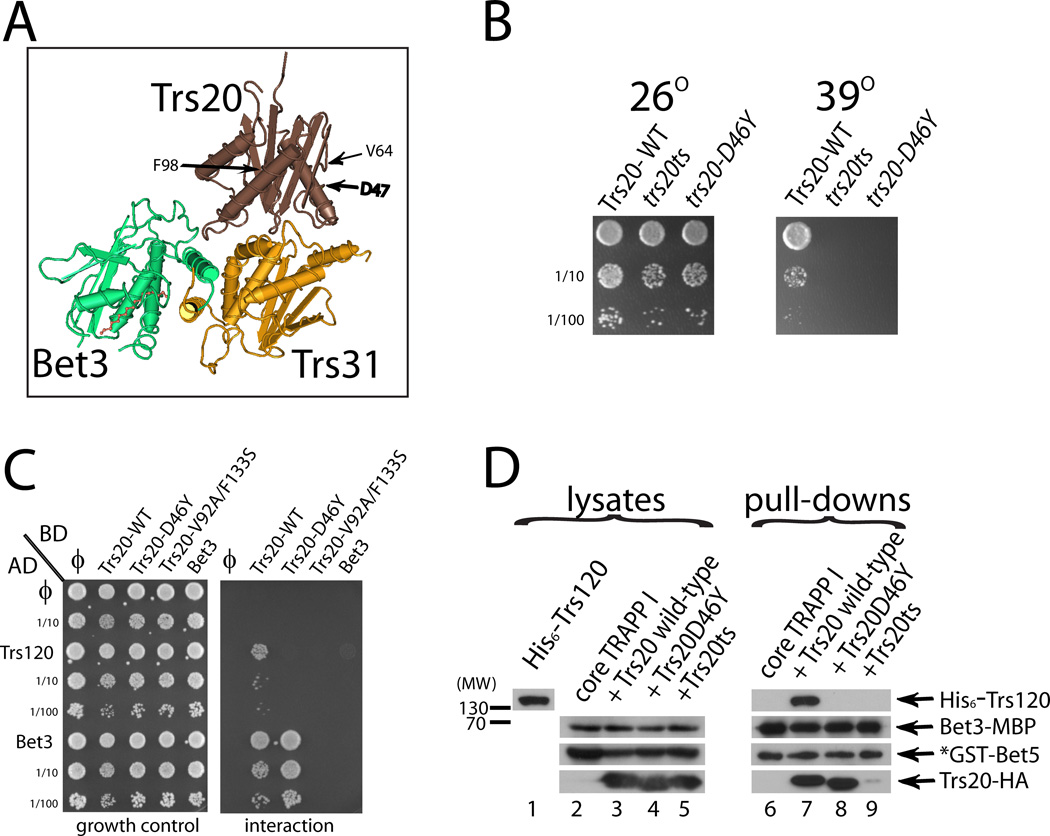
The X-linked SEDT disorder is caused by mutations in Sedlin, the human homolog of Trs20. The SEDT-linked Sedlin-D47Y mutation alters a residue conserved from yeast to humans and was suggested to affect Sedlin’s interactions with mTrs85 and mTrs120 (26). Furthermore, the mutant protein is stably expressed and localizes normally (22, 25). The D47 residue lies on the surface of Sedlin, but away from domains known to interact with other TRAPP subunits (Figure 5A), based on a crystal structure of the mouse proteins Sedlin, Trs31, and Bet3 in complex (15). Therefore, we hypothesized that the D47Y mutation disrupts interaction of Sedlin with hTrs120. To investigate the D47Y mutation of Sedlin in more detail, we created a yeast strain containing the corresponding mutation, D46Y in the yeast Trs20, as the sole copy of TRS20 in the cell. Growth analysis revealed that this strain is temperature sensitive (Figure 5B), confirming that this residue is important for Trs20 function in yeast.
Figure 5. The effect of the V92A-F133S and D46D mutations on Trs20 interactions.
A. The disease-causing mutation D47Y resides on the Trs20/Sedlin surface not in contact with TRAPP I. The positions of the Trs20/Sedlin mutations used in this study are shown on the crystal structure of the mouse Sedlin (brown) in complex with mTrs31 (yellow) and mBet3 (green), as published by (15). Shown are D47 and the two residues mutated in trs20ts: V92 and F133 in yTrs20 correspond to V64 and F98 in Sedlin, the latter two reside in two different Trs20/Sedlin domains. B. The trs20-D46Y mutation causes a growth defect in vivo. TRS20 was deleted from the chromosome in cells expressing Trs20 – wild type, Trs20ts or Trs20-D46Y – from CEN plasmids. Growth was determined on YPD plates at 26° and 39°C. Rows represent 10-fold serial dilutions from top to bottom. C. Wild type Trs20, but not the Trs20-D46Y or Trs20-V92A-F133S mutants, interacts with Trs120 in the yeast two-hybrid assay. Growth control is shown in the left panel, and interaction, measured by growth on the SD-Ura –Leu –His plate, is shown in the right panel. Trs20-V92A-F133S, but not Trs20-D46Y, is defective also in the interaction with the TRAPP I/II subunit, Bet3. D. His6-Trs120 interacts with a TRAPP I complex containing wild type Trs20, but not D46Y or Trs20-V92A-F133S. GST pull-down was done as described for Figure 1A. Lysate from bacteria expressing His6-Trs120 was added to glutathione resin bound to TRAPP complexes purified from bacteria expressing (from left to right): core TRAPP I, core TRAPP I plus wild type Trs20, core TRAPP I plus Trs20-D46Y, or core TRAPP I plus Trs20-V92A-F133S. Levels of proteins precipitated with GST-Bet5 were determined using immuno-blot analysis and antibodies against the tags. Results in panels B-D are representative of at least two independent experiments.
