Abstract
Both Metnase and Artemis possess endonuclease activities that trim 3′ overhangs of duplex DNA. To assess the potential of these enzymes for facilitating resolution of damaged ends during double-strand break rejoining, substrates bearing a variety of normal and structurally modified 3′ overhangs were constructed, and treated either with Metnase or with Artemis plus DNA-dependent protein kinase (DNA-PK). Unlike Artemis, which trims long overhangs to 4–5 bases, cleavage by Metnase was more evenly distributed over the length of the overhang, but with significant sequence dependence. In many substrates, Metnase also induced marked cleavage in the double-stranded region within a few bases of the overhang. Like Artemis, Metnase efficiently trimmed overhangs terminated in 3′-phosphoglycolates (PGs), and in some cases the presence of 3′-PG stimulated cleavage and altered its specificity. The nonplanar base thymine glycol in a 3′ overhang severely inhibited cleavage by Metnase in the vicinity of the modified base, while Artemis was less affected. Nevertheless, thymine glycol moieties could be removed by Metnase- or Artemis-mediated cleavage at sites farther from the terminus than the lesion itself. In in vitro end-joining systems based on human cell extracts, addition of Artemis, but not Metnase, effected robust trimming of an unligatable 3′-PG overhang, resulting in a dramatic stimulation of ligase IV- and XLF-dependent end joining. Thus, while both Metnase and Artemis are biochemically capable of resolving a variety of damaged DNA ends for the repair of complex double-strand breaks, Artemis appears to act more efficiently in the context of other nonhomologous end joining proteins.
1. Introduction
Artemis and Metnase are multifunctional proteins that have endonuclease activities specific for DNA ends [1,2]. In addition, Artemis is implicated in cell cycle checkpoint signaling [3], while Metnase has a histone methyltransferase function [4]. Nevertheless, there is evidence that the endonuclease activities of both enzymes are important for nonhomologous end joining (NHEJ) [5,6]. Moreover, DNA-dependent kinase catalytic subunit (DNA-PKcs), a core NHEJ protein, is required for Artemis nuclease activity [1], while Metnase interacts with DNA ligase IV, another essential NHEJ protein [7]. The basis of the apparent requirement for the endonucleolytic activity of these enzyme in end joining has never been rigorously determined, but one possible function of a DNA end-specific nuclease would be to trim DNA ends that have blocked termini and/or proximal base damage. Artemis, for example, efficiently trims overhangs bearing 3′-phosphoglycolate (PG) termini, yielding clean albeit slightly resected substrates bearing canonical 3′-hydroxyl termini, that could be readily patched and ligated to complete their repair by NHEJ [8].
DNA ends with oxidative base damage near the termini might also require similar trimming. Theoretical calculations of the track structure of γ-rays in aqueous solution predict that clusters of oxidative lesions in DNA, including DSBs accompanied by one or more nearby damaged bases, will be quite common [9,10], and there are extensive empirical data that support this view [11,12]. Two of the most common oxidative base damages are 8-oxoguanine (8-oxoG) and thymine glycol (Tg) [13,14]. While 8-oxoG can pair normally with cytosine, the pairing is less stable than a canonical G•C base pair, and the 8-oxoG can rotate around the glycosylic bond into a syn conformation and pair with adenine, making 8-oxoG highly mutagenic [15]. Tg results from oxidation of the 5–6 double bond of thymine, destroying its planarity and thereby introducing a severe distortion in DNA structure that can block rejoining when present near a DNA end [16,17]. In mammalian cells, 8-oxoG and Tg are removed primarily by 8-oxoguanine DNA glycosylase (OGG) and endonuclease III. Thus, base damages near DNA ends, even if not sequestered by DSB repair proteins, are likely to be poor substrates for these glycosylases, and their removal may instead require DSB-specific mechanisms such as end trimming.
To further assess the potential of Artemis and Metnase in resolving damaged DNA ends, their activities toward a variety of structurally modified DSB substrates were examined. The results show that while the specificities of the two enzymes differ in detail, they both have the potential to resolve both simple and complex DSB ends to produce substrates more suitable for further NHEJ processing.
2. Methods
2.1. Recombinant proteins
Metnase was purified from human 293 cells transfected with a pFLAG-CMV-2-based vector, as described [2]. Artemis and XRCC4 / DNA ligase IV (X4L4) complex were prepared from baculovirus-infected insect cells, as described [8,21]. X4L4 concentrations are expressed as moles of a presumed 2:2 heterotetramer of molecular weight 300,000 [22]. XLF was produced in E. coli and purified as described [23]. DNA-dependent protein kinase was prepared either from HeLa cells [24] (SMY laboratory) or from human placenta [25] (DAR laboratory). All other enzymes were from New England Biolabs.
2.2 Substrates
Oligomers bearing 3′-PG termini were prepared by treating appropriate longer oligomers with bleomycin, as described [8,26]. Preparation of internally labeled plasmid substrates, by ligation of these and other 5′-end-labeled oligomers into the 3′-resected ends of “trimmed” plasmid pRZ56, has been described [27,28]. The substrate used for end joining in cell extracts was constructed by sequentially ligating the unlabeled phosphorylated 13-mer CCGGACGCGTTT and the 5′-labeled 3′-PG-terminated 17-mer CGAGGAACGCGAAAACG to opposite ends of pRZ56. For preparation of substrates containing 8-oxoguanine (8-oxoG) the procedure was modified to minimize oxidation and degradation of the 8-oxoG residue during the purification process, as follows. The 14-mer CGAGGAACGC(8-oxoG)ACG was obtained from the Midland Certified Reagent Company, Inc., dissolved in 200 μl of 10 mM HEPES-NaOH / 1 mM EDTA pH 8 at a final oligomer concentration of 1.2 mM, and stored at −80°C. Twenty pmoles of the oligomer was labeled with [γ-32P]ATP (5 μl of 0.15 mCi/μl, 6000 Ci/mmole, Perkin-Elmer) using T4 PNK in a 12.5 μl reaction for 1 hr at 37°C. The reaction was extracted with 12.5 μl of phenol/chloroform. To 4 pmoles of the labeled 8-oxoG oligomer, 5 μg of trimmed pRZ56 plasmid was added followed by ethanol precipitation and brief vacuum drying. The DNA was dissolved in 70 μl of ligase buffer [50 mM Tris-HCl / 10 mM MgCl2 / 1 mM ATP / 10 mM dithiothreitol, pH 7.5] and treated with 500 units T4 ligase for 16 hr at 16°C, then ethanol- precipitated in the presence of 2 M ammonium acetate, leaving most of the unligated oligomer in the supernatant. The ligated DNA substrate was then purified on Qiaquick spin columns using the PCR purification protocol (Qiagen), eluted in 30 – 50 μl of elution buffer and stored at −80°C. Typically 0.5 μg – 1.2 μg was recovered.
