Summary
Enteropathy-associated T-cell lymphoma includes type I cases and distinctive type II cases that, according to 2008 and 2010 World Health Organization descriptions, are T-cell receptor β+. Although T-cell receptor γδ enteropathy-associated T-cell lymphomas are reported, it is unknown if they have distinctive features and if they should be categorized as enteropathy-associated T-cell lymphoma or as a mucocutaneous γδ T-cell lymphoma. To address these questions, the clinicopathologic, immunophenotypic, molecular, and cytogenetic features of 5 γδ-enteropathy-associated T-cell lymphomas were investigated. Only 1 patient had celiac disease and had type I enteropathy-associated T-cell lymphoma, and the others fulfilled the histopathologic criteria for type II enteropathy-associated T-cell lymphoma. All lacked cutaneous involvement. A celiac disease–associated HLA type was found in the patient with CD and one of four others. All were T-cell receptor γ+, T-cell receptor δ+, βF1−, CD3+, CD7+, CD5−, CD4−, and TIA-1+ with variable staining for CD2 (3/5), CD8 (2/5), Granzyme B (1/5), and CD56 (4/5). Fluorescence in situ hybridization demonstrated 9q34 gains in 4 cases, with 9q33-34 gains by single nucleotide polymorphism in 3 of these. Single nucleotide polymorphism analysis also demonstrated gains in 5q34-q35.1/5q35.1 (4/5), 8q24 (3/5), and in 32 other regions in 3 of 5 cases. Vδ1 rearrangements were identified in 4 of 4 cases with documented clonality showing the same clone in normal-appearing distant mucosa (3/3 tested cases). Thus, γδ-enteropathy-associated T-cell lymphomas share many features with other enteropathy-associated T-cell lymphoma and are mostly of type II. Their usual nonactivated cytotoxic phenotype and Vδ1 usage are features unlike many other mucocutaneous γδ T-cell lymphomas but shared with hepatosplenic T-cell lymphoma. These findings support the conclusion that a γδ T-cell origin at extracutaneous sites does not define a specific entity.
Keywords: γδ T-cells, Enteropathy-associated, T-cell lymphoma, Gastrointestinal, lymphoma
1. Introduction
T cells include a major αβ T-cell receptor (TCR) subset and a minor γδ TCR subset [1]. Although many B-cell lymphomas have been defined based on the precise “cell of origin” the neoplastic cells most closely resemble, T-cell lymphomas (TCLs) are often defined based on other clinicopathologic features. Currently, there is only 1 TCL that is defined in part based on its γδ T-cell origin (primary cutaneous γδ TCL) and 1 where a γδ origin excludes the diagnosis (subcutaneous panniculitis-like TCL) [2]. The other major TCL often but not always of γδ type is hepatosplenic TCL [3].
Enteropathy-associated TCL (EATL), as defined in the 2008 revision of the World Health Organization (WHO) Classification of Tumors of the Haematopoietic and Lymphoid Tissues, is divided into 2 types. Classical or type I EATL is more common in the West and often associated with celiac disease (CD) and CD risk factors. Type II EATL is less often associated with CD and has other distinctive morphologic, phenotypic, and genetic features. The WHO monographs on hematopoietic and lymphoid tumors and gastrointestinal tumors note that type I EATLs are TCR β+/− and that the monomorphic form, a synonym for cases of type II, is TCRβ+, in conflict with 2 recent studies [4-7]. The importance of TCR expression in EATL thus remains uncertain. Furthermore, because many of the γδ TCLs involve skin or mucosal sites, it has been suggested that they might all be designated “mucocutaneous γδ” TCL, although more recent studies suggest that nonhepatosplenic γδ TCLs are heterogeneous [4,8]. To help clarify the nature of intestinal γδ TCLs, 5 cases that fulfilled the WHO criteria for EATL were investigated to assess their clinicopathologic features, HLA (human leukocyte antigen) type, immunophenotype, Vδ type, and cytogenetic findings based on single nucleotide polymorphism (SNP) and fluorescence in situ hybridization (FISH) studies.
2. Materials and methods
2.1. Case selection and review
Five βF1-negative intestinal TCL resection specimens fulfilling WHO criteria for EATL were identified. Clinical data were obtained from deidentified medical records. Gross descriptions and histologic features of tumor and uninvolved bowel were examined. All available immunohistochemical (IHC) stains and flow cytometric immunophenotypic studies were reviewed. In 4 of 5 patients, the resection specimen studied was the original diagnostic material. In 1 patient (case 3), studies were performed on a postchemotherapy resection specimen obtained 1 month after the diagnostic biopsy, which was also reviewed. Limited data from 4 cases were included for comparison purposes in a study of extranodal natural killer (NK)/T cell lymphomas, nasal type [9].
2.2. HLA typing
DNA was isolated from formalin-fixed, paraffin-embedded (FFPE) tissue sections using a Qiagen (Valencia, CA) DNA extraction kit. HLA typing for HLA-DQA1 and HLA-DQB1 was performed using the LABType reverse SSO DNA typing method (One Lambda, Canoga Park, CA) per manufacturer’s specifications.
2.3. Immunohistochemical and in situ hybridization studies
Unless already available, FFPE IHC stains for the following were performed on the tumors (using a Bench-Mark XT automated immunostainer (Ventana, Tucson, AZ): CD20, CD2, CD3, CD4, CD5, CD7, CD8, CD56, CD25, CD279, FOXP3 (stained manually), TIA-1, Granzyme B, CD30, ALK-1, p53, and βF1 (Table 1). Staining for TCRγ (clone 3.20; Thermo Fisher Scientific, Rockford, IL) and TCRδ (clone 5A6.39; Thermo Fisher Scientific) was performed per published methods [4,10]. Sections of bowel away from the tumor were stained for CD3, CD5, CD8, CD56, TIA-1, and Granzyme B. Epstein-Barr virus–encoded RNA (EBER) in situ hybridization was performed using the Discovery XT automated stainer (Ventana).
Table 1.