The yeast two-hybrid assay was used to test whether the Trs20-D46Y mutant protein is defective in the interaction with Trs120. In this assay, wild type Trs20 can interact with both Trs120 and the TRAPP I/II subunit Bet3. The Trs20ts mutant protein, which contains the two mutations in the trs20ts strain, V92A/F133S, failed to interact with both Trs120 and Bet3, indicating that it is unable to bind its close neighbor in TRAPP I. Significantly, the Trs20-D46Y mutant protein interacted with Bet3 as well as the wild-type Trs20 protein, but completely lost interaction with Trs120 (Figure 5C). This result suggests that the Trs20-D46Y mutant protein is able to assemble with TRAPP I, but is unable to bring Trs120 to the complex.
This idea was tested using co-precipitation of recombinant proteins as described above (Figure 1A), using different alleles of TRS20. When wild type Trs20 is co-expressed with the core TRAPP I subunits, it precipitates with GST-Bet5 and allows the co-precipitation of Trs120. In contrast, the Trs20-92A/F133S mutant protein, which confers temperature sensitivity in trs20ts mutant cells, did not precipitate efficiently with the GST-Bet5 complex, which also did not pull down Trs120 (Figure 5D, lane 9). This result is in agreement with the yeast two-hybrid assay result and supports the idea that this double mutant Trs20 is defective in both its interaction with TRAPP I and in the assembly of Trs120 with TRAPP. As expected from the yeast two hybrid assay, the Trs20-D46Y mutant protein can assemble with TRAPP I as efficiently as wild type Trs20, but was unable to allow Trs120 join this TRAPP complex (Figure 5D, lane 8). These results support the idea that the D46 residue lies on the surface of Trs20 that interacts with Trs120, and the D46Y mutation abolishes this interaction, causing temperature sensitivity in yeast and maybe contributing to the SEDT disorder in humans.
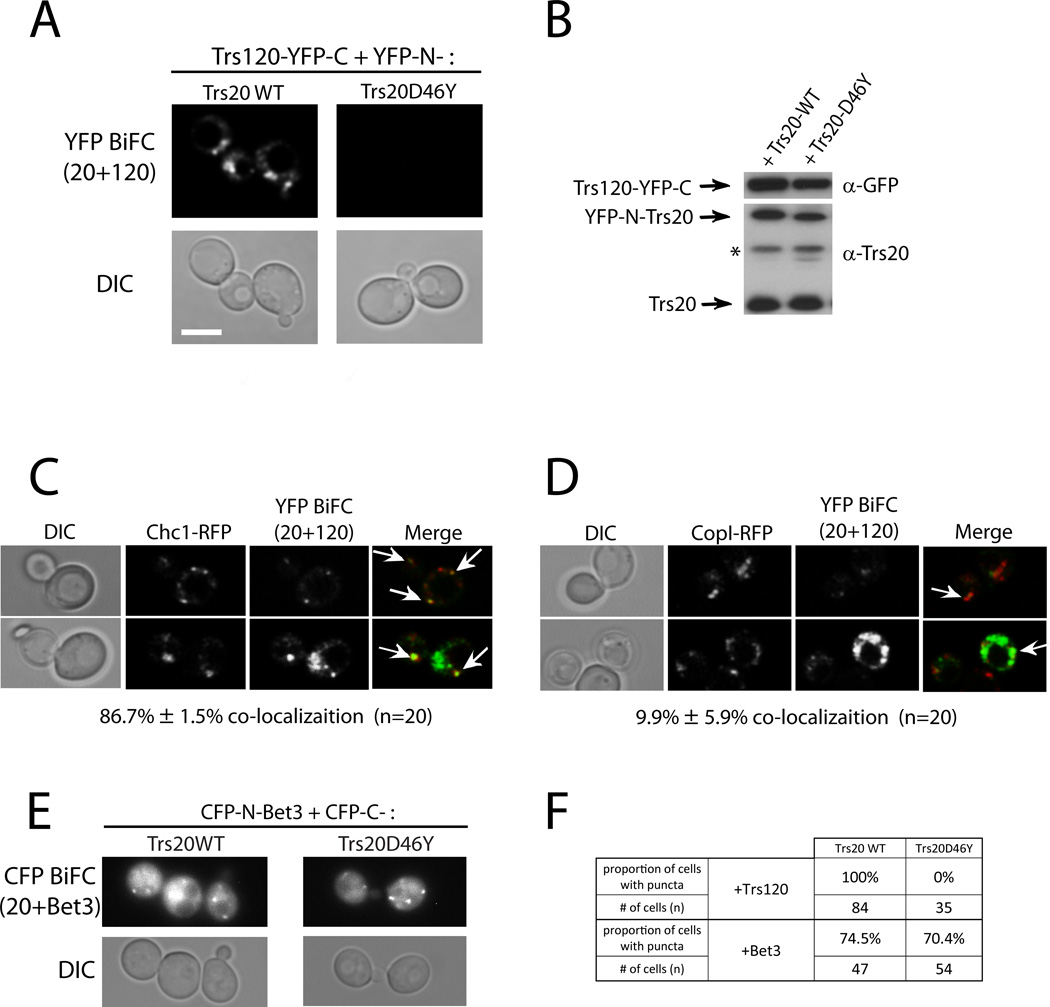
To verify the Trs20-Trs120 interaction and the effect of the D46Y mutation in vivo, we used the bimolecular-fluorescence complementation, BiFC, assay (31). Trs20 and Trs20-D46Y were tagged with the N-terminal fragment of YFP, and Trs120 was tagged with the C-terminal fragment of YFP. Yeast cells were transformed with plasmids expressing the two tagged proteins. Fluorescence in the YFP channel indicates that the two tagged proteins are physically interacting, thus bringing the two fragments of YFP into close proximity. BiFC can be observed in cells expressing Trs120 with wild-type Trs20, but not with the Trs20-D46Y mutant protein (Figure 6A). The expression of tagged Trs120, Trs20-WT, and Trs20-D46Y was confirmed using immuno-blot analysis (Figure 6B). The co-localization of the Trs20-Trs120 BiFC puncta with the cis and trans Golgi markers, COPI and Chc1, respectively, was determined in cells expressing red Golgi markers. If the two subunits interact to form TRAPP II, Trs20 and Trs120 should interact mostly on the trans Golgi. Indeed, the Trs20-Trs120 BiFC puncta co-localize mostly with the trans Golgi marker Chc1 (~87%), with very little co-localization with the cis Golgi marker COPI (~10%; Figure 6C–D). In contrast to the Trs20-Trs120 BiFC interaction, both Trs20-WT and Trs20-D46Y interact with Bet3 in the BiFC assay. In this case Bet3 was tagged with the N-terminus of CFP, and Trs20 and Trs20-D46Y were tagged with the C-terminus of CFP. The BiFC of Bet3 with Trs20 and Trs20-D46Y could be observed in the CFP channel (Figure 5E). This analysis further supports the idea that Trs20 interacts with Trs120 in vivo to form TRAPP II on the trans-Golgi, and that the D46Y mutation disrupts this interaction, but not the interaction of Trs20 with TRAPP I.