To generate substrates containing thymine glycol (Tg), the 23-mer ACCGCGAGACTCACGCTCA-Tg-ACG was obtained from Midland Certified Reagents and dissolved in 10 mM Tris-HCl pH 8 / 0.1 mM EDTA. Fifty pmoles of the Tg oligomer or a corresponding normal oligomer was 5′-end-labeled as above, and the kinase inactivated at 65°C for 10 min. Each oligomer was then annealed to an equal quantity of a complementary 5′-phosphate oligomer (Supplementary Figure 1), either ATGAGCGTGAGTCTCG (to generate a 3-base 3′ overhang) or GCGTGAGTCTCG (to generate a 7-base overhang), by first heating to 80°C followed by slow cooling to 16°C. The duplexes were extracted with phenol/chloroform, and ~10 pmoles were ethanol-precipitated in the presence of 4 μg of the purified 2.3-kb BsaI/ScaI fragment of pUC19, and then the oligomeric duplex was ligated to the plasmid (which bears a nonpalindromic CGGT- 5′ overhang), as described above for the 8-oxoG substrate. The ligation product was purified on an agarose gel and electroeluted as described [27].
2.3. Enzyme reactions
Artemis reactions, usually 10 μl, contained 25 mM Tris-HCl pH 8, 25 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.25 mM ATP and 50 μg/ml BSA, and were prepared at 22°C. Substrate (10 ng) was added and the sample mixed. Ku (25 nM), DNA-PKcs (65 nM) and Artemis (90 nM) were then added in rapid succession and the sample mixed by pipetting. Following incubation at 37°C for various times, the reactions were stopped by addition of 70 μl of digestion mix (0.3 M NaCl, 0.5 mg/ml proteinase K, 10 mM Tris pH 8, 1% SDS). Following incubation for 3 hr at 55°C, samples were extracted with phenol and then with chloroform and ethanol-precipitated. For pRZ56-based substrates, DNA was redissolved in 50 μl and cut with 40 units TaqI for 3 hr at 65°C. For pUC19-based substrates, DNA was redissolved in 50 μl and treated with 4 units BsrDI for 3 hr at 65°C. Samples were denatured and subjected to electrophoresis on 20% polyacrylamide sequencing gels. Phosphorimaging screens were exposed to the frozen gels for 2 days and then analyzed using a Typhoon 9100 imager (GE Healthcare) and ImageQuant 3.1 or 5.1 software.
Metnase reactions contained 25 mM Tris-HCl pH 8, 25 mM KCl, 10 mM magnesium acetate, 5 mM dithiothreitol, 0.25 mM ATP and 50 μg/ml BSA, and were similarly prepared, incubated and processed. For S1 nuclease reactions, the same buffer without ATP was used. Samples were preincubated at 37°C for 15 min with end joining proteins as indicated. S1 nuclease (0.3 U in a 10-μl reaction) was then added and the samples incubated at 25°C for 5 min. The reaction was stopped by adding 20 μl of 10mM EDTA, 0.45M NaOAc and 100 μg/ml tRNA, followed immediately by phenol/chloroform extraction and ethanol precipitation.
2.4 End joining in cell extracts
Reactions with whole-cell extracts contained 50 mM triethanolammonium acetate pH 7.5, 1 mM ATP, 1 mM dithiothreitol, 50 μg/ml BSA, 1.3 mM Mg(OAc)2, and dNTPs at 100 μM each. Typically, a 16-μl reaction contained 10 μl of extract, resulting in a final concentration of 8 mg/ml protein, 66 mM potassium acetate and 16% glycerol, and an effective Mg++ concentration of 1 mM (taking into account ~0.3 mM EDTA from the extract). Buffer components were first mixed with cell extract on ice. In some cases 100 nM recombinant XLF, 90 nM Artemis and/or 100 nM Metnase was added, as indicated. Finally, substrate (10–100 ng) was added and the reaction again mixed by pipetting and incubated at 37°C for 6 hr. Samples were deproteinized and analyzed as above except that DNA was cut with 20 units BstXI for 3 hr at 37°C before addition of TaqI.
Reactions with HeLa nuclear extracts contained 3.2 mg/ml extract (Promega in vitro transcription grade, 5 μl extract in a 16 μl reaction), 50 mM triethanolammonium acetate pH 7.5, 1 mM Mg(OAc)2, 40 mM KOAc, 0.5 mM dithiothreitol, 1 mM ATP, 50 μM of each dNTP, 50 μg/ml BSA, and 10 ng DNA substrate [29]. In some cases 100 nM recombinant X4L4 tetramer, 90 nM Artemis and/or 100 nM Metnase was added, as indicated. Following substrate addition, samples were incubated at 37°C for 6 hr, then deproteinized and analyzed as for whole-cell extracts.
3. Results
3.1. Metnase endonucleolytically trims 3′ overhangs of a duplex DNA substrate
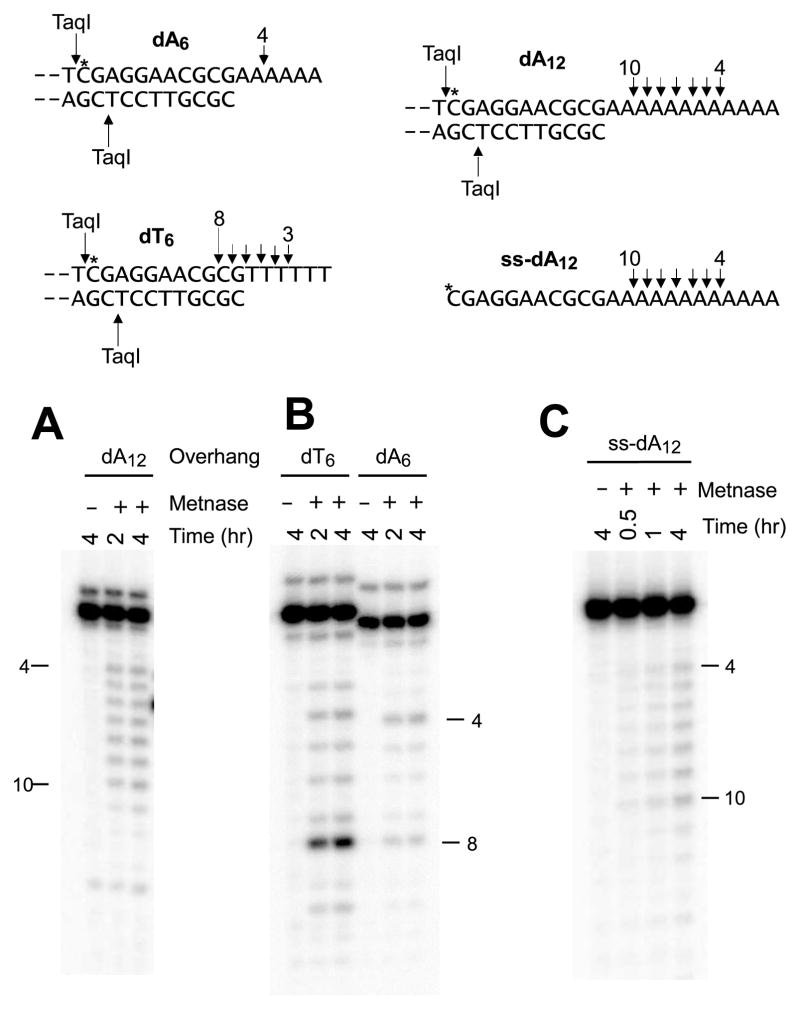
Studies with oligonucleotide substrates indicate that Metnase has ssDNA cleavage activity that can trim 3′ overhangs [2]. To investigate the potential of this activity for resolving DSB ends, longer plasmid-length substrates with 3′ overhangs of various length and structure were constructed by ligating 5′-labeled oligomers to a 3′-resected plasmid. Each substrate was treated with Metnase, and then the labeled oligomer was released from the 3′ end of the plasmid by TaqI digestion and analyzed on sequencing gels to assess cleavage.