Antibodies used for immunohistochemical studies
| Antibody | Clone | Host | Source | Dilution |
|---|---|---|---|---|
| Anaplastic lymphoma kinase (ALK1) |
ALK01 | Monoclonal mouse | Ventana (Tucson, AZ) | Predilute |
| βF1 T-cell receptor (βF1) | 8A3 | Monoclonal mouse | Thermo Fisher Scientific (Rockford, IL) |
1:25 |
| CD2 | AB75 | Monoclonal mouse | Novocastra (Leica Microsystems, Bannockburn, IL) |
1:100 |
| CD3 | n/a | Polyclonal rabbit | Dako (Carpinteria, CA) | 1:100 |
| CD4 | SP35 | Monoclonal mouse | Dako | 1:200 |
| CD5 | SP19 | Rabbit monoclonal | Ventana | Predilute |
| CD7 | LP15 | Monoclonal mouse | Vector Laboratories (Burlingame, CA) |
1:50 |
| CD8 | C8/144B | Monoclonal mouse | Dako | 1:40 |
| CD20/L26 | L26 | Monoclonal mouse | Ventana | Predilute |
| CD25/interleukin 2 | 4C9 | Monoclonal mouse | Novocastra | 1:400 |
| CD30 | Ber-H2 | Monoclonal mouse | Ventana | Predilute |
| CD56 | 123C3.D5 | Monoclonal mouse | Cell Marque (Rocklin, CA) | Predilute |
| FOXP3 | FOXP3 | Monoclonal mouse | Abcam Inc. (Cambridge, MA) | 1:50 |
| Granzyme B | GrB-7 | Monoclonal mouse | Dako | 1:25 |
| P53 tumor suppressor protein (p53) |
DO-7 | Monoclonal mouse | Dako | 1:100 |
| CD279 | NAT | Monoclonal mouse | Abcam | 1:100 |
| T-cell intracellular antigen-1 (TIA-1) | 2G9A10F5 | Monoclonal mouse | Beckman Coulter (Brea, CA) | 1:400 |
2.4. SNP oligonucleotide microarray
DNA was extracted from FFPE tissue from tumor and uninvolved bowel for SNP analysis in cases 1, 2, 4, and 5 and from the tumor only in case 3. Genomic DNA (1 μg) was digested with Sty I (New England Biolabs, Ipswich, MA). A modified Affymetrix 500K restriction digest protocol was used, with an excess of both Sty I enzyme and buffer 3 used to digest the genomic DNA [11]. Digested samples were concentrated to 20 μL, and the Affymetrix 500K protocol for the 250K Sty array was followed, with 4 polymerase chain reaction (PCR) reactions instead of 3, followed by purification of product using Agencourt AmpPure beads (Affymetrix Cytogenetics Copy Number Assay User Guide). Seventy to ninety micrograms of labeled, fragmented, purified PCR product was loaded onto 250K Sty arrays. All cases passed Affymetrix AGCC array quality control (QC) metrics. SNP data were analyzed with Partek software, version 6.5 (St Louis, MO) using the tumor and, in 4 cases, matched uninvolved bowel samples. In case 3 without corresponding uninvolved bowel, a pooled sample of the 3 uninvolved bowel sections with the highest QC call rates was used as the control. Case 3 results should be interpreted with some caution as use of a pooled sample from all 4 case controls lead to overlapping but varied results. The QC call rates ranged from 72.29 to 89.29 (mean, 80.46) in the tumor and from 76.93 to 87.71 (mean, 82.84) in the apparently uninvolved bowel. Copy number variations involving at least 50 SNP markers were analyzed.
2.5. Fluorescence in situ hybridization
FISH studies were performed on 4-μm-thick FFPE tissue sections using a BCR/ABL dual color/dual fusion probe and a MYC break apart probe (Vysis, Abbott Park, IL). Cases with ≥3% abnormal nuclei were considered positive. Normal controls did not show gains.
2.6. Competitive PCR for TCR gene rearrangements
Competitive PCR was performed as previously described, with minor modification using DNA from FFPE tissue of the tumor mass in all cases and uninvolved bowel in 3 cases [12]. Oligonucleotide primers used were in part previously described (see Table 2) [13]. Recombinase Activating Gene (RAG2) amplification served as a competitor and positive control. PCR products were visualized by 2% agarose gel electrophoresis containing ethidium bromide. The sensitivity of this assay is approximately 10% of cells with the same TCR delta (TCRD) rearrangement. Under these conditions, only the clonally rearranged, lymphoma-derived TCRD genes could be detected, but not the polyclonal TCRD rearrangements of normal T lymphocytes infiltrating the tumor. A more sensitive PCR assay using 50% more TCRD primers, more efficient polymerase, and 40 cycles of PCR was also performed, with a sensitivity of approximately 1%. The amplification products were directly sequenced using the same 5′ TRDV1 and 3′ TRDJ1 primers (Oligo, Warsaw, Poland).
Table 2.
Oligonucleotides used in TCR gene rearrangement studies
| 5′ TCRD primers | |
| Vδ1b | 5′-GCA AAG TAC TTT TGT GCT CTT G |
| Vδ2b | 5′-GCA CCA TCA GAG AGA GAT GA |
| Vδ3 | 5′-ACA GCA GAT CAG AAG GTG CA |
| Vδ5 | 5′-CTG AAG GTC CTA CAT TCC TG |
| Dδ2 | 5′-AGA GGG TTT TTA TAC TGA TGT |
| 3′ TCRD primer | |
| Jδ1c | 5′- GAG TTA CTT ACT TGG TTC CAC |
| Control primers | |
| RAG2b(5′) | 5′-GCA ACA TGG GAA ATG GAA CTG |
| RAG2b(3′) | 5′-GGT GTC AAA TTC ATC ATC ACC ATC |
3. Results
3.1. Clinical features
All patients were adults. Only 1 had documented CD (Table 3). All patients presented with abdominal pain, 3 acutely with intestinal perforation, and 1 with perforation after the first course of chemotherapy. No patients had hepatomegaly, splenomegaly, or skin involvement, although the patient with CD had dermatitis herpetiformis. One patient developed pulmonary involvement 10 months after diagnosis. All patients died of disease with a mean survival of 5.6 months (0-12 months). The 2 with the shortest survivals did not receive treatment following surgery.
Table 3.
Clinical and gross pathologic features of each case
| Case | Without CD |
With CD |
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Clinical features | |||||
| Age, y | 72 | 73 | 31 | 80 | 63 |
| Sex | Male | Female | Male | Female | Male |
| History of celiac disease | No | No a | No | No | Yes b |
| Anti-TTG IgA | Not done | Not done | Not done | Not done | Positive |
| Treatment (CHOP) | Yes | No | Yes | No | Yes |
| Dead of disease | Yes | Yes | Yes | Yes | Yes |
| Survival (months) | 12 | 2 | 7 | 0 | 7 |
| Tumor site and gross features | |||||
| Location | Ileum and jejunum | Jejunum | Jejunum | Small intestine | Small intestine |
| Mass lesion | Multifocal | Single | Multifocal | Multifocal | Single |
| Perforation | No | Yes | No | Yes | Yes |
| Extraintestinal sites of involvement | |||||
| Adenopathy | No | No | Yes, distant | No | Yes, regional |
| Bone marrow | Not biopsied | No | No | Not biopsied | No |
| Hepatosplenomegaly | No | No | No | No | No |
| Skin lesions | No | None described | None described | No | Dermatitis herpetiformis |
| Other sites | Lungs | No | Ascites fluid | No | No |
After the diagnosis of EATL, this patient began prednisone therapy for “CD,” despite the lack of a diagnostic biopsy and positive serology. In our study, this patient was also shown to lack the celiac-associated HLA genotype.
The CD was diagnosed 58 years before the lymphoma. The patient had been treated intermittently with steroids in the past and 3 months before his lymphoma began following a gluten-free diet after which his symptoms improved.
3.2. HLA typing
Two patients, including the patient with CD, were HLA-DQA*0501/DQB1*0201+ (Table 4).
Table 4.
HLA-DQA1 and HLA-DQB1 genotypes in each case
| Case | HLA- DQA1* |
HLA- DQA1* |
HLA-DQB1* | HLA-DQB1* |
|---|---|---|---|---|
| 1 | 0102 | 0301/02/ 03 |
0302 | 0602 |
| 2 | 0505/09 | 0505/09 | 0301/16/19/ 21/22 |
0301/13/16/19/ 21/22 |
| 3 | 0102 | 0505/09 | 0301/19/22 | 0501 |
| 4 | 0302/03 | 0501 | 0201/05 | 0301/19/21/22 |
| 5 | 0301 | 0501 | 0201/03/05 | 0302 |
3.3. Gross and microscopic pathologic features
All patients had small intestinal disease, with discrete tumor masses, 5 to 28 cm in greatest dimension, in 4 cases (Table 3). The entire bowel wall was thickened and edematous in case 3 (resection followed by chemotherapy). Three patients had grossly identified multifocal disease, but otherwise, where described, the mucosa was unremarkable.