Figure 6. In Vivo interactions with Trs20 on the trans-Golgi.
A. Trs120 interacts with wild type Trs20, but not with Trs20-D46Y, in the BiFC assay. Wild type cells (NSY128) were transformed with two plasmids, one expressing Trs120-YFP-C and the other expressing YFP-N-Trs20 or YFP-N-Trs20-D46Y. Interaction was determined by YFP fluorescence, which was seen only in cells co-expressing Trs120 and wild type Trs20, but not Trs20-D46Y. Representative cells are shown: YFP (top), DIC (bottom) (see quantification in F). B. Expression of Trs120-YFP-C was confirmed using immuno-blot analysis and anti-GFP antibody (top), and expression of YFP-N-Trs20, wild type and D46Y, was confirmed using immuno-blot analysis and anti-Trs20 antibody (bottom; endogenous Trs20, bottom band; * nonspecific band). C–D. Co-localization of the Trs120-Trs20 BiFC puncta with the trans-Golgi marker Chc1 (C), and the cis-Golgi marker CopI (D). Cells expressing RFP-tagged Golgi markers from their endogenous locus were transformed with Trs120-Trs20 (wt) BiFC plasmids (panel A). Whereas 87% of the BiFC puncta co-localize with Chc1, about 10% co-localize with CopI. E. Bet3 interacts with both Trs20 and Trs20-D46Y in the BiFC assay. Wild type cells (NSY128) were transformed with two plasmids, one expressing CFP-N-Bet3 And the other expressing CFP-C-Trs20 Or CFP-C-Trs20-D46Y. Interaction was determined by CFP fluorescence seen in cells co-expressing Bet3 and Trs20 or Trs20-D46Y. F. Quantification of the BiFC interaction of Trs20, wt and D46Y, with Trs120 (panel A), or with Bet3 (panel E). Scale bar, 5 µm, arrows show co-localization, n = number of cells. Results in are representative of at least two independent experiments.
The effect of the trs20 mutations on the localization of the Golgi Ypts
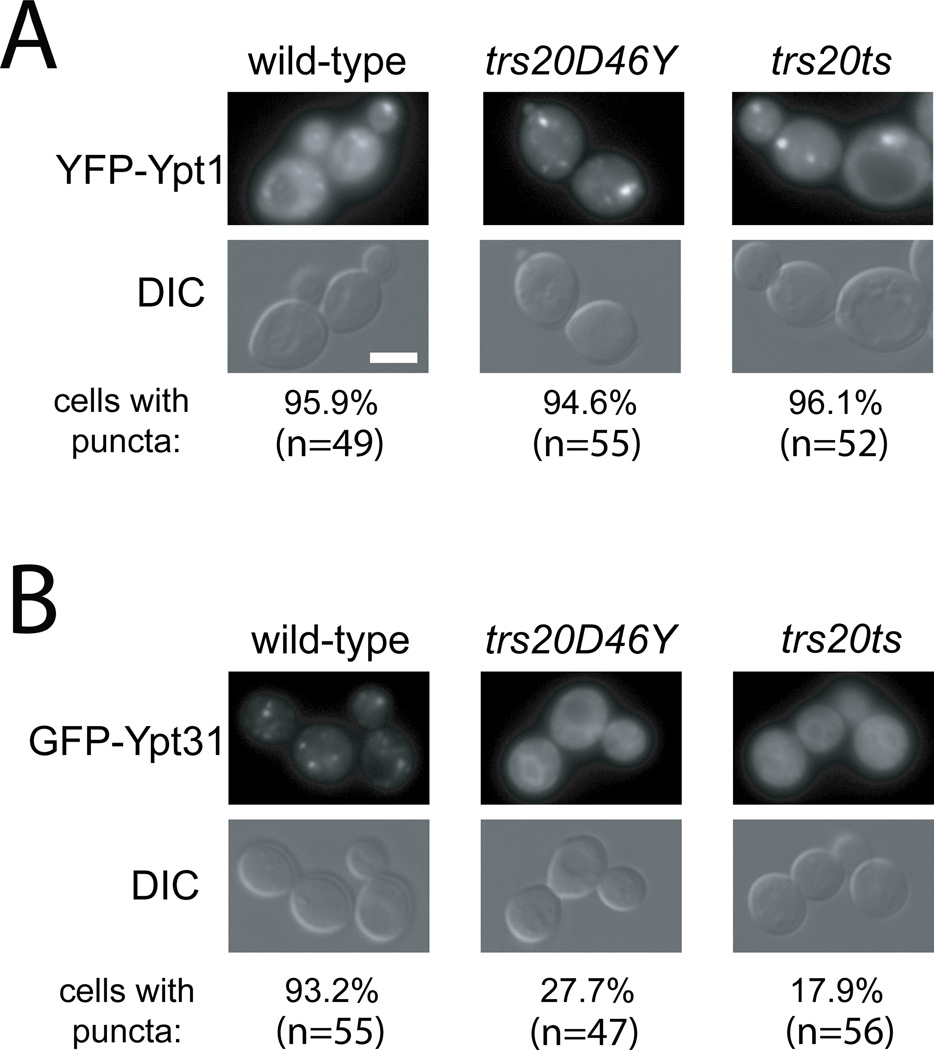
We have previously shown that mutations that disrupt the TRAPP II complex, e.g., trs130ts, trs65ts and trs33ts, result in diffuse Ypt31, but not Ypt1 (7, 17, 18). Because we show here that Trs20 is required for TRAPP II assembly and Ypt32 GEF activity, we wished to test whether the trs20 mutations, trs20-D46Y and trs20ts (V92A and F133S), also affect Ypt31 Golgi localization. GFP-Ypt1 or YFP-Ypt31 was expressed from CEN plasmids from their own promoter and terminator in wild type, trs20-D46Y or trs20ts mutant cells. Whereas Ypt1 puncta could be seen in all three strains, the Ypt31 puncta could only be seen in wild type cells, but not in trs20 mutants (Figure 7). This result supports the idea that both trs20 mutant strains are defective in TRAPP II assembly, and, therefore, in the Ypt31/Ypt32 GEF function in vivo.
Figure 7. The trs20ts and trs20-D46Y mutations affect Ypt31, but not Ypt1, cellular localization.
Wild type, trs20ts, and trs20-D46Y mutant cells (NSY1555–1557) were transformed with a CEN plasmid expressing yEVenus-Ypt1 (A) or yEGFP-Ypt31 (B) from their own promoter and terminator, both functional as a single copy. Cells were visualized by live-cell deconvolution microscopy. Ypt1 puncta can be seen in wild type, trs20-D46Y and trs20ts mutant cells. In contrast, Ypt31 puncta are disrupted in trs20ts and trs20-D46Y mutant cells. Fluorescence in the YFP/GFP channel is shown at the top and DIC at the bottom. Scale bar 5 µM; n: number of cells visualized in two independent experiments.
