Abstract
Melanoma is the most highly malignant skin cancer, and nucleotide excision repair (NER) is involved in melanoma susceptibility. In this analysis of 1042 melanoma patients, we evaluated whether genetic variants of NER genes may predict survival outcome of melanoma patients. We used genotyping data of 74 tagging single nucleotide polymorphisms (tagSNPs) in eight core NER genes from our genome-wide association study (including 2 in XPA, 14 in XPC, 3 in XPE, 4 in ERCC1, 10 in ERCC2, 8 in ERCC3, 14 in ERCC4, and 19 in ERCC5) and evaluated their associations with prognosis of melanoma patients. Using the Cox proportional hazards model and Kaplan-Meier analysis, we found a predictive role of XPE rs28720291, ERCC5 rs4150314, XPC rs2470458 and ERCC2 rs50871 SNPs in prognosis of melanoma patients (rs28720291: AG vs. GG, adjusted hazard ratio [adjHR] = 11.2, 95% confidence interval [CI] 3.04–40.9, P = 0.0003; rs4150314: AG vs. GG, adjHR = 4.76, 95% CI 1.09–20.8, P = 0.038; rs2470458: AA vs. AG/GG, adjHR = 2.11, 95% CI 1.03–4.33, P = 0.040; and rs50871: AA vs. AC/CC adjHR =2.27, 95% CI 1.18–4.35, P = 0.015). Patients with an increasing number of unfavorable genotypes had dramatically increased death risk. Genetic variants of NER genes, particularly XPE rs28720291, ERCC5 rs4150314, XPC rs2470458 and ERCC2 rs50871, may independently or jointly modulate survival outcome of melanoma patients. Because our results were based on a median follow-up of 3 years without multiple test corrections, additional large prospective studies are needed to confirm our findings.
Keywords: melanoma, nucleotide excision repair, survival, association
INTRODUCTION
Melanoma is the most lethal skin cancer, ranking the sixth most common cancer in the United States. There were estimated 76,250 new melanoma cases, in addition to 55,560 melanoma in situ, in 2012 (Siegel et al. 2012). Although surgery remains the mainstay treatment, biochemotherapy and radiotherapy are also considered in an attempt to improve local control and overall survival. Despite aggressive treatment, patient prognosis varies substantially between individuals, with a 5-year survival rate ranging from over 80% in early stages to less than 10% in patients with distant metastasis (Buettner et al. 2005).
Some important tumor morphological and biological characteristics are known to be associated with patients’ survival, including primary tumor thickness, ulceration, mitotic activity, lymph node infiltration and distant metastasis (Spatz et al. 2010). However, these histopathological features of primary tumors do not provide sufficient information for assessing tumor malignancy. For example, a subset of “thin” melanoma (tumor thickness < 0.76 mm) can be lethal due to undetected metastasis (Woods et al. 1983). Although the underlying mechanisms are unclear, tumor genetic heterogeneity and interactions among the host and tumor factors may be responsible for rapid evolution and development of malignancies in these patients. Some somatic mutations (e.g. BRAF and p16) are commonly implicated in melanoma progression (Chin et al. 2006), whereas an enhanced host’s immune system can efficiently suppress cancer cell spreading, contributing to prolonged survival (DiFronzo et al. 2002). Nevertheless, it is possible that some other unknown genetic factors, by interacting with the known clinicopathological factors, may modulate survival outcomes of melanoma patients, thus uncovering biomarker for patients’ long-term survival.