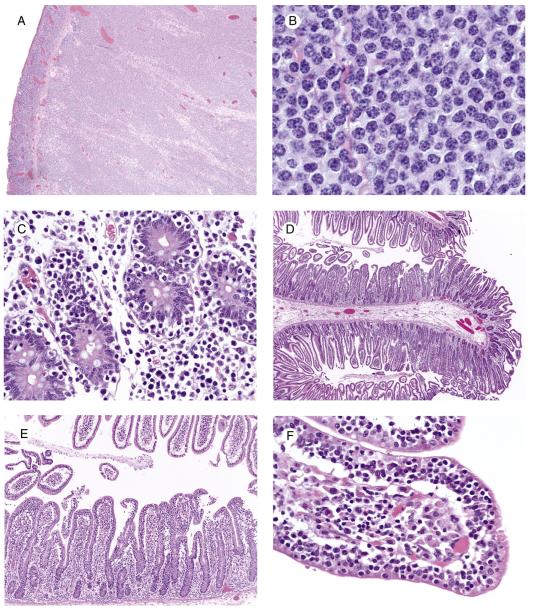
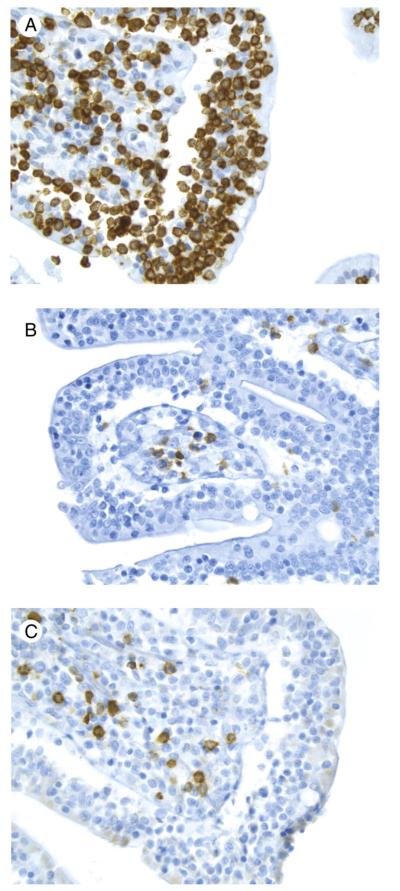
The major tumor masses were ulcerated and showed transmural infiltration (Fig. 1A) by a monotonous population of small- to medium-sized cells with mostly inconspicuous nucleoli (Fig. 1B) and occasional admixed large cells. There was epitheliotropism of the overlying mucosa (Fig. 1C) and few to absent admixed inflammatory cells except in the previously treated resection specimen, which had relatively few neoplastic cells. This patient’s diagnostic small mucosal biopsy demonstrated a markedly epitheliotropic infiltrate that initially had been diagnosed as a MALT lymphoma. The epitheliotropism in the remaining cases was mild (3) or moderate (1). One of the 4 cases without CD showed 1 focus with markedly increased IEL without villous blunting (Fig. 1D-F), and the other 3 showed multifocal areas with increased IEL and variable villous blunting. The bowel away from the main tumor mass in the patient with CD showed diffuse enteropathic change (Fig. 2). The biopsy demonstrating lung involvement showed patchy infiltration of cells similar to those in the main tumor mass (Fig. 3).
Fig. 1.
EATL without CD (case 4). A, Note the ulceration and diffuse transmural infiltrate in the main tumor mass (original magnification ×4). B, The infiltrate is composed of a monotonous population of medium sized cells with round nuclei and relatively scant cytoplasm. There are very few admixed reactive cells (original magnification ×100). C, The overlying mucosa shows prominent epitheliotropism (original magnification, ×40). D, The bowel distant to the main tumor mass demonstrates 1 area with very focal increased IEL visible at low magnification and an adjacent segment with unremarkable intact villi (original magnification ×4). E, At slightly higher magnification, the contrast in the number of IEL is striking between the 2 areas (original magnification ×10). F, Note the numerous small- to medium-sized IEL with relatively condensed chromatin in the abnormal area (original magnification ×40).
Fig. 2.
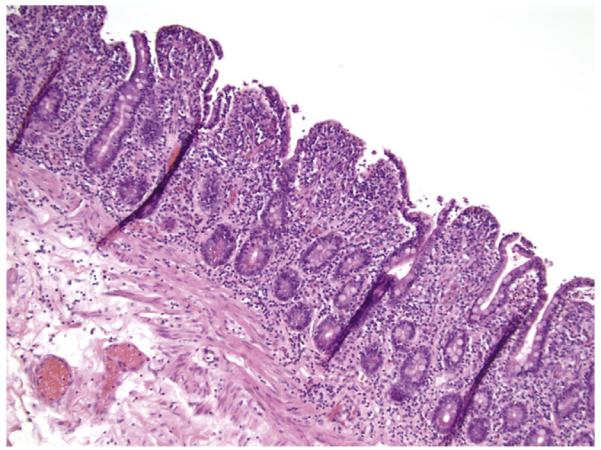
EATL with associated CD (case 5). The bowel distant to the tumor shows diffuse enteropathic changes (original magnification ×10).
Fig. 3.
Intestinal γδ TCL involving lung (case 1). Note the patchy lymphoid infiltrate within the alveolar septae (original magnification ×10) (A) composed of medium-sized lymphocytes with round to angulated nuclear contours (original magnification ×40) (B). CD3 highlights the infiltrating lymphocytes (C), but there are only scattered CD5+, presumably normal T cells (original magnification ×10) (D).
3.4. Immunophenotypic and in situ hybridization studies
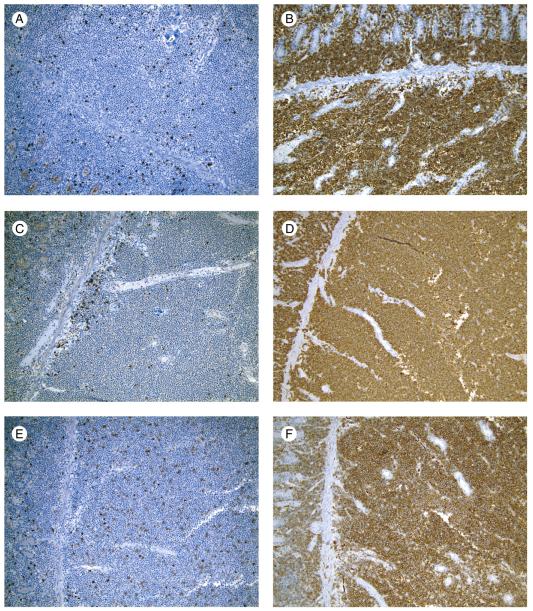
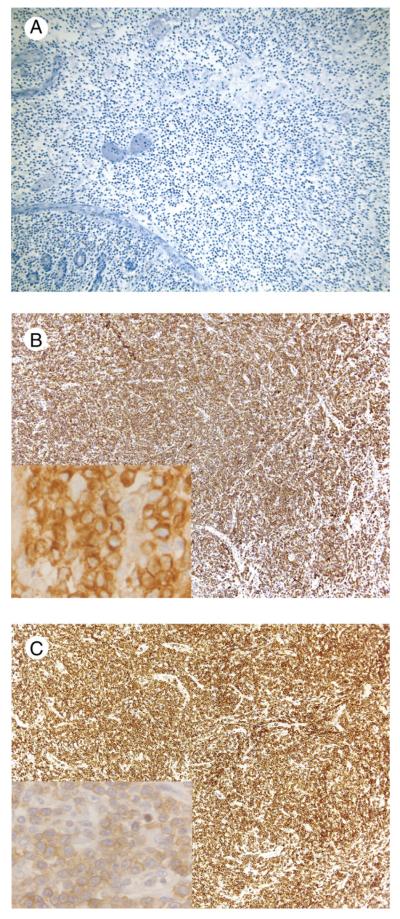
All tumors were βF1−, TCRγ+, TCRδ+, CD3+, CD7+, CD5−, CD4−, TIA-1+, CD30−, CD25−, FOXP3−, EBER−, and CD20− (Table 5, Figs. 4 and 5). There was variable staining for CD2, CD8, CD56, CD279, and Granzyme B. P53 was positive in most cells in 1 case and negative in all others. Flow cytometric immunophenotypic studies performed in 2 cases were concordant with the IHC (cases 3 and 5). The IEL away from the main tumor mass in all cases were CD3+, CD5−, TIA-1+, and Granzyme B– by IHC (Fig. 6). One case was positive for CD56. Three cases had CD8+ IEL, including 2 cases in which the tumor was negative.