Discussion
Here we show that Trs20 interacts with Trs120 and is required for incorporation of the latter into the TRAPP complex. Furthermore, an alteration analogous to a SEDT-causing mutation on the surface of Sedlin, D47Y, disrupts the interaction of Trs20 with Trs120 and the attachment of Trs120 to TRAPP. The Trs20-Trs120 association and its disruption by the SEDT-associated mutation were shown using three different approaches: yeast-two hybrid, co-precipitation of recombinant proteins, and BiFC in live cells. The requirement of the Trs20-Trs120 interaction for the assembly of TRAPP II was shown both in vitro and in vivo using co-precipitation analyses from bacterial and yeast lysates as well as localization of GFP-tagged TRAPP subunits and Golgi Ypts. Together, these results indicate that Trs20 plays a key role in the assembly of the TRAPP II complex in yeast. Moreover, the facts that Sedlin is a functional homolog of Trs20 and that the SEDT-associated mutation specifically affects the Trs20-Trs120 interaction suggest that this role is conserved.
TRAPP II assembly
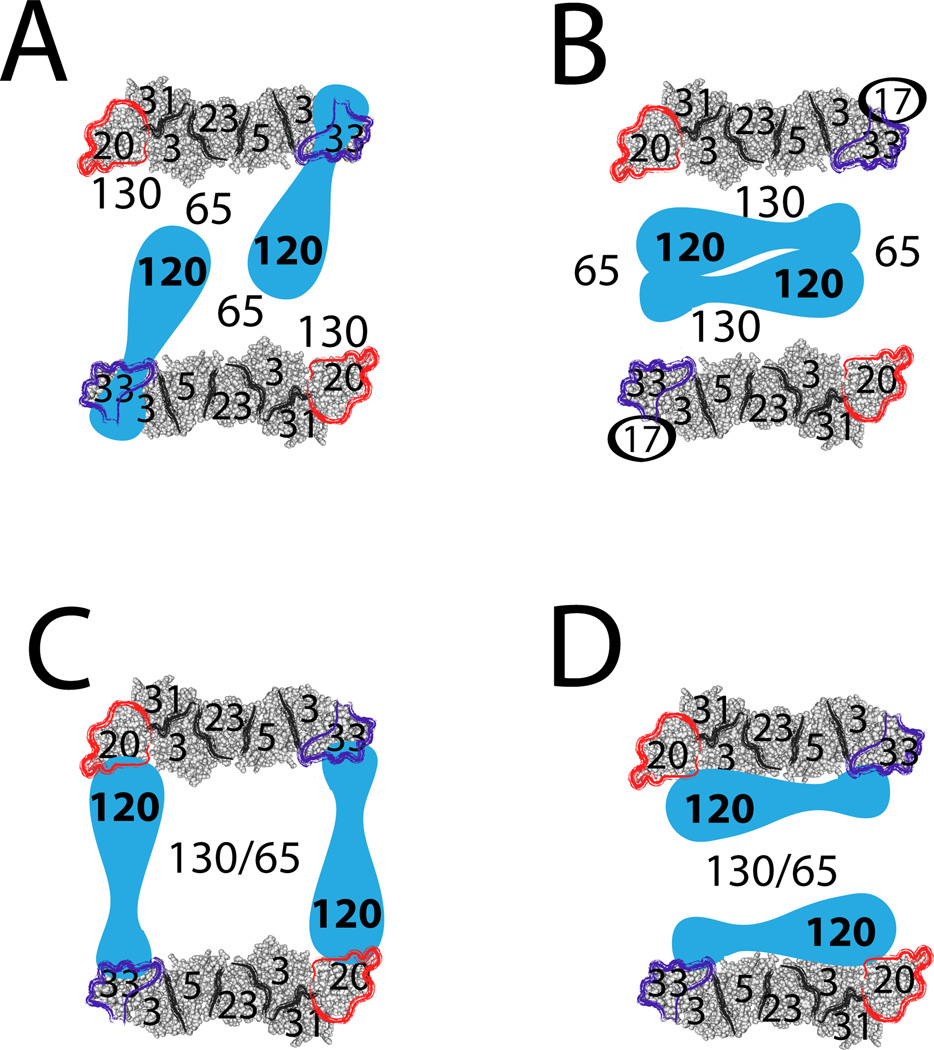
Whereas the assembly and structure of the TRAPP I core complex are well documented, not much is known about the larger TRAPP II complex. Currently, molecular structures of the TRAPP II-specific subunits are not available. Electron microscopy (EM) and gel filtration analyses revealed that TRAPP II complexes purified from yeast lysates form dimers in which the TRAPP II-specific subunits are sandwiched between two TRAPP I complexes. However, the importance of TRAPP II dimerization is not clear, since it depends on the non-essential TRAPP subunit Trs65 (27, 32), and currently there is no proof that this dimer exists in vivo. Regardless, based on EM data, a recent space-filling model of TRAPP II depicts Trs120 attached to the Trs33-end of TRAPP I, away from Trs20, as part of the bridge between the two TRAPP I monomers (32) (Figure 8A). Another model assigns more importance to the Tca17-Trs130 interaction in bridging the two TRAPP I monomers (27) (Figure 8B). Both models are of TRAPP purified from yeast cell lysates, whose construction depends on salt concentration and subunit composition (27, 33).
Figure 8. Models of TRAPP II complex architecture.
Shown are schemes of four possible models for TRAPP II complex structure. In all schemes, TRAPP II is depicted as a dimer, with TRAPP II-specific subunits sandwiched between two TRAPP I complexes (32). The structure of Trs20-containing TRAPP I was manually outlined loosely based on crystal structures of TRAPP sub-complexes (15). We have shown that Trs65 (17), Trs33 (18), and now Trs20, are important for assembly of the TRAPP II-specific subunits Trs120 and Trs130 with TRAPP I. A. Subunit organization in this scheme is based on space-filling EM images (32). It depicts Trs120 interacting with Trs33 and Bet3, whereas Trs130 interacts with Trs20, and Trs65 bridges the two TRAPP I monomers. B. This scheme is drawn based on Choi et al. (27), and includes