First, to determine the positional specificity of cleavage while minimizing the influence of sequence preferences, trimming of 6- and 12-base oligo(dA) overhangs (designated dA6 and dA12) was examined. Trimming of dA12 by Metnase was spread evenly over most of the length of the overhang, excluding the 3 bases nearest the terminus and the 2 bases nearest the single-strand / double-strand (ss/ds) junction (Fig. 1A). A similar product distribution was observed when the unligated 23-mer used to construct the plasmid-length duplex substrate was treated with Metnase, but the intensity of cleavage products was diminished (Fig. 1C), suggesting that a partial duplex structure stimulates but is not essential for Metnase cleavage activity. Trimmed fragments accumulated over time without any detectable change in the distribution of products, suggesting endo- rather than exonucleolytic cleavage.
Figure 1.
Trimming of 3′ overhangs by Metnase. Internally labeled substrates bearing homopolymer dA6, dA12, and dT6 overhangs were constructed and treated with 50 nM Metnase for the indicated times, then cut with TaqI and the products analyzed on sequencing gels (A, B). Similar experiments were performed with a simple 23-base oligomer, ss-dA12 (C). Labeled bands and arrows above sequences show the major cleavage sites and the corresponding number of bases removed. Total cleavage of ss-dA12 was 7.1% at 4 hr, vs. 15.9% for the dA12 overhang.
For the dA6 overhang, the predominant cleavage site was 4 bases from the terminus, resulting in a 2-base overhang (Fig. 1B). For a dT6 overhang, only 2 bases at the terminus were excluded from cleavage, and significant cleavage was detected at all positions up to and including the ss/ds junction. In addition, very strong cleavage was induced within the duplex region, 2 bases from the ss/ds junction, and there was some cleavage at this site in the dA6 substrate as well (Fig. 1B). Thus, overall, Metnase trims 3′ overhangs in a sequence-dependent manner, over the length of the ss and nearby ds regions, excluding a few bases at the terminus.
To assess possible NHEJ-specific interactions, trimming of the dA12 substrate was also examined in the presence of Ku, either with or without DNA-PKcs. Ku reduced cleavage over the length of the overhang by about ~15% (quantitation not shown), while inhibition by Ku plus DNA-PKcs was ~25% (Fig. 2). To determine whether this cleavage reflected specific interactions between Metnase and DNA-PK to allow cleavage of DNA that was otherwise sequestered, cleavage by nuclease S1, which presumably has no DNA-PK-specific interactions, was examined. Cleavage of the overhang by S1 was also only moderately inhibited (~40%) by DNA-PK, even in the absence of ATP, a condition that should maximize persistent sequestration of the DNA end. Overall, these data show no evidence that Metnase is recruited by or has any specific interactions with Ku and DNA-PKcs.
Figure 2.
Effect of Ku and DNA-PKcs on trimming of the dA12 overhang by Metnase or S1 nuclease. The internally labeled dA12 substrate shown in Fig. 1 was treated with 50 nM Metnase for 2 hr or with S1 nuclease for 15 min in the presence or absence of 25 nM Ku and 65 nM DNA-PKcs, then cut with TaqI prior to analysis on sequencing gels. ATP at 0.25 mM was present in Metnase- but not S1-treated samples. ND = not determined due to low cleavage level or inadequate resolution from the full-length band.
3.2. Metnase trims 3′-PG-terminated overhangs with altered efficiency and specificity
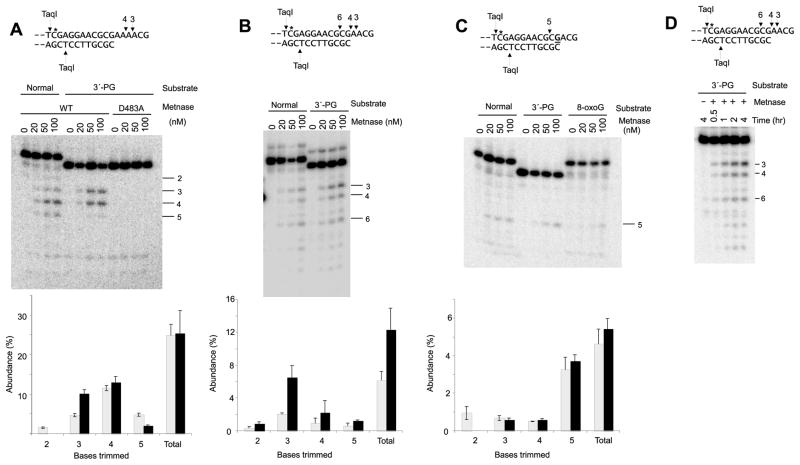
Previous work showed that Artemis plus DNA-PK efficiently trims 3′-PG-terminated overhangs ≥3 bases in length, leaving 2- to 5-base 3′-hydroxyl-terminated overhangs [8]. Although Metnase also trims ss regions of partial duplexes, it appears to require a free end for entry [6] and thus might be blocked by terminal modifications. However, Metnase efficiently trimmed PG-terminated 4- and 6-base overhangs, and in both cases the predominant cleavage sites were 3 and 4 bases from the terminus (Fig. 3A–B). Total cleavage was about twice as great for the 6-base as for the 4-base overhang, and there was no cleavage by an identically prepared endonuclease-deficient mutant Metnase [2] harboring a D483A substitution in the transposase domain (Fig. 3A). Comparison with the corresponding 3′-hydroxyl substrates showed that in both cases the presence of 3′-PG resulted in enhanced cleavage 3 bases from the terminus, shifting the cleavage pattern slightly toward the PG terminus (Fig. 3A–B). For the 4-base overhang this increase was quite dramatic and cleavage at other sites was increased as well. Again, the extent of cleavage increased with time but the cleavage pattern did not change, suggesting that cleavage was endonucleolytic (Fig. 3D).
Figure 3.