Previous epidemiologic studies have supported the notion that DNA-damaging UV irradiation causes cutaneous melanoma by inducing genetic abnormality (von Thaler et al.). The well-studied nucleotide excision repair (NER) pathway consists of at least 23 genes/proteins that act to remove UV-induced DNA lesions. Several SNPs of the NER genes have been shown to be associated with melanoma susceptibility (Li et al. 2006). However, their influence on patients’ survival has not been thoroughly investigated. In a recent study of eight non-synonymous SNPs of DNA repair genes (i.e., XPC p.Ala499Val, XPCp.Lys939Gln, ERCC2 p.Lys751Gln and ERCC5 p.Asp1104His of NER; APEX1 p.Asp148Glu, XRCC1 p.Arg399Gln of base excision repair; and XRCC3 p.Thr241Met and NBS1 p.Glu185Gln of the homologous recombination repair), only ERCC5 p.Asp1104His (rs17655) and ERCC2 p.Lys751Gln (rs13181) were found to have an effect on prognosis of melanoma (Schrama et al. 2011), suggesting that the NER genes may be involved in melanoma outcomes, although genes involved in cell cycle checkpoint are also found to be important (Kauffmann et al. 2008).
Here we report our results of an analysis of prognosis of 1042 melanoma patients in association with 74 tagging SNPs of the NER genes available to us in a previously published genome-wide association study of melanoma (Amos et al. 2011). In the present analysis, we evaluated the association between these SNPs and survival and explored their interactions with clinicopathological risk factors in determining melanoma patient prognosis.
RESULTS
Patient Characteristics
The analysis consisted of 1042 patients with primary cutaneous melanoma (Table 1), who had available data from questionnaire, genotyping, and survival. The patients were aged between 18 and 84 years at diagnosis with a mean of 50.8 years and standard deviation of 13.1 years. There were slightly more women than men (58.8% vs. 41.2%); 83.1% of the patients had early-stage melanoma (in situ and stages I and II), and 16.9% had later-stage melanoma (stages III and IV). We also collected complete information about tumor morphology, including primary tumor thickness, ulceration, metastasis to local lymph nodes, mitotic rate (mitoses/mm2) of tumor cells [because there was no association with mitotic rate by the American Joint Committee on Cancer (AJCC) staging system of mitoses ≥1/mm2 versus <1/mm2, we used 3/mm2 as the cutoff as shown in Table 1], anatomic site of the tumor and patient biological characteristics, including colors of the skin, hair and eyes, tanning ability after sun exposure, lifetime sunburns with blistering, moles and family history of skin cancer. The median follow-up time was 35.7 months, during which 52 (5.0%) of the 1042 patients had died at the last follow-up.
Table 1.
Associations of patient demographics and tumor related characteristics with overall survival
| Parameter1 | Patient
|
Death
|
Univariate Analysis
|
Multivariate Analysis2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | HR | 95% CI | P-value | HR | 95% CI | P-VALUE | |
| Age | ||||||||||
| <=50 | 481 | 46.2 | 14 | 26.9 | 1.00 | 1.00 | ||||