Tca17. Here, Trs130 interacts with Tca17 in TRAPP I, Trs65 bridges between the two TRAPP I complexes, and Trs120 is sandwiched in the center. C. In this model Trs120 interacts with Trs20 and Trs33 on two different TRAPP I monomers. Together with Trs65 and Trs130, Trs120 bridges between the two parts of the TRAPP II dimer. D. In this model Trs120 interacts with Trs20 and Trs33 on the same TRAPP I monomer. In this scenario, Trs65 and Trs130 are important for the formation of the TRAPP II dimer. Data presented here and in our previous paper (18) are in agreement with models C and D, but not with Models A and B.
Our current data does not agree with either model. Therefore, we propose two other alternative models for TRAPP II architecture. In both models, Trs120 interacts with Trs20 and Trs33. In one model, Trs120 bridges two anti-parallel TRAPP I complexes, directly contacting Trs20 on one monomer and Trs33 on the other monomer. In this model, Trs120, Trs130 and Trs65 could stabilize the dimer (Figure 8C). In the second model, Trs120 contacts Trs20 and Trs33 on one TRAPP I monomer; in this model a TRAPP II dimer could be formed via Trs130 and Trs65 (Figure 8D). Both models are based on the Trs20-Trs120 interaction shown here, a previously shown interaction of Trs120 with the non-essential subunit Trs33 (18), the dependence of Trs120 interaction with TRAPP I on Trs20, and the enhancement of this interaction by Trs33. We have previously shown that the attachment of the second essential TRAPP II-specific subunit, Trs130, requires Trs120, but not vice versa (7). Therefore, we interpret the effect of the trs20ts mutation on the attachment of Trs130 to TRAPP as an indirect effect of Trs120 absence. Trs130 also interacts with two non-essential subunits, Trs65 and Tca17 (27). We suggest that whereas the Trs120-dependent attachment of Trs130 to the complex is essential for TRAPP II function, its interaction with Trs65 and Tca17 is not. With at least four models for TRAPP II architecture and the question of whether dimerization is important for TRAPP II function, future studies should include both high-resolution structure and in vivo analyses.
We suggest that TRAPP II is assembled by the addition of the TRAPP II-specific subunits to TRAPP I. Based on our current and previous studies, we propose a mechanism for TRAPP II assembly in which each step depends on the previous. The first step is association of Trs20 and Trs33 to the TRAPP I core complex. The second step is the assembly of Trs120 with the TRAPP I complex, which is dependent on its interaction with Trs20 and is stabilized by Trs33. This association, in turn, is necessary for the interaction of Trs130 with the complex and thus for the assembly of TRAPP II (7). This third step in TRAPP II assembly requires one of the non-essential subunits, Trs33 or Trs65 (18). An alternative possibility is that a TRAPP II sub-complex that contains Trs120 and Trs130 attaches to TRAPP I through the Trs20-Trs120 interaction and Trs33 stabilization. While a role for the fully assembled TRAPP II complex in the trans-Golgi has been shown (5), it is not clear whether any intermediate complexes play an independent role.