Trimming of 3′-PG-terminated 3′ overhangs by Metnase. Internally labeled (*) plasmid substrates bearing 3′-PG or normal 3′-hydroxyl termini on 6-base (A), 4-base (B, D) or 3-base (C) 3′ overhangs were treated with the indicated concentrations of Metnase for 2 hr, then cut with TaqI to release labeled oligomers which were analyzed on sequencing gels. Graphs in bottom panels show quantification of cleavage by 100 nM Metnase in three independent experiments (mean ± SEM); grey and black bars indicate 3′-hydroxyl and 3′-PG substrates, respectively. Labeled bands and arrows above sequences show the major cleavage sites and the corresponding number of bases removed. In (A), some samples were treated with an identically prepared nuclease-deficient D483A mutant Metnase. In (C), some samples contained a substrate with an 8-oxoG base substituted at the underlined bolded G; cleavage of this substrate was 2.8 ± 0.8% for 5-base trimming and 5.4 ± 1.4% total cleavage, similar to the PG- and hydroxyl-terminated substrates. In (D), the 4-base overhang substrates were treated for various times with 100 nM Metnase. Metnase treatment of the analogous 1-base overhang substrates yielded only weak cleavage (<1% at any single site, Supplemental Fig. 2).
A PG-terminated 3-base overhang was trimmed much less efficiently than the 4- and 6-base overhangs, with most of the cleavage occurring 5 bases from the terminus, within the duplex region of the substrate (Fig. 3C). Metnase had even less activity toward a 1-base overhang, with very weak cleavage detected at 4–5 bases from the terminus (<1% cleavage at each site in both substrates, Supplemental Fig. 2). For these shorter substrates, the PG terminus had little or no effect on the efficiency or specificity of cleavage. There was no apparent trimming of a 3′-PG-terminated blunt end by Metnase (Supplemental Fig. 3). X4L4, which has been reported to interact with Metnase [7], had no apparent effect on Metnase-mediated trimming of the PG-terminated 3-base overhang (Supplemental Fig. 4). Ku also had no effect, while DNA-PKcs (in the absence of ATP) had an inhibitory effect. Overall, the activities of Metnase toward these substrates suggest that it could play a role in resolving PG termini for DSB repair, particularly on overhangs >3 bases. However, there is again no evidence that other NHEJ components recruit Metnase to DNA ends or stimulate its activity.
3.3 Thymine glycol inhibits cleavage by Artemis and Metnase at adjacent sites
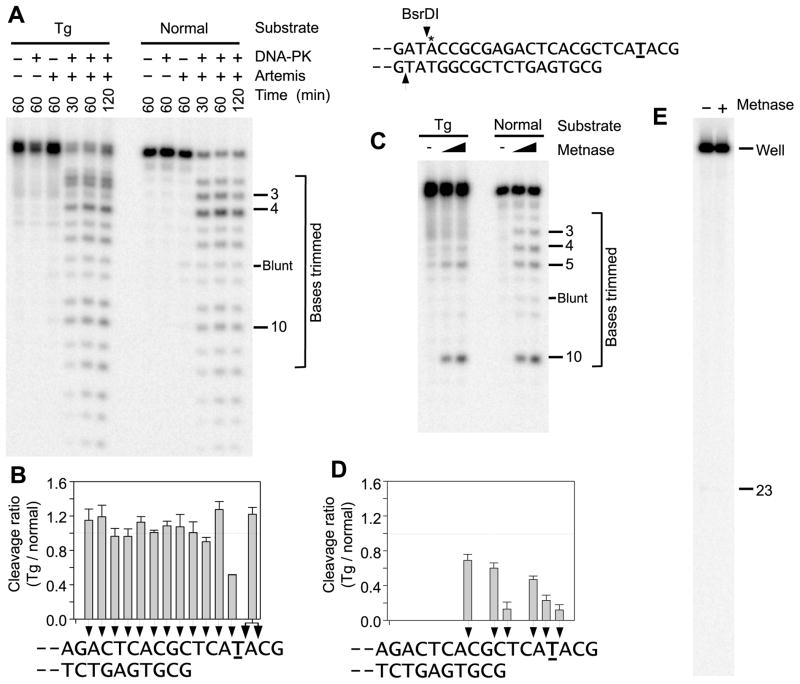
Many ionizing radiation-induced DSBs are expected to be accompanied by oxidative base damage near the DSB ends [30,31]. To assess the possible roles of Artemis and Metnase in resolving such complex DSBs, internally labeled DSB substrates were constructed containing site-specific oxidatively modified residues in various contexts. One of the most frequent oxidized bases, thymine glycol (Tg), is nonplanar and highly disruptive of DNA structure. To generate 3′ overhangs with Tg near the terminus, a labeled Tg-containing oligomer was annealed to a complement and ligated to BsaI-cut pUC19. Following Artemis or Metnase treatment, the 23-base fragment (or its cleavage products) was released by treatment with BsrDI. When placed as the central base in a 7-base 3′ overhang, Tg inhibited cleavage by both Artemis and Metnase at sites near the modified base, but the effect was greater and more widespread for Metnase (Fig. 4). In the corresponding unmodified substrate, Artemis induced prominent cleavage 4 bases from the terminus, leaving a 3-base overhang (Fig. 4A). Cleavage at this site was inhibited about twofold by the presence of Tg, but cleavage at interior sites, which became more prominent with longer incubations, was not inhibited by the presence of the modified base (Fig. 4B). Cleavage at sites near the terminus was difficult to quantitate due to apparent spontaneous breakdown of a portion of the Tg substrate and its tendency to run as a doublet (presumably reflecting glycol isomers). However, the sum of cleavage at the sites 2 and 3 bases from the terminus was nearly equal for the modified and unmodified substrates. Thus, Artemis-mediated trimming can release Tg when it is formed in 3′ overhangs, potentially facilitating subsequent gap filling and ligation.
Figure 4.
Trimming of a Tg-containing 3′ overhang by Artemis plus DNA-PK or by Metnase. The internally labeled (*) substrate (top right) consisted of an oligomeric partial duplex ligated to a 2.3-kb fragment of pUC19. Bolded underlined T indicates the site of Tg substitution. A. Trimming by Artemis. Samples were treated for the indicated times with 90 nM Artemis in the presence of 25 nM Ku and 65 nM DNA-PKcs, then cut with BsrDI and analyzed on a sequencing gel. B. Quantitation of Artemis-mediated cleavage, plotted as the ratio of cleavage of the Tg-containing substrate to that of the normal substrate, at each position. Dotted line indicates equal cleavage in normal and Tg substrates. Error bars represent mean ± SEM for the three time points shown in (B.); a replicate experiment yielded similar results. C. Trimming by Metnase. The same substrate was treated with 100 or 200 nM Metnase for 4 hr, cut with BsrDI, and similarly analyzed. D. Quantitation of Metnase-mediated cleavage. Error bars show the mean ± SEM for two Metnase concentrations from each of two experiments. E. Localization of Metnase-mediated cleavage to DNA ends. The normal substrate was treated with 200 nM Metnase for 4 hr and run on a sequencing gel without BsrDI cleavage. Lack of detectable fragmentation indicates that there was essentially no Metnase-mediated cleavage in ds regions of the plasmid beyond the BsrDI site.