| >50 | 561 | 55.7 | 38 | 73.1 | 2.65 | 1.43 – 4.89 | 0.002 | 1.90 | 0.86 – 4.18 | 0.110 |
| Sex | ||||||||||
| Female | 613 | 58.8 | 36 | 69.2 | 1.00 | 1.00 | ||||
| Male | 429 | 41.2 | 16 | 30.8 | 0.56 | 0.31 – 1.01 | 0.053 | 0.90 | 0.41 – 2.04 | 0.825 |
| Skin color | ||||||||||
| Fair | 938 | 90.1 | 47 | 9.6 | 1.00 | 1.00 | ||||
| Dark and brown | 103 | 9.9 | 5 | 90.4 | 0.98 | 0.39 – 2.46 | 0.961 | 4.55 | 1.28 – 16.70 | 0.019 |
| Hair color | ||||||||||
| Blond or red | 355 | 34.1 | 10 | 19.2 | 1.00 | 1.00 | ||||
| Black or brown | 686 | 65.9 | 42 | 80.8 | 2.13 | 1.08 – 4.35 | 0.030 | 0.86 | 0.37 – 2.00 | 0.735 |
| Eye color | ||||||||||
| Not blue | 604 | 58.0 | 29 | 55.8 | 1.00 | 1.00 | ||||
| Blue | 437 | 42.0 | 23 | 44.2 | 1.08 | 0.63 – 1.87 | 0.781 | 0.62 | 0.30 – 1.27 | 0.192 |
| Tanning ability after prolonged sun exposure | ||||||||||
| Good (high) | 651 | 63.0 | 36 | 69.2 | 1.00 | 1.00 | ||||
| Poor (low) | 382 | 37.0 | 16 | 30.8 | 0.77 | 0.43 – 1.39 | 0.39 | 1.26 | 0.58 – 2.77 | 0.560 |
| Life time sunburns with blistering | ||||||||||
| 0 | 302 | 29.0 | 18 | 35.3 | 1.00 | 1.00 | ||||
| >=1 | 738 | 71.0 | 33 | 64.7 | 0.65 | 0.37 – 1.15 | 0.140 | 0.97 | 0.47 – 2.01 | 0.927 |
| Freckling in the sun as a child | ||||||||||
| No | 600 | 57.6 | 17 | 32.7 | 1.00 | 1.00 | ||||
| Yes | 441 | 42.4 | 35 | 67.3 | 3.33 | 1.89 – 5.88 | <0.0001 | 2.90 | 1.33 – 6.33 | 0.008 |
| Moles | ||||||||||
| No | 242 | 23.2 | 11 | 21.1 | 1.00 | 1.00 | ||||
| Yes | 800 | 76.8 | 41 | 78.9 | 1.03 | 0.53 – 2.01 | 0.925 | 1.22 | 0.53 – 2.83 | 0.639 |
| Dysplastic nevi | ||||||||||
| No | 939 | 90.1 | 51 | 98.1 | 1.00 | 1.00 | ||||
| Yes | 103 | 9.9 | 1 | 1.9 | 0.18 | 0.02 – 1.27 | 0.085 | 0.56 | 0.07 – 4.24 | 0.575 |
| Family history of skin cancer | ||||||||||
| No | 385 | 36.9 | 19 | 36.5 | 1.00 | 1.00 | ||||
| Yes | 657 | 63.1 | 33 | 63.5 | 1.10 | 0.62 – 1.93 | 0.745 | 1.15 | 0.56 – 2.37 | 0.704 |
| AJCC stages | ||||||||||
| IN SITU, I and II | 866 | 83.1 | 24 | 46.1 | 1.00 | 1.00 | ||||
| III and IV | 176 | 16.9 | 28 | 53.9 | 6.87 | 3.97 – 11.9 | <0.0001 | 5.60 | 2.69 – 11.64 | <0.0001 |
| Primary tumor thickness | ||||||||||
| <1 mm | 425 | 49.0 | 8 | 17.0 | 1.00 | 1.00 | ||||
| >0.99 mm | 443 | 51.0 | 39 | 83.0 | 5.17 | 2.42 – 11.1 | <0.0001 | 2.14 | 0.72 – 6.35 | 0.169 |
| Ulceration | ||||||||||
| No | 846 | 87.2 | 28 | 59.6 | 1.00 | 1.00 | ||||
| Yes | 124 | 12.8 | 19 | 40.3 | 5.71 | 3.19 – 10.2 | <.00001 | 2.72 | 1.28 – 5.78 | 0.0009 |
| SLNB | ||||||||||
| No | 333 | 32.1 | 13 | 25.5 | 1.00 | 1.00 | ||||
| Yes | 705 | 67.9 | 38 | 74.5 | 1.31 | 0.69 – 2.49 | 0.417 | 0.44 | 0.19 – 1.03 | 0.058 |
| Mitotic rate (mitoses/mm2) | ||||||||||
| ≤ 3 | 589 | 78.6 | 20 | 47.6 | 1.00 | 1.00 | ||||
| > 3 | 160 | 21.4 | 22 | 52.4 | 4.70 | 2.56 – 8.63 | <0.0001 | 1.54 | 0.74 – 3.23 | 0.251 |
| Primary tumor anatomic site | ||||||||||
| Face, head, and neck | 117 | 11.5 | 9 | 17.6 | 1.00 | 1.00 | ||||
| Trunk, extremities, and others | 902 | 88.5 | 42 | 82.4 | 1.75 | 0.85 – 3.61 | 0.152 | 0.52 | 0.17 – 1.57 | 0.244 |
Abbreviations: HR, hazard ratio; CI, Confidence Interval.