Table 5.
Immunophenotypic features of the tumor and IEL in bowel distant to tumor
| Without CD (n = 4) |
With CD (n = 1) |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Tumor, IHC | |||||
| βF1 | − | − | − | − | − |
| TCRδ | + | + | + | + | + |
| TCRγ | + | + | + | + | + |
| CD2 | + | − | + | − | + |
| CD3 | + | + | + | + | + |
| CD5 | − | − | − | − | − |
| CD7 | + | + | + | + | + |
| CD4 | − | − | − | − | − |
| CD8 | − | − | + | − | + |
| CD56 | + | + | + | + | − |
| TIA-1 | + | + | + | + | + |
| Granzyme B | − | − | − | + | − |
| CD30 | − | − | − | − | − |
| CD25 | − | − | − | − | − |
| PD1 | + | − | − | − | − |
| FOXP3 | − | − | − | − | − |
| CD20 | − | − | − | − | − |
| EBER | − | − | − | − | − |
| p53 | − | − | + | − | − |
| Tumor, flow cytometry | |||||
| αβ−, γδ+ | Not done | Not done | + a | Not done | + b |
| CD103+ | Not done | Not done | + | Not done | + b |
| Intraepithelial lymphocytes in bowel distant to tumor, IHC | |||||
| CD3 | + | + | + | + | + |
| CD5 | − | − | − | − | − |
| CD8 | + | + | − | − | + |
| CD56 | − | − | − | + | − |
| TIA-1 | + | + | + | + | + |
| Granzyme B | − | − | − | − | − |
Cells were probably γδ+, but interpretation was somewhat difficult. CD103 expression was weak.
Cells were apparently γδ+ and possibly CD103+, but interpretation was somewhat difficult.
Fig. 4.
EATL without CD, main tumor mass (case 4). Immunohistochemical stains showed the following phenotype: CD2 negative (A), CD3 positive (B), CD5 negative (C), CD7 positive (D), CD8 negative (E) in at least most neoplastic cells, and CD56 positive (F) (immunohistochemical studies with hematoxylin counterstain, original magnification ×10).
Fig. 5.
EATL with associated CD, main tumor mass (case 5). Immunohistochemical stains demonstrate a βF1 negative (A), TCRγ positive (B), and TCRδ positive phenotype (C) (immunohistochemical studies with hematoxylin counterstain, original magnification, ×10; Fig 5B and 5C insets, ×50 oil).
Fig. 6.
EATL without CD, IEL in bowel distant to tumor (case 4). A, CD3 demonstrates numerous positive cells within the intestinal epithelium and scattered positive cells in the lamina propria. B, CD5 shows few scattered positive cells in the lamina propria; intraepithelial cells are essentially negative. C, CD8 is similar to CD5 (immunohistochemical studies with hematoxylin counterstain, original magnification ×40).
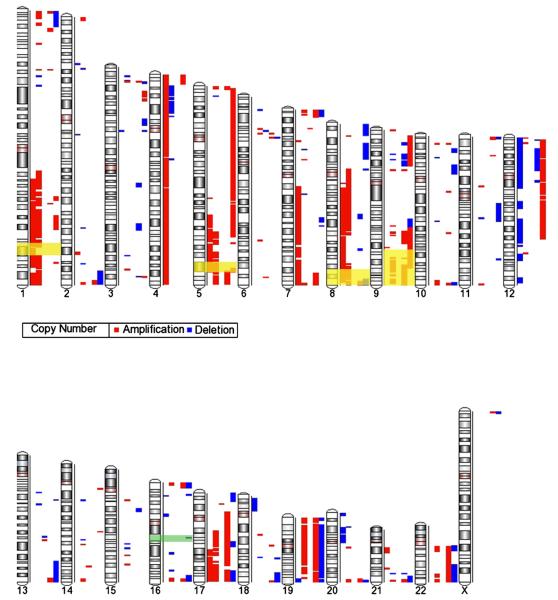
3.5. SNP analysis
A total of 363 copy number variations were identified, with a mean of 72.6 ± 61.4 genomic alterations per sample (range, 7-150). The percentage of the genome altered per case ranged from 0.1% to 22.2% (mean, 13.7% ± 7.8%). Abnormalities were seen in all chromosomes except Y, for which there were no probes in the SNP array. There were 238 total segmental gains and 125 total deletions (Fig. 7). +5q34-5q35.1 and +5q35.1 were identified in 4 of 5 cases. In addition, SNP studies revealed 33 additional DNA segment gains in 3 of 5 cases involving 7 additional chromosomal regions (1q24.2-1q42.13, 17q22-25.3, 5p15.33, 5q31.1-35.1, 7q36.3, 8q24.3, 9q22.2-33.3) (Table 6). Identical deletions were never found in more than 2 cases.
Fig. 7.
SNP analysis. This karyogram demonstrates the amplifications (red bars) and deletions (blue bars) seen in each of the cases. The regions with recurrent genetic abnormalities as reported in the 2008 WHO monograph are highlighted, with gains in yellow and losses in green. Because of variations in the literature, the chromosomal region highlighted on chromosome 9q is slightly larger (9q31-34) than what is cited in the WHO monograph. Each column represents a case, going from case 1 through 5 from left to right.
Table 6.