Metnase produced cleavage throughout the unmodified 7-base overhang, with the strongest cleavage near the middle of the overhang (Fig. 4C). Presence of Tg at the center of the overhang appeared to completely block cleavage at all sites between Tg and the terminus, and cleavage at sites between Tg and the duplex was partially blocked. In addition, there was a hotspot of cleavage just inside the duplex region, 3 bases from the ss/ds junction, and cleavage at this site was inhibited only slightly by the presence of Tg in the overhang (Fig. 4D). Thus, in principle Metnase, like Artemis, could release Tg from this complex DSB and thereby facilitate end joining. When BsrDI cleavage was omitted, essentially all of the labeled DNA remained in the well of the sequencing gel, implying that Metnase-mediated cleavage was restricted to the region near the DNA end (Fig. 4E).
Given its potential to disrupt DNA structure, it was postulated that Tg at or near the end of a DNA duplex might promote single-strandedness and thereby stimulate Artemis- and Metnase-mediated trimming of very short overhangs, which are otherwise poor substrates for both enzymes. On the contrary however, substitution of Tg for thymine as the last base preceding a 3-base 3′ overhang further inhibited cleavage of these substrates within the ds region by Artemis, by about 4- to 5-fold at most sites (Fig. 5A, quantitation not shown). For Metnase, cleavage of the unmodified substrate in the ds region was barely above background, so that the effect of Tg was difficult to quantitate, but clearly Tg did not significantly enhance cleavage by promoting single-strandedness (Fig. 5B).
Figure 5.
Effect of Tg and 8-oxoG at the ss/ds junction on trimming of a 3-base 3′ overhang by Artemis or Metnase. A. Normal and Tg-substituted substrates shown were treated with 90 nM Artemis in the presence of Ku and/or DNA-PK, then cut with BsrDI and analyzed on sequencing gels. B. The same substrates were treated with the indicated concentrations of Metnase for 4 hr, then cut with BsrDI. Underlined T shows the site of Tg substitution. C. Normal or 8-oxoG-susbstituted substrates were treated with Artemis plus Ku and DNA-PKcs as indicated, then deproteinized, cut with TaqI and analyzed on a sequencing gel. Quantitation of cleavage (Supplemental Figure 5) showed that the presence of 8-oxoG increased 2-base trimming by ~2-fold. Trimming at other sites was not affected. For trimming of this substrate by Metnase, see Fig. 3C.
3.4. Presence of 8-oxoG stimulates trimming by Artemis but not Metnase
When positioned as the last paired base preceding a 3-base 3′ overhang, 8-oxoG had no effect on Metnase-mediated trimming (Fig. 3C), but it increased Artemis-mediated cleavage 2 bases from the terminus by approximately twofold (Fig. 5C and Supplemental Fig. 5). However, such trimming does not remove the damaged base, and trimming at interior sites that would remove the 8-oxoG was not stimulated by its presence.
3.5 Artemis trims damaged 3′ overhangs and promotes end joining in in vitro NHEJ models
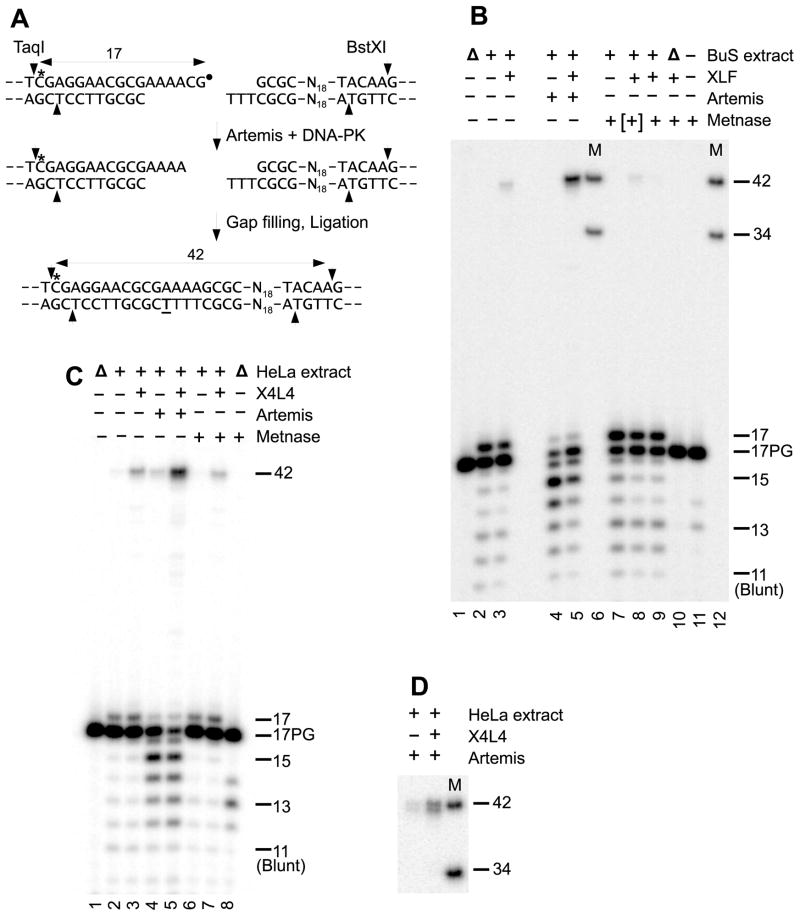
The experiments described above show that both Artemis (plus DNA-PK) and Metnase are capable of trimming either damaged or undamaged 3′ overhangs, an activity that can potentially facilitate end joining. Moreover, previous work indicated that Metnase can enhance joining of incompatible ends in cell extracts, presumably by end trimming [6]. To assess whether the end trimming activities of Artemis and Metnase can trim damaged ends in the context of classical NHEJ, each enzyme was added to an in vitro end joining assay based on whole-cell extracts of BuS cells, which harbor a homozygous nonsense mutation in the XLF gene at base 178 [32,33]. Previous work showed that in these extracts, end joining of substrates requiring gap filling is completely dependent on addition of exogenous recombinant XLF [23], implying that this end joining is attributable exclusively to classical NHEJ. The substrate chosen for the assay (Fig. 6A) is a plasmid terminated at one end with a 3-base -TTT 3′ overhang. The other end is internally labeled and has a 6-base PG-terminated 3′ overhang which, upon trimming by Artemis or Metnase, would yield an -AA, -AAA or -AAAA overhang suitable for annealing with the -TTT overhang, followed by gap filling by polymerase λ or μ, and ligation by X4L4. Following incubation in the extract, processing and end joining was assessed by cutting the labeled DNA with BstXI and TaqI prior to analysis on sequencing gels.
Figure 6.