The numbers of subjects in some of the strata were less than the total number of subjects included in our study, because some subjects did not provide complete information in their screening questionnaires
Multivariate Cox regression analyses were adjusted for all factors listed in Table 1.
To determine if there was any confounding factor influencing patients’ death or survival time, we performed Cox proportional hazards regression analysis to assess the association between overall survival (OS) and clinicpahtological characteristics. In the univariate analysis, older age, dark color of hair, freckling in the sun as a child, advanced tumor stages, thick tumor, presence of tumor ulceration, and increased primary tumor mitotic rate were significant predictors for poor survival. When all of these variables were included in a Cox proportional hazards regression model for adjustment to calculate hazards ratio (HR), only dark color of the skin (HR = 4.55), freckling in the sun as a child (HR = 2.90), advanced AJCC stage (HR = 5.60), and presence of tumor ulceration (HR = 2.72) remained statistically significant predictors for poor survival (Table 1).
Determination of melanoma survival prediction model
We performed the stepwise multivariate Cox proportional hazards regression analysis to further screen for optimal predictors of survival in melanoma patients, using covariates listed in Table 1 and the 74 selected SNPs of the 8 NER core genes (i.e., two SNP for XPA, 14 SNPs for XPC, three SNPs for XPE/DDB1, four SNPs for ERCC1, 10 SNPs for ERCC2/XD, eight SNPs for ERCC3/XPB, 14 SNPs for ERCC4/XPF, and 19 SNP for ERCC5/XPG). As shown in Table 2, clinicopathological factors of age (≤50 vs. >50), stage (in situ/I/II vs. III/IV), ulceration (no vs. yes) and mitotic rate (≤3 vs. >3/mm2), and SNPs of rs28720291 (GG vs. AG), rs4150314 (GG vs. AG), rs2470458 (AG+GG vs. AA) and rs50871 (AC+CC vs. AA) were selected as the most significant predictors of survival, among which covariates of later stages (III/IV) (HR = 6.34; 95% CI 3.11–11.9), rs28720291 GG genotype (HR = 6.69; 95% CI 1.83–23.7), and rs4150314 GG genotype (HR= 6.15; 95% CI 1.46–28.5) were of the strongest predictors. Older age, ulceration, increased mitotic rate, rs2470458 AG/GG genotypes, and rs50871 AC/CC genotypes were of low or moderate risk factors (1< HR <3).
Table 2.
Predictors of overall survival in melanoma patients obtained from stepwise multivariate cox regression analysis of selected variables1
| Selected variables | P-value | HR | 95% CI |
|---|---|---|---|
| Age (≤50 vs. >50) | 0.003 | 1.05 | 1.01 – 1.07 |
| Stage (IN SITU, I, II vs. III, IV) | <0.0001 | 6.34 | 3.11 – 11.9 |
| Ulceration (no vs. yes) | 0.013 | 2.52 | 1.25 – 5.35 |
| Mitotic rate (≤3 vs. >3) | 0.012 | 2.55 | 1.06 – 4.61 |
| rs28720291 (GG vs. AG) | 0.004 | 6.69 | 1.83 – 23.7 |
| rs4150314 (GG vs. AG) | 0.017 | 6.15 | 1.46 – 28.5 |
| rs2470458 (AG+GG vs. AA) | 0.025 | 2.42 | 1.08 – 4.76 |
| rs50871 (AC+CC vs. AA) | 0.020 | 2.23 | 1.18 – 4.50 |
Abbreviations: HR, hazard ratio; CI, Confidence Interval.