Identical chromosomal abnormalities identified by SNP analysis in at least 3 cases
| Chromosome no. | Start nucleotide |
Stop nucleotide |
Chromosomal cytoband |
Total no. of cases with amplifications |
Specific cases with amplification |
|---|---|---|---|---|---|
| 5 | 167636997 | 169349402 | 5q34-5q35.1 | 4 | 1,2,4,5 |
| 5 | 169363515 | 169658176 | 5q35.1 | 4 | 1,2,4,5 |
| 1 | 169554263 | 171104078 | 1q24.2-1q24.3 | 3 | 1,2,5 |
| 1 | 171237165 | 172499758 | 1q24.3 | 3 | 1,2,5 |
| 1 | 174928236 | 175968783 | 1q25.1 | 3 | 1,2,4 |
| 1 | 185495183 | 198911301 | 1q25.3-1q32.1 | 3 | 1,2,5 |
| 1 | 199052764 | 199471865 | 1q32.1 | 3 | 1,2,5 |
| 1 | 228653365 | 229363835 | 1q42.13 | 3 | 1,2,5 |
| 5 | 2325664 | 3420288 | 5p15.33 | 3 | 2,4,5 |
| 5 | 131969143 | 133326559 | 5q31.1 | 3 | 1,2,5 |
| 5 | 133472722 | 134671479 | 5q31.1 | 3 | 1,2,5 |
| 5 | 134771119 | 137416280 | 5q31.1-5q31.2 | 3 | 1,2,5 |
| 5 | 143411749 | 143604652 | 5q31.3 | 3 | 1,2,5 |
| 5 | 165329750 | 167535353 | 5q34 | 3 | 1,2,5 |
| 5 | 167592470 | 167636997 | 5q34 | 3 | 1,2,5 |
| 5 | 169349402 | 169363515 | 5q35.1 | 3 | 1,4,5 |
| 5 | 169658176 | 169668330 | 5q35.1 | 3 | 1,4,5 |
| 7 | 157057643 | 159086805 | 7q36.3 | 3 | 1,2,4 |
| 8 | 142330315 | 143786829 | 8q24.3 | 3 | 1,2,4 |
| 9 | 92153796 | 93522010 | 9q22.2 | 3 | 2,4,5 |
| 9 | 93575308 | 93592336 | 9q22.2 | 3 | 2,4,5 |
| 9 | 93638631 | 93899271 | 9q22.2 | 3 | 2,4,5 |
| 9 | 95378915 | 96691486 | 9q22.31-9q22.32 | 3 | 2,4,5 |
| 9 | 116451489 | 117090546 | 9q32 | 3 | 2,4,5 |
| 9 | 122556547 | 126119221 | 9q33.2-9q33.3 | 3 | 2,4,5 |
| 17 | 70049123 | 71412197 | 17q24.3-17q25.1 | 3 | 1,2,4 |
| 17 | 71412197 | 72077647 | 17q25.1 | 3 | 1,2,4 |
| 17 | 72077647 | 80685522 | 17q25.1-17q25.3 | 3 | 1,2,4 |
| 17 | 51094261 | 52976257 | 17q22 | 3 | 1,2,4 |
| 17 | 53848596 | 57324904 | 17q22 | 3 | 1,2,4 |
| 17 | 61546534 | 63575337 | 17q23.3-17q24.1 | 3 | 1,2,4 |
| 17 | 64622048 | 66662025 | 17q24.2 | 3 | 1,2,4 |
| 17 | 66662025 | 66810240 | 17q24.2 | 3 | 1,2,4 |
| 17 | 69884518 | 70049123 | 17q24.3 | 3 | 1,2,4 |
| 17 | 80685522 | 81012186 | 17q25.3 | 3 | 1,2,4 |
3.6. Fluorescence in situ hybridization
FISH studies demonstrated gains in 9q34 in 4 cases (Fig. 8), 3 of which showed gains in 9q33-q34 by SNP analysis. The seemingly discrepant case (patient 3) had prominent necrosis and only 6.9% cells with an extra ABL signal by FISH. In the 3 cases with gains in 8q24 by SNP analysis, FISH for MYC confirmed a gain in 1, failed in 1, and showed 2 normal signals in 1. The 2 cases without +8q24 by SNP analysis were also negative by FISH.
Fig. 8.
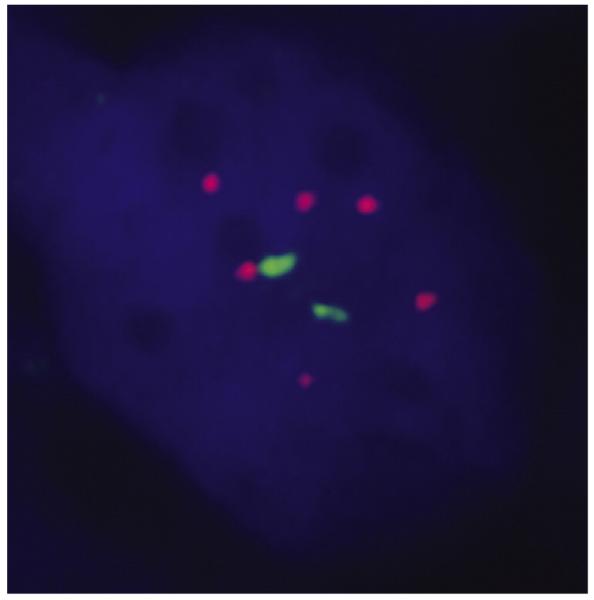
FISH using a BCR/ABL dual color, dual fusion probe demonstrates extra signals with 9q34 (ABL) (case 2). BCR is the green probe; ABL is the red probe.
3.7. TCR rearrangements
Using the less sensitive competitive PCR assay with the TRDV1 and TRDV2 5′ primers, distinct PCR bands of intensity comparable to the RAG2 competitor were observed in tumor samples from cases 1, 2, and 4, suggesting the presence of a rearrangement in most cells (Fig. 9). In the tumor sample from case 3, the rearrangement band was weaker than the RAG2 band, indicating a lower content of the cells harboring the amplified rearrangements. No amplification products were obtained in either the tumor sample from case 5, the 4 tissue samples from bowel away from the tumors (cases 1, 2, 4, 5) or in peripheral blood leukocytes from healthy individuals with this assay. No rearrangements were amplified with Vδ3, Vδ5, and Dδ2 5′ primers. In the cases with a clone demonstrated in the tumor and with available material (cases 1, 2, 4), the more sensitive PCR assay demonstrated Vδ1-Jδ1 rearrangements in the bowel away from tumor, although the bands were weaker than in the corresponding tumor samples, consistent with a lower proportion of the clonal T cells. Direct sequencing analysis of the amplification products with the universal Jd1c primer revealed clonal in frame TRDV1-J1 rearrangements in tumor samples from cases 1, 2, 3, and 4 (Table 7). All of them contained the TCRD diversity segment (TRDD3). Two cases (1 and 3) additionally harbored the less frequent TRDD2 segment. The sequence analysis in apparently uninvolved bowel samples from cases 1, 2, and 4 demonstrated the same T-cell clone as in the corresponding tumor sample, as well as polyclonal TRDV1-J1 and TRDV2-J1 sequences from the normal T cells.
Fig. 9.
A, TCRD gene rearrangements using the less sensitive PCR assay, with paired tumor (T) and distant normal samples (designated with corresponding case number only). Tumor samples from cases 1 to 4 show rearrangements, with no rearrangements in samples from bowel distant to tumor in all cases. Case 5 showed no rearrangements. B, TCRD gene rearrangements using the more sensitive PCR assay. Rearrangements are seen in the tumor samples from cases 1 to 4. Rearrangements are seen in the samples from bowel away from main tumor mass samples in cases 1, 2, and 4, although the bands were weaker than in the corresponding tumor samples, suggesting a lower proportion of the clonal T cells. No rearrangements are seen in case 5.
Table 7.