Effect of Artemis and Metnase addition on end joining in cell extracts. A. Internally labeled substrate showing predicted trimming by Artemis and subsequent gap filling (underlined T) and ligation in extracts. Cleavage of the resulting product by BstXI and TaqI should yield a labeled 42-base fragment. B. End joining in whole-cell extracts of XLF-deficient BuS fibroblasts (8 mg/ml). Extracts were supplemented with 100 nM XLF, 90nM Artemis and/or 100 nM Metnase, as indicated. Lanes M contain a mixture of a labeled 42-base oligomer having the same sequence shown in (A), and a 34-base size marker. Δ = heat-inactivated extract. C. End joining in nuclear extracts of HeLa cells (3.2 mg/ml protein), supplemented with 100 nM X4L4 complex, 90 nM Artemis and/or 50 nM ([+]) or 100 nM (+) Metnase. D. Resolution of the Artemis-dependent end joining product into a doublet. Conditions are the same as in (C), but electrophoresis time was increased and the 42- and 34-base markers (M) were included.
In unsupplemented extracts, there was some conversion of PG to hydroxyl ends, presumably by TDP1 and PNKP, and a small degree of apparent exonucleolytic 3′→5′ resection, but no detectable end joining of the PG-terminated substrate (Fig 6B, lane 2). Extracts supplemented with only XLF (lane 3) showed a trace of end joining, whereas addition of Artemis alone resulted in highly efficient end trimming to a 4-base -AAAA overhang, but no detectable repaired products (lane 4). Addition of XLF plus Artemis resulted in conversion of most of the trimmed ends to a single product that comigrated with a 42-base marker having the predicted -CGAAAACG- repair joint (lane 5). In 2 experiments, Artemis addition increased the yield of this product by 12.7- and 14.5-fold, to about a quarter of the total substrate. These results indicate that Artemis can efficiently trim DNA ends in the context of classical NHEJ, and thereby facilitate their end joining. Moreover, the trimmed substrates appear to be efficiently shunted to other NHEJ proteins for patching and ligation, as indicated by the conspicuous absence of the trimming into ds regions that was seen upon extended incubation of similar substrates with DNA-PK and Artemis alone (as in Fig. 4A). In contrast, addition of Metnase did not increase the yield of repaired products in these extracts (lanes 7–9), even though it appeared to slightly increase resection of the overhang. Instead, addition of the highest concentration of Metnase appeared to substantially inhibit end joining (lane 9). The basis of this inhibition is unclear, but it is unlikely to be due to excess trimming, as trimming was essentially the same for the two Metnase concentrations (compare lanes 8 and 9).
Similar experiments were performed using a second in vitro model of classical NHEJ, based on HeLa nuclear extracts (Fig. 6C). The endogenous level of X4L4 in these extracts is insufficient to support efficient end joining and must be supplemented with exogenous recombinant X4L4 [29], thus providing a distinction between classical and alternative end joining pathways. In the absence of exogenous X4L4, there was again some 3′-PG removal and 3′→5′ resection, although even less than in the whole-cell extracts (Fig. 6C, lane 2; compare with lane 2 of Fig. 6B). Although the majority of the PG ends remained unprocessed, there was, in the sample supplemented with X4L4 alone, a small but detectable quantity of a ~42-base end joining product (lane 3). Addition of Artemis to the extracts resulted in trimming of most of the labeled ends, predominantly to a 4-base overhang (15-mer), accompanied by a dramatic increase in the yield of X4L4-dependent ~42-base repair product (fold increase 8.3 ± 2.4, N=3) (lanes 4–5). Longer electrophoresis revealed that this product was actually a doublet, consisting of a larger fragment exactly comigrating with a 42-base marker of the expected sequence, and a second one-base shorter product (Fig. 6D). Sequencing of 27 cloned plasmids from a replicate sample yielded 6 clones with the expected -CGAAAACG- repair joint and 16 clones with a -CGAAACG- joint; the remaining 5 clones had terminal deletions of various lengths (Supplementary Fig. 6). These results confirm that the observed end joining product was formed by trimming of the overhang followed by patching and ligation. (Given that X4L4-mediated ligation is relatively tolerant of mismatches in DSB overhangs [34,35], the unexpected predominance of -CGAAACG- vs. -CGAAAACG- repair joints in cloned plasmids may be due to joining events wherein the bottom strand was ligated without gap filling while the trimmed -AAAA overhang and ss break in the top strand was left intact and subsequently resolved in E. coli. Such events would not be detected in the gel assay.) Overall, these results confirm that Artemis can interact with the NHEJ complex in such a manner that it gains access to the overhang, trims it, and thereby facilitates end joining.
In the absence of extract Metnase was able to effect significant trimming in the end joining buffer (lane 8), primarily to a 2-base overhang. However, in the presence of HeLa nuclear extract (lanes 6–7) this Metnase-mediated trimming appeared to be suppressed, and the yield of ligated repair product, rather than being enhanced, was slightly reduced from the level seen with extract plus X4L4 alone.
4. Discussion
Both Artemis and Metnase, and their nuclease activities in particular, have been implicated in NHEJ [5,6,36,37], but the precise function of nucleolytic cleavage in end joining has not been clearly defined for either enzyme. The machinery of classical NHEJ, incorporating DNA-PK, XLF, X4L4 and polymerases λ and μ, has a remarkable capacity to anneal, patch and ligate DNA ends having minimal and imperfect complementarity [35]. Nevertheless, blocked termini and adjacent DNA base damage can constitute a significant barrier to end joining [17,38–40], and although a variety of enzymes are available for resolving these terminal modifications, ends sequestered by end joining proteins may not be accessible to those enzymes [41]. Moreover, even if accessible, heavily damaged ends with multiple modifications may not be recognized as substrates by repair enzymes. Thus, the purpose of the present study was to assess whether the DNA end-trimming activities of Artemis and Metnase could provide a means of generating undamaged ends suitable for patching and ligation, bypassing and removing blocked termini as well as damaged bases at or near the termini.
Artemis endonuclease activity, which is dependent on association with autophosphorylated DNA-PK, is essential for the hairpin-opening step of V(D)J recombination [1]. Artemis is also required, along with ATM kinase and MRN, for repair of a subset of radiation-induced DSBs [36], and rescue of this repair function as well as restoration of radioresistance in Artemis-deficient cells requires its endonuclease activity [5]. Experiments with ATM-deficient cells and ATM inhibitors suggest that this subset of DSBs consists mostly of DSBs in heterochromatin, and that ATM is needed primarily to phosphorylate KAP-1, resulting in a local decondensation of chromatin that allows access to DSB ends by the NHEJ machinery [42,43]. Why Artemis endonuclease activity should be required for repair of these DSBs, but not the repair of structurally similar DSBs in euchromatin, is puzzling, but resolution of damaged ends by trimming off the damaged nucleotides remains the most plausible function for Artemis in this context.