Age, sex, tumor Breslow thickness, tumor stages, ulceration of the tumor, tumor cell mitotic rate, involvement of lymph nodes, primary tumor anatomic site and the 74 selected SNPs of the 8 NER core genes [i.e., XPA (rs1800975 and rs2808667), XPC (rs1350344, rs2227999, rs2228000, rs2228001, rs2470458, rs2607772, rs2733533, rs2733537, rs3731062, rs3731125, rs3731127, rs3731146, rs3731149, and rs3731151), XPE (rs2230356, rs4939513, and rs28720291), ERCC1 (rs11615, rs1007616, rs2298881, and rs3212955), ERCC2 (rs13181, rs50871, rs171140, rs238406, rs238416, rs1052555, rs1618536, rs1799786, rs1799787, and rs1799793), ERCC3 (rs1566823, rs1803541, rs4150403, rs4150436, rs4150496, rs4150523, rs4662718, rs9282675), ERCC4 (rs254942, rs1799801, rs1800067, rs1800124, rs2276464, rs2276465, and rs2276466, rs3136146, rs3136166, rs3136187, rs3136189, rs3136195, rs3743538, and rs16963255), and ERCC5 (rs17655, rs751402, rs873601, rs1047768, rs1047769, rs2227869, rs2296147, rs2296148, and rs4150260, rs4150275, rs4150314, rs4150330, rs4150339, rs4150342, rs4150355, rs4150383, rs4771436, rs8002276, and rs11069498)] genotypes were included in the stepwise multivariate Cox proportional hazards regression analysis.
NER genetic polymorphisms as independent survival risk factors
The initial stepwise Cox proportional hazards regression analysis suggested four SNPs (XPE rs28720291, ERCC5 rs4150314, XPC rs2470458 and ERCC2 rs50871) as important and independent predictors for survival of melanoma patients. We further performed univariate and multivariate Cox proportional hazards regression analyses to evaluate their effects on risk of death or in the presence of other clinicopathological covariates. In the univariate analysis, XPE rs28720291AG and ERCC2 rs50871AA genotypes were associated with increased hazards of early death (AG vs. GG: HR = 4.92, 95% CI 1.77–13.70, P = 0.002; and AA vs. AC+CC: HR = 2.18, 95% CI 1.26–3.77, P = 0.005, respectively). In the multivariate analyses performed with adjustment for age, sex, tumor Breslow thickness, tumor stage, ulceration, tumor cell mitotic rate, involvement of lymph nodes, and tumor anatomic site, the four SNPs remained significantly associated with survival outcome of melanoma patients [i.e., rs28720291: AG (no AA was observed) vs. GG 11.2, 95% CI 3.04–40.9, P = 0.0003; rs4150314: AG (no AA was observed) vs. GG 4.76, 95% CI 1.09–20.8, P = 0.038; rs2470458: AA vs. AG+GG 2.11, 95% CI 1.03–4.33, P = 0.040; and rs50871: AA vs. AC+CC 2.27, 95% CI 1.18–4.35, P = 0.015] (Table 3).
Table 3.