Nucleotide sequence of the TRDV1-J1 rearrangements
| Germ link sequence |
TRDV1
|
TRDD2
|
TRDD3
|
TRDJ1
|
N |
|---|---|---|---|---|---|
| TGT GCT GGG GAA CT | CCTTTCCTAC | ACTGGGGATACG | AC ACC GAT AAA CTC ATC | ||
| Patients | |||||
| 1 | TGT GCT CTT GGG G | TC TACCTTTCCT |
TGGGGGATACG CTG CGG T | AC ACC GAT AAA CTC ATC | +27 |
| 2 | TGT GCT CTT GGG GAA C | GG CCA GGG AGC TACTGGGGGATAC | CC GAT AAA CTC ATC | +21 | |
| 3 | TGT GCT CTT GGG GAA CT | C CCTACG | TGGGGGATA CTG G | AC ACC GAT AAA CTC ATC | +21 |
| 4 | TGT GCT CTT GGG GAA | GCT CAG TAC TGGGGGATC G | CC GAT AAA CTC ATC | +15 | |
NOTE: The nucleotide sequences are divided into amino acid coding triplets. All rearrangements are in frame and functional. N: length of the third complementarity determining region (CDR3). Germ line sequences of the TRD segments are indicated in the upper row. Bold: TRD variable (TRDV1) and joining (TRDJ1) segments; single underlined: TRD diversity segment 3 (TRDD3); double underlined: TRD diversity segment 2 (TRDD2); normal characters: random nucleotides inserted by the terminal deoxynucleotidyl transferase.
4. Discussion
EATL is an intestinal TCL with intraepithelial T cells together with villous atrophy and crypt hyperplasia in the adjacent small intestinal mucosa [6]. However, “the degree of enteropathy is highly variable,” and there may only be “an increase in intraepithelial lymphocytes” [6]. Many, but not all cases, are associated with CD. Two distinct, but overlapping “subtypes” of EATL, have been recognized[13-15]. Type I or classic cases have “nonmonomorphic” cytomorphology, frequent gains in chromosomes 1q and 5q, and are usually CD56− and CD8− [15]. They have an association with CD and share genetic alterations and HLA-DQB1 genotype patterns with “refractory” CD [15]. Type II cases have monomorphic small- to medium-sized neoplastic cells, frequent CD56 and CD8 expression, and gains in 8q24 (MYC) (73% vs 25% of type I cases), but only “rare” gains in 1q and 5q (21% of cases) and a HLA-DQB1 genotype pattern that resembles the normal white population [16]. In contrast to “pleomorphic” EATL, “monomorphic” cases are reported to have biallelic TCRγ rearrangements [18]. Both types express cytotoxic granules, are usually CD5− [5,16-18], and frequently have gains in 9q (more common) or loss of 16q21.1 [5,16-18]. The 2008 WHO monograph suggested that type II EATL might represent a distinct disease entity [6]. The 2010 WHO monograph on gastrointestinal tumors segregates these cases as a subtype of “TCL of the small intestine” termed monomorphic CD56+ intestinal TCL [7]. Others have suggested the name “monomorphic intestinal TCL” [5]. According to the 2008 WHO monograph on hematopoietic/lymphoid tumors and the 2010 WHO monograph on gastrointestinal tumors, EATL type I is TCRβ+/− and type II TCRβ+ [6,7]. However, many cases of both types have been reported to be TCRβ− and with many of those cases also TCRδ− (“receptor silent”) [17]. A recent study reported that most type II EATLs are of γδ origin [5]. The current study was performed to study the detailed clinical and biologic features of intestinal TCLs of γδ T-cell type without preselection of only type II EATL and with inclusion of features not previously analyzed in this specific subset of patients, including the HLA type, cytogenetic copy number variations, and Vδ gene use. It also addresses the issue of their categorization.
Demonstration of a γδ T-cell origin in lymphomas can be very problematic, in the absence of fresh cells or frozen tissue. Negative staining with the βF1 antibody does not establish a γδ T-cell origin because many T-cell neoplasms can be TCR silent, including intestinal TCL [4,17]. Conventional TCR PCR studies are also inadequate to make this assessment because TCRγ gene rearrangements are frequently seen in αβ T cells, and TCRβ gene rearrangements can be present in γδ T cells [19,20]. Recently, however, paraffin reactive antibodies for TCRγ and δ have been reported with a sensitivity and specificity for TCRγδ of 95% and 86%, respectively, and a sensitivity of 60% for TCRδ [4,10]. The functional nature of the TCRD rearrangements that could be sequenced in 3 of the cases also strongly supports a TCRγδ origin [21].
Of the 5 TCRγδ cases studied here, 4 were typical type II EATL with small intestinal disease, monomorphic cytology, at least some epitheliotropism, CD56 positivity, variable CD8, and no definite CD, and with only 1 having a CD-associated HLA-DR type. Others have also noted variation in CD8 expression in type II EATL (with 1 study reporting no statistical differences between type I and type II EATL) as well as variation in CD8 (and CD56) staining between the tumor and other IEL [5,16,17,22]. Also consistent with the diagnosis, the neoplastic cells were CD3+, CD2 variable, CD7+, CD5−, and TIA-1+. Most were Granzyme B–. This nonactivated cytotoxic T-cell phenotype is similar to hepatosplenic TCL but in contrast to the activated cytotoxic phenotype found in CD, in EATL in general, and in other mucocutaneous γδ TCL [3,7,22]. All demonstrated at least focal epitheliotropic involvement at sites distant from the main tumor mass, with 3 showing villous blunting and 1 case showing a single striking focus with marked epitheliotropism in villi of normal height. Others have described mild villous blunting in a minority of type II EATL [5]. The molecular studies demonstrated a small clonally related T-cell population in microscopically unremarkable bowel supporting the presence of widespread dissemination, even when not grossly or histologically apparent. A recent series specifically looking at type II EATL found 61% γδ TCR+, 17% αβ TCR+, 17% αβ/γδ TCR+, and 6% αβ/γδ TCR− [5]. The 11 γδ TCR+ only cases were CD3+, 9 of 11 CD2+, CD8+, CD56+ and with 1 focally CD20+. At least 2 other definite type II EATLs of γδ TCR type have been reported [4]. Limited other γδ TCR+ intestinal lymphomas with an absence of any documented enteropathic changes have also been reported, including some pleomorphic cases and 1 that might represent an extranodal T/NK cell lymphoma [4,23-26].
One of our small intestinal γδ TCL, in spite of a monotonous appearance, was best classified as a type I EATL as the patient had well-documented CD, a CD-associated HLA type, dermatitis herpetiformis, diffuse enteropathic change, and it was CD56−. One other CD56−, CD8− small intestinal γδ TCL associated with CD but lacking villous atrophy has been reported as well as 1 other γδ type I EATL [5,23]. The previously documented minimal megakaryocyte-associated tyrosine kinase (MATK) expression in this case, in contrast to the much more marked staining in the 3 type II EATL cases tested here, also supports its categorization as a type I EATL [9,27].
Cytogenetic studies also support the inclusion of these 5 γδ TCR+ as EATL. Gains in 9q33-34, present in 4 cases here, are the most common abnormality in EATL, reported in 58% to 80% of cases [16,18,28,29]. Similar gains are reported in 0% to ~20% of other types of TCL [29,30]. Loss of 16q12.1, reported in 23% of EATL, usually in the absence of gains in 9q, was seen in 1 of our cases, but this patient also had a gain in 9q. One case of type II βF1+ EATL with both loss of 16q and gain of 9q33.1-q34.12 has been reported [28]. Other commonly found cytogenetic abnormalities reported in EATL were seen in a variable number of our cases (Table 8).