Previous work showed that Artemis could efficiently trim PG-terminated 3′ overhangs [8], and could even trim PG-terminated blunt ends, albeit much more slowly [44]. The present work shows that Artemis can also trim 3′ overhangs containing oxidized bases such as Tg, whose nonplanar ring imparts a severe structural distortion to DNA (Fig. 4). Tg inhibits Artemis-mediated cleavage of the bond directly 5′ to the Tg residue, but cleavage at interior sites is not affected, so that Tg can be removed by Artemis to expose undamaged DNA for further NHEJ processing. Crystallographic structural studies of Tg as a template base for DNA replication have revealed a steric clash with the base immediately to the 5′ side that disrupts the normal conformation of that base [16]. A similar perturbation may underlie inhibition of Artemis-mediated cleavage between these two bases, although the conformation of DNA in complex with Artemis is not yet known.
Tg at the ss/ds junction, rather than stimulating Artemis by promoting melting of the ends, strongly inhibited Artemis-mediated 3′ trimming into the ds region (Fig. 5A). Previous work showed that this 3′ trimming was preceded and enabled by prior 5′ trimming that yielded a 3′ overhang [44], and it may be the required 5′ trimming that is directly blocked by the Tg. The Artemis protein used in these experiments does contain 5′→3′ exonuclease activity, recently reported to be a contaminant rather than intrinsic [45]. However, in the presence of DNA-PK, this activity appears to be completely suppressed at double-strand DNA ends and Artemis instead acts endonucleolytically at both 5′ and 3′ termini [44]. Thus, exonuclease activity should not affect the trimming results or their interpretation.
Metnase is implicated in NHEJ by its interaction with DNA ligase IV [7], and by experiments showing that it increases the efficiency of end joining of DSB substrates having noncohesive overhangs, both 3′ and 5′, when added to various whole-cell extracts [6]. In those experiments, the size of terminal deletions was positively correlated with cellular Metnase levels, suggesting that Metnase trimmed the ends prior to religation, and that trimming penetrated well into ds regions of the substrates. Metnase also has methyltransferase activity, which specifically phosphorylates histone H3 at Lys36 near a site-specific DSB, thereby promoting recruitment of Ku and NBS1, and facilitating end joining [4]. Thus, Metnase is likely to be present at or near DSB ends during NHEJ. In the present study, Metnase was found to efficiently trim 3′ overhangs >3 bases in length, a specificity very similar to that of Artemis. However, whereas Artemis shortens overhangs up to at least 25 bases in length to ~5 bases in a single step [8], Metnase-mediated cleavage was spread more evenly over the full length of the overhang, excluding 2–3 terminal bases (Fig. 1). For many substrates a cleavage hotspot was found in the ds region, just 2–3 bases from the ss/ds junction. Although Metnase was more strongly inhibited by the nonplanar base Tg than was Artemis, it nevertheless could trim off this modified base centered in a 7-base overhang (Fig. 4). Trimming at the hotspot just inside the ds region, in particular, was only slightly affected by the modified base.
Although Metnase appears to require a free DNA end for entry and (unlike Artemis) does not cleave hairpins or other ss loops [6], a 3′-PG terminus did not interfere with this requirement. Metnase efficiently trimmed PG-terminated 3′ overhangs, leaving ligatable 3′-hydroxyl ends, and indeed for some substrates, the PG terminus stimulated trimming (Fig. 3). Thus, overall, Metnase has appropriate enzymatic activities for resolving damaged DNA ends during NHEJ.
However, even if nucleases have biochemical activities capable of resolving damaged DSB ends, they may not necessarily act to do so in vivo, as DSB ends are likely to be sequestered by NHEJ proteins and access to them by other enzymes stringently controlled. In X4L4-supplemented HeLa nuclear extracts, for example, substrates with canonical termini and bearing partially complementary overhangs are efficiently patched and ligated [46], yet protruding 3′-PG termini persist for many hours (Fig. 6C), despite the presence of sufficient concentrations of TDP1 to efficiently remove them [40]. These findings suggest that only enzymes that interact appropriately with the NHEJ machinery will be allowed access to DNA ends. Addition of purified recombinant Artemis to cell extracts containing a DSB substrate with a PG-terminated 6-base 3′-overhang resulted in efficient trimming of almost all of the available substrate, dramatically enhancing end joining to a partially complementary end (Fig. 6). This end joining was dependent on X4L4 (in HeLa nuclear extracts) as well as XLF (in BuS whole-cell extracts), implying that it was carried out by classical, DNA-PK-based NHEJ. While Artemis is known to interact cooperatively with DNA-PK and other core end joining proteins in the context of V(D)J recombination, that process, involving hairpin ends and RAG proteins that may persist at the DSB site [47], is rather a special case. The present results clearly show that, in the presence of the full complement of core NHEJ proteins, Artemis can form a complex with DNA-PK, trim a damaged DSB overhang, and then efficiently handoff the substrate for subsequent patching and ligation, arresting any further trimming by Artemis into ds regions.
In the same experiments with either of the two in vitro models (Fig. 6), Metnase did not promote end joining, even with a substrate it is clearly capable of trimming. The simplest explanation of this result is that Metnase, unlike Artemis, cannot gain access to DNA termini bound by the full complement of NHEJ proteins. Thus, overall the data do not support a role for the endonuclease activity of Metnase in NHEJ. However, it is difficult to exclude the possibility that endonucleolytic trimming by Metnase in the context of NHEJ could require conditions that were not reproduced or cofactors that were not recovered in the extracts. Another recent study showed that Metnase can facilitate end joining in cell extracts, by trimming DNA ends bearing incompatible 5′ and 3′ overhangs (formed by restriction enzyme cleavage) [6]. A distinguishing feature of the two in vitro systems used in the present work (Fig. 6) is a very high ratio of cell protein to DNA substrate, i.e, 3.2 mg/ml nuclear or 8 mg/ml whole-cell protein and 0.6 μg/ml DNA, vs. 0.3 mg/ml protein and 10 μg/ml DNA used in previous work [6]. The higher protein/DNA ratios result in highly efficient end joining, allowing direct biochemical detection of specific repair products and eliminating the need for cloning or PCR amplification. DNA ends in these extracts appear to be highly sequestered, resulting in a spectrum of end joining events that is highly conservative, at least as conservative as typically observed in vivo [48,49], if not moreso. In vitro models employing a lower protein/DNA ratio might reasonably be expected to be more permissive of repair events involving more extensive resection, due to greater accessibility of the DNA ends, as was reported [6].
The finding that Artemis can dramatically enhance end joining of a terminally blocked substrate in cell extracts raises the question of why endogenous Artemis in the extracts was not equally effective in doing so. In fibroblasts, endogenous levels of Artemis are extremely low, typically below the level of detection by western blot, yet are clearly sufficient to function in DSB repair in vivo [5]. Although Artemis has never been shown to co-localize with H2AX to DSB repair foci, its phosphorylated forms do localize to discrete nuclear sites [50]. Thus, in the ordered environment of the cell nucleus, there may be some mechanism, that was not retained in crude extracts, for locally concentrating Artemis at repair sites as needed.