Association between selected NER genetic variants1 and overall survival of melanoma patients
| Genotypes1 | Patient
|
Death
|
Univariate Analysis
|
Multivariate Analysis†
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| XPE | ||||||||||
| rs28720291 | ||||||||||
| GG | 1019 | 97.8 | 48 | 92.3 | 1.00 | 1.00 | ||||
| AG | 23 | 2.2 | 4 | 7.7 | 4.92 | 1.77 – 13.7 | 0.002 | 11.2 | 3.04 – 40.9 | 0.0003 |
| ERCC5 | ||||||||||
| rs4150314 | ||||||||||
| GG | 1021 | 98.2 | 50 | 96.1 | 1.00 | 1.00 | ||||
| AG | 19 | 1.8 | 2 | 3.9 | 2.39 | 0.58 – 9.83 | 0.227 | 4.76 | 1.09 – 20.8 | 0.038 |
| XPC | ||||||||||
| rs2470458 | ||||||||||
| AA | 646 | 62.1 | 37 | 71.1 | 1.00 | 1.00 | ||||
| AG | 345 | 33.1 | 12 | 23.1 | 0.60 | 0.31 – 1.15 | 0.126 | 0.42 | 0.19 – 0.92 | 0.031 |
| GG | 50 | 4.8 | 3 | 5.8 | 1.06 | 0.33 – 3.43 | 0.927 | 0.89 | 0.21 – 3.78 | 0.879 |
| AG+GG | 395 | 37.9 | 15 | 28.9 | 0.66 | 0.36 – 1.20 | 0.175 | 0.47 | 0.23 – 0.97 | 0.040 |
| AG+GG | 395 | 37.9 | 15 | 28.9 | 1.00 | 1.00 | ||||
| AA | 646 | 62.1 | 37 | 71.1 | 1.52 | 0.83 – 2.77 | 0.172 | 2.11 | 1.03 – 4.33 | 0.040 |
| ERCC2 | ||||||||||
| rs50871 | ||||||||||
| AA | 286 | 27.4 | 23 | 44.2 | 1.00 | 1.00 | ||||
| AC | 529 | 50.8 | 18 | 34.6 | 0.41 | 0.22 – 0.76 | 0.005 | 0.45 | 0.22 – 0.90 | 0.024 |
| CC | 227 | 21.8 | 11 | 21.2 | 0.57 | 0.28 – 1.17 | 0.125 | 0.44 | 0.17 – 1.15 | 0.093 |
| AC+CC | 756 | 72.6 | 29 | 55.8 | 0.46 | 0.27 – 0.79 | 0.005 | 0.44 | 0.23 – 0.85 | 0.015 |
| AC+CC | 756 | 72.6 | 29 | 55.8 | 1.00 | 1.00 | ||||
| AA | 286 | 27.4 | 23 | 44.2 | 2.18 | 1.26 – 3.77 | 0.005 | 2.27 | 1.18 – 4.35 | 0.015 |
Abbreviations: HR, hazard ratio; CI, Confidence Interval.
Only listed the four SNPs from the stepwise multivariate Cox proportional hazards regression analysis model shown in Table 2.
Adjusted by age, sex, tumor Breslow thickness, tumor stage, ulceration of the tumor, tumor cell mitotic rate, involvement of lymph nodes, and primary tumor anatomic site.
Survival of melanoma patients and combined genetic risk factors
To assess the joint effect of the four SNPs on patient prognosis, we combined their unfavorable genotypes (i.e., XPE rs28720291AG, ERCC5 rs4150314AG, XPC rs2470458AA, and ERCC2 rs50871AA genotypes). The frequencies of patients with 0, 1, 2 and 3 unfavorable genotypes were 276, 566, 188 and 10, respectively; no patient had all four unfavorable genotypes. Patients with an increasing number of unfavorable genotypes had dramatically increased risk of death by over 30-fold (HR = 34.3; 95% CI 7.48–157.2; P < 0.0001) in patients with any three unfavorable genotypes, compared with those without any unfavorable genotypes (Table 4 and Figure 1). Since there were only ten patients carrying three unfavorable genotypes, we next grouped all patients into a low-risk group (patients with <=1 unfavorable genotypes) and a high-risk group (patients with 2 or 3 unfavorable genotypes) for further stratified analysis (Table 4).
Table 4.