One open question concerning TRAPP II is the location of its assembly. TRAPP assembly could occur either on the Golgi or before the TRAPP complexes attach to membranes. We have proposed TRAPP conversion on the Golgi as a mechanism for coordination of entry into and exit from the Golgi (7). Alternatively, TRAPP I and TRAPP II could assemble in the cytoplasm independently before they attach to the appropriate Golgi cisterna.
Another unresolved question is the Ypt GEF substrate of TRAPP II. Currently there is controversy whether TRAPP II acts as a GEF for Ypt1 or Ypt31/Ypt32 (14, 16). We have shown that TRAPP II can act as a GEF for Ypt31/Ypt32 in vitro, and the function of Trs20, Trs120 and Trs130 are required for this activity (this study and (7)). The effects of TRAPP mutations on the cellular localization of Ypts and genetic interactions support this idea (this study and (7, 17, 18, 34)). However, direct demonstration as to which Ypt TRAPP activates in vivo is still pending.
Implications for Sedlin and SEDT
We propose that the role of Trs20 in TRAPP II assembly is conserved with its human counterpart, and that the SEDT-associated mutation D47Y in Sedlin causes a critical defect in this role. This idea is based on the findings that the D47Y analogous mutation in Trs20 results in specific disruption of the Trs20-Trs120 interaction, but not of the interaction of Trs20 with the TRAPP I core complex (Figures 5 and 6).
The idea that the role of Sedlin in TRAPP II assembly is conserved is further supported by two pieces of evidence. First, Sedlin is a functional homolog of Trs20 (19), suggesting conservation of essential interactions. Second, based on co-IP from transfected mammalian cell lysates, a role for Sedlin was proposed in the association of the large TRAPP II and TRAPP III subunits, TRAPPC9/mTrs120 and TRAPPC8/mTrs85, respectively, (26). Two observations support the idea that the D47 residue of Sedlin is required for its interaction with mTrs120, and therefore for mTRAPP II assembly. First, the finding that Sedlin with the D47Y mutation cannot complement trs20Δ (19) suggests that this mutation disrupts a conserved essential interaction. Second, the D47Y mutation causes a reduction in the co-IP of Sedlin with TRAPPC9/mTrs120 (26).
The SEDT symptoms associated with of the D47Y substitution are similar to those of Sedlin truncations (25). Therefore, if Sedlin-D47Y is defective in TRAPP II assembly, the most straightforward explanation for the SEDT-related intracellular trafficking defects is that they are caused by a TRAPP II assembly defect. However, it is possible that the D47 residue of Sedlin is also required for interaction with a yet unknown Sedlin partner. While this paper was in revision, a role for Sedlin in ER exit of large cargo, e.g., procollagen (PC), was shown through its interactions with the PC receptor TANGO1 and the COPII vesicle GTPase Sar1. The role of Sedlin information of “megacarriers”, but not of normal COPII vesicles, is thought to precede its function in TRAPP-dependent Ypt/Rab activation, and may explain the tissue-specific effects of the SEDT mutations (24). However, the effects of the Sedlin-D47Y mutation on the interactions of Sedlin with either TANGO1 or Sar1 were not determined.
Future Trs20/Sedlin Questions
Trs20 was originally identified as a TRAPP I/II subunit. However, to date there is no evidence that it functions in the context of TRAPP I and it is not clear for which transport step it is required. While a GALp shut-off analysis suggested that Trs20 is required for ER-to-Golgi (5), it does not provide direct proof. We show here that Trs20 is required for TRAPP II assembly and Ypt31 localization. Therefore, Trs20 is expected to play a role in the trans Golgi. Evidence for a direct role for Trs20 in a specific transport step in yeast is still missing.
Trs20 is essential for yeast cell viability and Sedlin is expressed ubiquitously in human cells. Therefore, it is not clear why the effect of disruption of Sedlin function in humans is cargo and tissue specific, affecting transport of large molecules like collagen (23). If the essential function of Sedlin was to facilitate association of mTrs120 with mTRAPP, it would be expected that disruption of Sedlin and mTrs120 would result in similar phenotypes. However, whereas Sedlin mutations cause a cartilage-specific disease, mutations in hTrs120/TRAPPC9 result in mental retardation (35–37). Therefore, additional roles for Sedlin and TRAPC9 are expected. Interestingly, both yeast and humans express a Trs20/Sedlin paralog, Tca17/TRAPPC2L, important for TRAPP II assembly (27, 29). Future studies should clarify the reason for diverse specific effects of TRAPP II subunit disruption in humans.
Materials and Methods
Reagents
Yeast strains and plasmids used in this study are summarized in Tables S1 and S2, respectively, and their construction is described in Supplementary Information. All reagents were purchased from Thermo Fisher Scientific unless otherwise noted. Yeast nitrogen base and yeast extract were purchased from US Biological. Amino acids, Triton X-100, and antibiotics were purchased from Sigma. DTT was purchased from Biorad. Protogel for acrylamide gels was purchased from National Diagnostics. EDTA-free Protease Inhibitor Cocktail (PIC) was purchased from Roche. Antibodies used for this study include α-HA (mouse, Cell Signaling), α-myc (mouse, Santa Cruz), α-GFP (mouse, Roche), α-His (mouse, Clontech), α-G6PDH (rabbit, Sigma) α-GST (rabbit, Invitrogen), α-Gal4 binding domain (rabbit, Santa Cruz), α-Bet3 and α-Trs20 (rabbit, generously provided by M. Sacher).