In summary, two nucleases, Artemis and Metnase, show remarkably similar specificities in trimming undamaged and damaged 3′ overhangs. In principle, either could act to remove both blocked termini and nearby base damage, to produce substrates suitable for patching and ligation, although Artemis appears better able to function in the presence of other core NHEJ proteins.
Supplementary Material
Acknowledgments
This work was supported by grants CA40615 (SM, KA, LFP), CA151367 (SHL) and CA84442 (DAR) from the National Institutes of Health, US Public Health Service, and by contract DE-AC02-05CH11231, US Department of Energy Office of Science (SMY).
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflicts of interest.
References
- 1.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 2.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, Lee SH. Biochemical characterization of a SET and transposase fusion protein, Metnase: Its DNA binding and DNA cleavage activity. Biochemistry (N Y) 2007;46:11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Zhang X, Geng L, Teng L, Legerski RJ. Artemis regulates cell cycle recovery from the S phase checkpoint by promoting degradation of cyclin E. J Biol Chem. 2009;284:18236–18243. doi: 10.1074/jbc.M109.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohapatra S, Kawahara M, Khan IS, Yannone SM, Povirk LF. Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient Artemis. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck BD, Lee SS, Williamson E, Hromas RA, Lee SH. Biochemical characterization of Metnase’s endonuclease activity and its role in NHEJ repair. Biochemistry. 2011;50:4360–4370. doi: 10.1021/bi200333k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hromas R, Wray J, Lee SH, Martinez L, Farrington J, Corwin LK, Ramsey H, Nickoloff JA, Williamson EA. The human set and transposase domain protein Metnase interacts with DNA ligase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair (Amst) 2008;7:1927–1937. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 9.Nikjoo H, O’Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int J Radiat Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 10.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. Wolters Kluwer / Lippincott, Williams & Wilkins; 2012. [Google Scholar]
- 11.Cadet J, Ravanat JL, Tavernaporro M, Menoni H, Angelov D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Magnander K, Elmroth K. Biological consequences of formation and repair of complex DNA damage. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Acids Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 14.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: Identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 15.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: An enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 16.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou RZ, Blanco L, Garcia-Diaz M, Bebenek K, Kunkel TA, Povirk LF. Tolerance for 8-oxoguanine but not thymine glycol in alignment- based gap filling of partially complementary double-strand break ends by DNA polymerase λ in human nuclear extracts. Nucleic Acids Res. 2008;36:2895–2905. doi: 10.1093/nar/gkn126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 19.Parsons JL, Zharkov DO, Dianov GL. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali MM, Hazra TK, Hong D, Kow YW. Action of human endonucleases III and VIII upon DNA-containing tandem dihydrouracil. DNA Repair (Amst) 2005;4:679–686. doi: 10.1016/j.dnarep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KJ, Huang J, Takeda Y, Dynan WS. DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell- free end-joining system. J Biol Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 23.Akopiants K, Zhou RZ, Mohapatra S, Valerie K, Lees-Miller SP, Lee KJ, Chen DJ, Revy P, de Villartay JP, Povirk LF. Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases λ and μ for nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2009;37:4055–4062. doi: 10.1093/nar/gkp283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu H, Yannone SM, Chen DJ. Defining the interactions between DNA-PK and ligase IV/XRCC4. DNA Repair. 2001;1:225–235. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 25.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA- dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Hannis JC, Flora JW, Muddiman DC, Charles K, Yu Y, Povirk LF. Homogeneous preparations of 3′-phosphoglycolate-terminated oligodeoxynucleotides from bleomycin-treated DNA as verified by electrospray ionization fourier transform ion cyclotron resonance mass spectrometry. Anal Biochem. 2001;289:274–280. doi: 10.1006/abio.2000.4936. [DOI] [PubMed] [Google Scholar]
- 27.Bennett RAO, Gu XY, Povirk LF. Construction of a vector containing a site-specific DNA double-strand break with 3′-phosphoglycolate termini and analysis of the products of end-joining in CV-1 cells. Intl J Radiat Biol. 1996;70:623–636. doi: 10.1080/095530096144509. [DOI] [PubMed] [Google Scholar]
- 28.Povirk LF, Zhou RZ, Ramsden DA, Lees-Miller SP, Valerie K. Phosphorylation in the serine/threonine 2609-2647 cluster promotes but is not essential for DNA-dependent protein kinase- mediated nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2007;5:3869–3878. doi: 10.1093/nar/gkm339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end- joining reaction reveals a requirement for an Mre11/Rad50/NBS1- containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Semenenko VA, Stewart RD. A fast Monte Carlo algorithm to simulate the spectrum of DNA damages formed by ionizing radiation. Radiat Res. 2004;161:451–457. doi: 10.1667/rr3140. [DOI] [PubMed] [Google Scholar]
- 32.Wu PY, Frit P, Malivert L, Revy P, Biard D, Salles B, Calsou P. Interplay between Cernunnos-XLF and nonhomologous end-joining proteins at DNA ends in the cell. J Biol Chem. 2007;282:31937–31943. doi: 10.1074/jbc.M704554200. [DOI] [PubMed] [Google Scholar]
- 33.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, Le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Povirk LF. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Molecular Biology. 2012;2012:345805. doi: 10.5402/2012/345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Löbrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 37.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta K, Purkayastha S, Neumann RD, Pastwa E, Winters TA. Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat Res. 2011;175:97–112. doi: 10.1667/RR2332.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double-strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;276:24323–24330. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 40.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JW, Inamdar KV, Hannah MF, Lees-Miller SP, Povirk LF. DNA end sequestration by DNA-dependent protein kinase and joining of sterically constrained substrates in whole-cell extracts. Environ Mol Mutagen. 2003;42:279–287. doi: 10.1002/em.10197. [DOI] [PubMed] [Google Scholar]
- 42.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 44.Yannone SM, Khan IS, Zhou R, Zhou T, Valerie K, Povirk LF. Coordinate 5′ and 3′ endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawelczak KS, Turchi JJ. Purification and characterization of exonuclease-free Artemis: Implications for DNA-PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair (Amst) 2010;9:670–677. doi: 10.1016/j.dnarep.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JW, Blanco L, Zhou T, Bebenek K, Garcia-Diaz M, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 47.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P, de Villartay JP. V(D)J and immunoglobulin class switch recombinations: A paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26:7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 48.Adams BR, Hawkins AJ, Povirk LF, Valerie K. ATM-independent, high-fidelity nonhomologous end joining predominates in human embryonic stem cells. Aging (Albany NY) 2010;2:582–596. doi: 10.18632/aging.100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Soubeyrand S, Pope L, De Chasseval R, Gosselin D, Dong F, de Villartay JP, Hache RJ. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J Mol Biol. 2006;358:1200–1211. doi: 10.1016/j.jmb.2006.02.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.