Table 8.
Comparison of SNP findings with published comparative genomic hybridization (CGH) findings in EATL
| Region involved |
Published data for EATL |
SNP findings |
||||||
|---|---|---|---|---|---|---|---|---|
| Zettl et al [16] |
Deleeuw et al [15] |
Zettl et al [29] |
Without CD |
With CD |
||||
| 1 | 2 | 3a | 4 | 5 | ||||
| +9q33.2-q33.3 | 67% | − | + | − | + | + | ||
| +9q33-q34 | 64% | 58% | − | + | − | + | + | |
| +5q34-q35.2 | 21% | 50% | 18% | + | + | − | + | + |
| +1q32.2-q41 | 26% | 47% | + | + | − | − | − | |
| +7q33-7q34 | 47% | + | − | − | − | − | ||
| +1q23.1-q23.3 | 46% | + | + | − | − | − | ||
| +1q25.3-q31.2 | 43% | + | + | − | − | + | ||
| +7q22.2-q31.1 | 43% | + | − | − | − | − | ||
| +7q36.1-qter | 43% | + | − | − | + | − | ||
| +8q13.3-q21. | 11 43% | + | + | − | − | − | ||
| +8q22.2-q24.3 | 18% | 43% | 11% | + | + | − | + | − |
| +5p15.33 | 37% | − | + | − | + | + | ||
| +7q11.22-q11.23 | 37% | + | − | − | − | − | ||
| +7q11.23-q21.3 | 37% | + | − | − | − | − | ||
| −8p22-p23.2 | 21% | 37% | − | − | − | − | + | |
| +7q21-q22 | 31% | 24% | + | − | − | − | − | |
| −10q26.2-q26.3 | 30% | − | − | − | − | + | ||
| +7p22.3 | 27% | − | + | − | + | − | ||
| +12p13.31-p13.33 | 27% | − | + | − | − | + | ||
| −13q22 | 25% | − | − | + | − | − | ||
| −13q21 | 24% | − | − | − | − | − | ||
| −8pcen-p21 | 24% | − | − | − | − | + | ||
| −16q12.1 | 23% | 23% | − | − | − | − | + | |
| +6p25.2-pter | 23% | − | − | − | − | − | ||
| +16p13.3 | 23% | − | + | + | + | − | ||
| +17q25.3 | 23% | + | − | − | + | − | ||
| +21q22.3 | 23% | − | + | − | + | − | ||
| +22q12.3-q13.2 | 23% | − | − | − | + | − | ||
| −9p21.2-p21.3 | 15% | 23% | − | + | − | + | − | |
| −17p12-p13.2 | 21% | − | − | − | − | + | ||
NOTE: The published abnormal regions do include some areas thatoverlap but were reported separately. If there was a genomic alteration identified by SNP analysis in any portion of the chromosomal cytoband as published, it was considered positive for that alteration
This is the case with extensive posttherapy tumor necrosis that also lacked a paired normal sample for direct comparison.
Cytogenetic differences have been documented between type I and type II EATL [15]. Classic type I cases are reported to more frequently show gains in 1q32.2-q41 and in 5q34-q35.2, with both of these gains tending to occur together, although they are also found in 20% to 27% of type II cases [15]. Both abnormalities were found in 2 of our cases including 2 type II EATLs, and the gain in 5 was present in 2 additional cases. In contrast, gains in 8q22.2-q24.3, found in 3 type II cases, are more common in type II EATL (73%) but are also present in 27% of type I cases [15]. A recent study reports 50% of type II EATL with MYC gains [31].
In addition to a lack of cutaneous involvement in any of our cases, the Vδ data reported here also support separating the intestinal from the cutaneous γδ TCL and suggest a closer relationship to hepatosplenic TCL. Although there are 6 Vδ gene segments, more than 95% of γδ T cells express either Vδ1 or Vδ2. Vδ1 T cells are preferentially found in the spleen, thymus, and intestinal epithelium, whereas Vδ2 T cells are found more frequently in the peripheral blood, tonsils, and skin [12]. Hepatosplenic TCLs are usually of Vδ1 type, whereas cutaneous γδ TCLs are of Vδ2 type [12]. The γδ EATLs in this series were also distinct from the cutaneous γδ TCL in that the 4 informative γδ EATLs all had Vδ1 rearrangements, like normal intestinal γδ T cells. Also unlike our γδ EATL, nasal γδ TCLs have also been reported to be of Vδ2 type as well as 1 laryngeal and 1 EBV+ ileal case [23]. Two Vδ3+ gastrointestinal γδ TCLs, including 1 associated with CD, have also been reported [23].
In summary, these findings support that categorization of intestinal TCLs cannot be based on the type of TCR that they express and that at least many are very distinct from cutaneous γδ TCLs. Therefore, γδ-TCR+ EATL should not be placed in a category of “mucocutaneous γδ TCL.” Furthermore, TCR expression cannot be used to distinguish type I from type II EATL, even if many of the TCRγδ+ cases fall in the latter category. TCR αβ+ type II cases, which are common in some series, exist, and TCR γδ type I cases are well recognized, along with TCR silent cases [5,17,29,32]. The findings also support the concept that although type I and type II EATL have significant overlapping features, the type II cases have some very distinctive features and probably should be separated from classic EATL in the next revision of the WHO classification of hematopoietic and lymphoid tumors. From a practical point of view, however, this distinction may be difficult when only biopsies are obtained, given the heterogeneity seen in both types of EATL.
Acknowledgment
The late Kevin Salhany was instrumental in the detailed evaluation of the index case that led to this more complete study.