Microscopy
Strains expressing Trs120, Trs130, or Bet3 tagged with GFP on the COOH-terminus on the chromosome, as well as strains expressing GFP-tagged Ypt1 and Ypt31 from CEN plasmids, were grown at 26° in synthetic dextrose medium to OD600 ≈1.5. For 37°C microscopy, cells were shifted to a shaking 37°C water bath for 90 minutes. Cells were visualized using a deconvolution Axioscope microscope (Carl Zeiss). For the Bi-molecular Fluorescence Complementation (BiFC) assay, cells (NSY128) were transformed with two plasmids expressing Trs120-YFP-C and YFP-N-Trs20 or YFP-N-Trs20-D46Y; or CFP-N-Bet3 with CFP-C-Trs20 or CFP-C-Trs20-D46Y. Independent transformants were grown to mid-log phase, and YFP or CFP fluorescence were visualized using a Zeiss Confocal LSM 700microscope.
Pull-down of yeast TRAPP complexes
Plasmids encoding either GST or GST-Bet5 were transformed into either wild type or trs20ts mutant yeast strains. Cells were grown in synthetic media to OD600 ≈1 and induced with 0.5 mM copper sulfate. Upon induction, cells were incubated at 26°C for 50 minutes, followed by 70 minutes at 26° or 37°C. Cells were then harvested and frozen. For the purification, cell pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 10% glycerol, 4 mM MgCl2, 5 mM DTT, 2X PIC) at a concentration of ≈140 OD600 units/ml lysis buffer. 0.5 mm glass beads (Biospec Products) were then added to ≈30% total volume, and cells lysed by vortexing. Triton X-100 was then added to 0.1%, and lysates were incubated on ice for 15 minutes. Lysates were cleared at 3000 g for 10 minutes, and 1 ml of lysate was added to 120 µl Glutathione Sepharose 4B (GE) resin. Proteins were allowed to bind for 2 hours at 4°C. The resin was then washed three times in wash buffer (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 4 mM MgCl2, 10% glycerol, 3 mM DTT). Proteins were eluted by boiling with 120 µl Laemmli buffer, and samples were run on 10% acrylamide gels.
Protein level in yeast lysates
To compare the expression of yeast proteins for this study, 5 OD600 units of cells were harvested for each sample. Each was resuspended in 10% TCA, and lysaed with glass beads in the presence of urea, as described previously (38), except that TCA was used at a final concentration of 10%, and samples were vortexed with glass beads for 3 minutes.
Pull-down of recombinant TRAPP subunits
Plasmids encoding recombinant proteins were transformed into BL21 (DE3) E. coli. After growing to OD600 ≈0.8, cells were induced with 0.4 mM IPTG at 26°C for 3 hours. Cell pellets were re-suspended in lysis buffer (PBS, 10% glycerol, 1.5 mM DTT, PIC) at a concentration of ≈70 OD600 units/ml lysis buffer and lysed by sonication. Lysates were cleared at 2500 g for 15 minutes, and, when indicated, were further cleared at 100,000 g. Three hundred µl of the cleared lysate was added to 50 µl of either Glutathione Sepharose 4B (GE), Ni-NTA Agarose (Qiagen), or Amylose Resin (NEB), depending on the experiment. Proteins were bound for 60 minutes, and the resin was washed once with wash buffer (PBS, 10% glycerol, 1 mM DTT), spinning at 400 g for 1 minute. For His-pull-downs, imidazole was added to 10 mM in the lysis buffer, and 20 mM in the wash buffer. 400 µl of lysates containing binding partners or controls, prepared the same way as described above, were then added to the resin and allowed to bind for 90 minutes, after which the resin was washed 3 more times. Proteins were eluted by boiling in 40 µl Laemmli buffer and samples were run on 10% acrylamide gels.
GDP-release assay
GDP-release assay, to determine GEF functionality, was done as described previously (39), except using 11 pmol Ypt with 10 µg yeast pull-down product.
Yeast 2-hybrid
The pGBDU-C2 plasmids were transformed into NSY752; pACT2 plasmids were transformed into NSY468. The haploid transformants were then mated, and the resulting diploids spotted onto SD – Ura –Leu (for growth control), and SD –Ura -Leu –His + 3 mM 3AT (for detection of interaction), with one or two 10-fold serial dilutions, and incubated for 5 days at 26°C. Expression was confirmed by western blot analysis, probing with α-HA for activation domain constructs and α-Gal4 for binding domain.
Cell growth assay
Strains of TRS20Δ expressing Trs20 wild type, Trs20-D46Y, or Trs20ts from pRS315 were spotted onto YPD plates with two 10-fold serial dilutions, and incubated for 3 days at 26° or 39°C.
Supplementary Material
Acknowledgments
We thank M. Sacher for strains and antibodies, and Y. Liang for Y2H plasmids. This research was supported by grant GM-45444 from NIH to N. Segev.
References
- 1.Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. The Journal of cell biology. 1997;137(3):563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. The Journal of cell biology. 1995;131(3):583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science (New York, NY. 1991;252(5012):1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Molecular biology of the cell. 2000;11(12):4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Molecular cell. 2001;7(2):433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. The Journal of cell biology. 2000;151(2):289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozova N, Liang Y, Tokarev AA, Chen SH, Cox R, Andrejic J, Lipatova Z, Sciorra VA, Emr SD, Segev N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nature cell biology. 2006;8(11):1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 8.Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. The Journal of cell biology. 2005;171(5):823–833. doi: 10.1083/jcb.200505145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Molecular biology of the cell. 2005;16(1):178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(18):6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. The Journal of biological chemistry. 2005;280(39):33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 13.Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005;1(1):37–45. doi: 10.4161/auto.1.1.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S, Reinisch KM. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133(7):1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127(4):817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 16.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. The TRAPP complex: insights into its architecture and function. Traffic (Copenhagen, Denmark) 2008;9(12):2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Morozova N, Tokarev AA, Mulholland JW, Segev N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Molecular biology of the cell. 2007;18(7):2533–2541. doi: 10.1091/mbc.E07-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokarev AA, Taussig D, Sundaram G, Lipatova Z, Liang Y, Mulholland JW, Segev N. TRAPP II complex assembly requires Trs33 or Trs65. Traffic (Copenhagen, Denmark) 2009;10(12):1831–1844. doi: 10.1111/j.1600-0854.2009.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gecz J, Shaw MA, Bellon JR, de Barros Lopes M. Human wild-type SEDL protein functionally complements yeast Trs20p but some naturally occurring SEDL mutants do not. Gene. 2003;320:137–144. doi: 10.1016/s0378-1119(03)00819-9. [DOI] [PubMed] [Google Scholar]
- 20.Jang SB, Kim YG, Cho YS, Suh PG, Kim KH, Oh BH. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. The Journal of biological chemistry. 2002;277(51):49863–49869. doi: 10.1074/jbc.M207436200. [DOI] [PubMed] [Google Scholar]
- 21.Gedeon AK, Colley A, Jamieson R, Thompson EM, Rogers J, Sillence D, Tiller GE, Mulley JC, Gecz J. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nature genetics. 1999;22(4):400–404. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 22.Choi MY, Chan CC, Chan D, Luk KD, Cheah KS, Tanner JA. Biochemical consequences of sedlin mutations that cause spondyloepiphyseal dysplasia tarda. The Biochemical journal. 2009;423(2):233–242. doi: 10.1042/BJ20090541. [DOI] [PubMed] [Google Scholar]
- 23.Tiller GE, Hannig VL, Dozier D, Carrel L, Trevarthen KC, Wilcox WR, Mundlos S, Haines JL, Gedeon AK, Gecz J. A recurrent RNA-splicing mutation in the SEDL gene causes X-linked spondyloepiphyseal dysplasia tarda. American journal of human genetics. 2001;68(6):1398–1407. doi: 10.1086/320594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venditti R, Scanu T, Santoro M, Di Tullio G, Spaar A, Gaibisso R, Beznoussenko GV, Mironov AA, Mironov A, Jr, Zelante L, Piemontese MR, Notarangelo A, Malhotra V, Vertel BM, Wilson C, et al. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science (New York, NY. 2012;337(6102):1668–1672. doi: 10.1126/science.1224947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeyabalan J, Nesbit MA, Galvanovskis J, Callaghan R, Rorsman P, Thakker RV. SEDLIN forms homodimers: characterisation of SEDLIN mutations and their interactions with transcription factors MBP1, PITX1 and SF1. PloS one. 2010;5(5):e10646. doi: 10.1371/journal.pone.0010646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zong M, Wu XG, Chan CW, Choi MY, Chan HC, Tanner JA, Yu S. The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PloS one. 2011;6(8):e23350. doi: 10.1371/journal.pone.0023350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi C, Davey M, Schluter C, Pandher P, Fang Y, Foster LJ, Conibear E. Organization and assembly of the TRAPPII complex. Traffic (Copenhagen, Denmark) 2011;12(6):715–725. doi: 10.1111/j.1600-0854.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- 28.Montpetit B, Conibear E. Identification of the novel TRAPP associated protein Tca17. Traffic (Copenhagen, Denmark) 2009;10(6):713–723. doi: 10.1111/j.1600-0854.2009.00895.x. [DOI] [PubMed] [Google Scholar]
- 29.Scrivens PJ, Shahrzad N, Moores A, Morin A, Brunet S, Sacher M. TRAPPC2L is a novel, highly conserved TRAPP-interacting protein. Traffic (Copenhagen, Denmark) 2009;10(6):724–736. doi: 10.1111/j.1600-0854.2009.00906.x. [DOI] [PubMed] [Google Scholar]
- 30.Reck-Peterson SL, Novick PJ, Mooseker MS. The tail of a yeast class V myosin, myo2p, functions as a localization domain. Molecular biology of the cell. 1999;10(4):1001–1017. doi: 10.1091/mbc.10.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annual review of biophysics. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nature structural & molecular biology. 2010;17(11):1298–1304. doi: 10.1038/nsmb.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunet S, Noueihed B, Shahrzad N, Saint-Dic D, Hasaj B, Guan TL, Moores A, Barlowe C, Sacher M. The SMS domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in Saccharomyces cerevisiae. Cellular logistics. 2012;2(1):28–42. doi: 10.4161/cl.19414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou S, Liu Y, Zhang XQ, Chen Y, Ye M, Zhu X, Yang S, Lipatova Z, Liang Y, Segev N. Modular TRAPP Complexes Regulate Intracellular Protein Trafficking Through Multiple Ypt/Rab GTPases in Saccharomyces cerevisiae. Genetics. 2012 doi: 10.1534/genetics.112.139378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, Kahrizi K, Rafiq MA, Weksberg R, Nasr T, Naeem F, Tzschach A, Kuss AW, Ishak GE, Doherty D, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. American journal of human genetics. 2009;85(6):909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, Bodell A, Crosier M, Straussberg R, Walsh CA. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. American journal of human genetics. 2009;85(6):897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippe O, Rio M, Carioux A, Plaza JM, Guigue P, Molinari F, Boddaert N, Bole-Feysot C, Nitschke P, Smahi A, Munnich A, Colleaux L. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. American journal of human genetics. 2009;85(6):903–908. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. The Journal of cell biology. 1991;114(2):207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones S, Richardson CJ, Litt RJ, Segev N. Identification of regulators for Ypt1 GTPase nucleotide cycling. Molecular biology of the cell. 1998;9(10):2819–2837. doi: 10.1091/mbc.9.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.