References
- [1].Jaffe ES, Harris NL, Stein H, et al. Introduction and overview of the classification of the lymphoid neoplasms. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. pp. 158–66. [Google Scholar]
- [2].Gaulard P, Berti E, Willemze R. Primary cutaneous peripheral T-cell lymphomas, rare subtypes. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. pp. 302–5. [Google Scholar]
- [3].Gaulard P, Jaffe ES, Krenacs L. Hepatosplenic T-cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. pp. 292–3. [Google Scholar]
- [4].Garcia-Herrera A, Song JY, Chuang SS, et al. Nonhepatosplenic gammadelta T-cell lymphomas represent a spectrum of aggressive cytotoxic T-cell Lymphomas with a mainly extranodal presentation. Am J Surg Pathol. 2011;35:1214–25. doi: 10.1097/PAS.0b013e31822067d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chan JK, Chan AC, Cheuk W, et al. Type II enteropathy-associated T-cell lymphoma: a distinct aggressive lymphoma with frequent gammadelta T-cell receptor expression. Am J Surg Pathol. 2011;35:1557–69. doi: 10.1097/PAS.0b013e318222dfcd. [DOI] [PubMed] [Google Scholar]
- [6].Isaacson PG, Chott A, Ott G. Enteropathy-associated T-cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. pp. 289–91. [Google Scholar]
- [7].Müller-Hermelink HK, Delabie J, Ko YH, Jaffe ES, Nakamura S. T-cell lymphoma of the small intestine. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. 4th ed IARC; Lyon: 2010. pp. 112–4. [Google Scholar]
- [8].Miyazaki K, Yamaguchi M, Imai H, et al. Gene expression profiling of peripheral T-cell lymphoma including gamma-delta T-cell lymphoma. Blood. 2009;113:1071–4. doi: 10.1182/blood-2008-07-166363. [DOI] [PubMed] [Google Scholar]
- [9].Pongpruttipan T, Sukpanichnant S, Assanasen T, et al. Extranodal NK/T-cell lymphoma, nasal type, includes cases of natural killer cell and αβ, γδ, and αβ/γδ T-cell origin: a comprehensive clinicopathologic and phenotypic study. Am J Surg Pathol. 2012;36:481–99. doi: 10.1097/PAS.0b013e31824433d8. [DOI] [PubMed] [Google Scholar]
- [10].Roullet M, Gheith SM, Mauger J, Junkins-Hopkins JM, Choi JK. Percentage of γδ T cells in panniculitis by paraffin immunohistochemical analysis. Am J Clin Pathol. 2009;131:820–6. doi: 10.1309/AJCPMG37MXKYPUBE. [DOI] [PubMed] [Google Scholar]
- [11].Lyons-Weiler M, Hagenkord J, Sciulli C, Dhir R, Monzon FA. Optimization of the Affymetrix GeneChip Mapping 10K 2.0 Assay for routine clinical use on formalin-fixed paraffin-embedded tissues. Diagn Mol Pathol. 2008;17:3–13. doi: 10.1097/PDM.0b013e31815aca30. [DOI] [PubMed] [Google Scholar]
- [12].Przybylski GK, Wu H, Macon WR, et al. Hepatosplenic and subcutaneous panniculitis-like gamma/delta T cell lymphomas are derived from different Vdelta subsets of gamma/delta T lymphocytes. J Mol Diagn. 2000;2:11–9. doi: 10.1016/s1525-1578(10)60610-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Isaacson P, Wright DH. Malignant histiocytosis of the intestine. Its relationship to malabsorption and ulcerative jejunitis. Hum Pathol. 1978;9:661–77. doi: 10.1016/s0046-8177(78)80049-5. [DOI] [PubMed] [Google Scholar]
- [14].Isaacson P, Wright DH. Intestinal lymphoma associated with malabsorption. Lancet. 1978;1:67–70. doi: 10.1016/s0140-6736(78)90004-1. [DOI] [PubMed] [Google Scholar]
- [15].Deleeuw RJ, Zettl A, Klinker E, et al. Whole-genome analysis and HLA genotyping of enteropathy-type T-cell lymphoma reveals 2 distinct lymphoma subtypes. Gastroenterology. 2007;132:1902–11. doi: 10.1053/j.gastro.2007.03.036. [DOI] [PubMed] [Google Scholar]
- [16].Zettl A, deLeeuw R, Haralambieva E, Mueller-Hermelink HK. Enteropathy-type T-cell lymphoma. Am J Clin Pathol. 2007;127:701–6. doi: 10.1309/nw2bk1dxb0eqg55h. [DOI] [PubMed] [Google Scholar]
- [17].Chott A, Haedicke W, Mosberger I, et al. Most CD56+ intestinal lymphomas are CD8+CD5-T-cell lymphomas of monomorphic small to medium size histology. Am J Pathol. 1998;153:1483–90. doi: 10.1016/S0002-9440(10)65736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Baumgartner AK, Zettl A, Chott A, et al. High frequency of genetic aberrations in enteropathy-type T-cell lymphoma. Lab Invest. 2003;83:1509–16. doi: 10.1097/01.lab.0000090157.13040.58. [DOI] [PubMed] [Google Scholar]
- [19].Wertheim GRM, Jones D, et al. Sensitive and specific detection of gamma T-cell receptor in paraffin-embedded T-cell lymphomas. Mod Pathol. 2010;23:328A. [Google Scholar]
- [20].Joachims ML, Chain JL, Hooker SW, Knott-Craig CJ, Thompson LF. Human alpha beta and gamma delta thymocyte development: TCR gene rearrangements, intracellular TCR beta expression, and gamma delta developmental potential—differences between men and mice. J Immunol. 2006;176:1543–52. doi: 10.4049/jimmunol.176.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Delabie J, Holte H, Vose JM, et al. Enteropathy-associated T-cell lymphoma: clinical and histological findings from the International Peripheral T-Cell Lymphoma Project. Blood. 2011;118:148–55. doi: 10.1182/blood-2011-02-335216. [DOI] [PubMed] [Google Scholar]
- [22].Oberhuber G, Vogelsang H, Stolte M, et al. Evidence that intestinal intraepithelial lymphocytes are activated cytotoxic T cells in celiac disease but not in giardiasis. Am J Pathol. 1996;148:1351–7. [PMC free article] [PubMed] [Google Scholar]
- [23].Arnulf B, Copie-Bergman C, Delfau-Larue MH, et al. Nonhepatosplenic gammadelta T-cell lymphoma: a subset of cytotoxic lymphomas with mucosal or skin localization. Blood. 1998;91:1723–31. [PubMed] [Google Scholar]
- [24].de Bruin PC, Kummer JA, van der Valk P, et al. Granzyme B–expressing peripheral T-cell lymphomas: neoplastic equivalents of activated cytotoxic T cells with preference for mucosa-associated lymphoid tissue localization. Blood. 1994;84:3785–91. [PubMed] [Google Scholar]
- [25].Katoh A, Ohshima K, Kanda M, et al. Gastrointestinal T cell lymphoma: predominant cytotoxic phenotypes, including alpha/beta, gamma/delta T cell and natural killer cells. Leuk Lymphoma. 2000;39:97–111. doi: 10.3109/10428190009053543. [DOI] [PubMed] [Google Scholar]
- [26].Chiba H, Takimoto R, Sato Y, et al. Gamma/delta T-cell lymphoma of duodenal bulb: a case report. Gastrointest Endosc. 2003;58:616–20. [PubMed] [Google Scholar]
- [27].Tan SY, Ooi AS, Ang MK, et al. Nuclear expression of MATK is a novel marker of type II enteropathy-associated T-cell lymphoma. Leukemia. 2011;25:555–7. doi: 10.1038/leu.2010.295. [DOI] [PubMed] [Google Scholar]
- [28].Ko YH, Karnan S, Kim KM, et al. Enteropathy-associated T-cell lymphoma—a clinicopathologic and array comparative genomic hybridization study. Hum Pathol. 2010;41:1231–7. doi: 10.1016/j.humpath.2009.11.020. [DOI] [PubMed] [Google Scholar]
- [29].Zettl A, Ott G, Makulik A, et al. Chromosomal gains at 9q characterize enteropathy-type T-cell lymphoma. Am J Pathol. 2002;161:1635–45. doi: 10.1016/S0002-9440(10)64441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Soulier J, Pierron G, Vecchione D, et al. A complex pattern of recurrent chromosomal losses and gains in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer. 2001;31:248–54. doi: 10.1002/gcc.1141. [DOI] [PubMed] [Google Scholar]
- [31].Malamut G, Afchain P, Verkarre V, et al. Presentation and long-term follow-up of refractory celiac disease: comparison of type I with type II. Gastroenterology. 2009;136:81–90. doi: 10.1053/j.gastro.2008.09.069. [DOI] [PubMed] [Google Scholar]
- [32].Takeshita M, Nakamura S, Kikuma K, et al. Pathological and immunohistological findings and genetic aberrations of intestinal enteropathy-associated T cell lymphoma in Japan. Histopathology. 2011;58:395–407. doi: 10.1111/j.1365-2559.2011.03768.x. [DOI] [PubMed] [Google Scholar]