Association between combined NER variants and overall survival of melanoma patients
| No. of variant genotypes1 | Patient
|
Death
|
Univariate Analysis
|
Multivariate Analysis2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| No. of variant genotypes1 | 0.0005 | <0.00001 | ||||||||
| 0 | 276 | 26.5 | 9 | 17.3 | 1.00 | 1.00 | ||||
| 1 | 566 | 54.4 | 23 | 44.2 | 1.28 | 0.59 – 2.78 | 0.525 | 1.26 | 0.51 – 3.10 | 0.061 |
| 2 | 188 | 18.1 | 17 | 32.7 | 2.94 | 1.31 – 6.59 | 0.009 | 3.90 | 1.50 – 10.1 | 0.005 |
| 3 | 10 | 1.0 | 3 | 5.7 | 17.8 | 4.77 – 66.3 | <0.0001 | 34.3 | 7.48 – 157.2 | <0.0001 |
| Ptrend <0.0001 | ||||||||||
| Combined group | ||||||||||
| 0–1 | 844 | 81.0 | 32 | 61.5 | 1.00 | 1.00 | ||||
| 2–3 | 198 | 19.0 | 20 | 38.5 | 2.10 | 1.42 – 3.12 | 0.0002 | 4..01 | 2.04 – 7.86 | <0.0001 |
Abbreviations: HR, hazard ratio; CI, Confidence Interval.
rs28720291 AG, rs4150314 AG, rs2470458 AA, and rs50871AA.
Multivariate Cox proportional Hazards regression analysis with adjustment for age, sex, tumor Breslow thickness, tumor stage, ulceration of the tumor, tumor cell mitotic rate, involvement of lymph nodes, and primary tumor anatomic site.
Fig. 1.
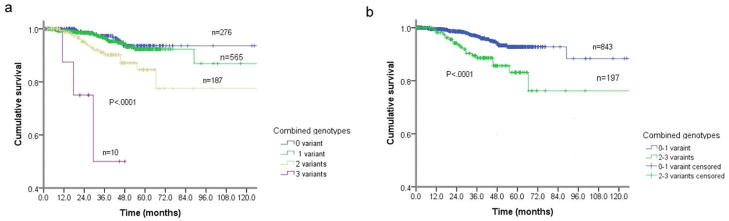
Kaplan-Meier overall survival for patients with primary melanoma by combined NER genotypes (i.e., rs28720291 GG, rs4150314 GG, rs2470458 AG+GG, and rs50871 AC+CC). (a) By 0, 1, 2,3 NER variant genotypes (P < 0.0001); and (b) By 0–1 and 2–3 NER variant genotypes (P = 0.0001).
Stratification analysis between the unfavorable genotypes and melanoma survival
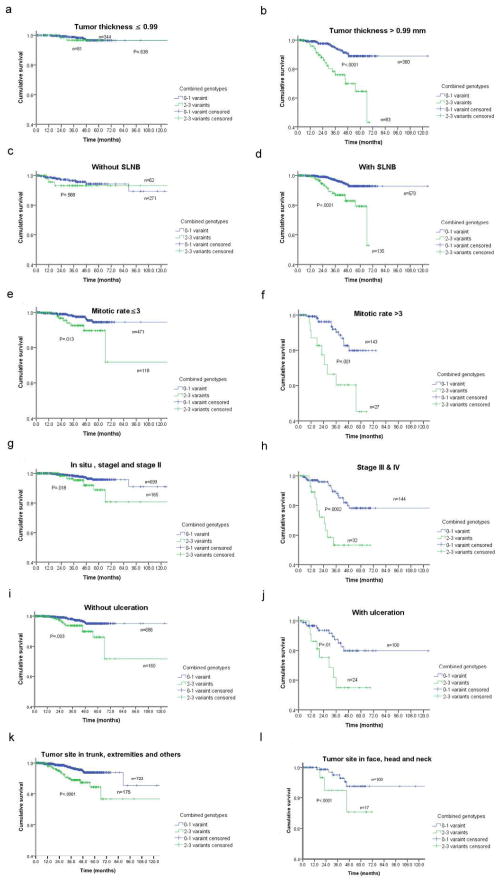
We further performed stratified analysis to investigate if the combined effect of unfavorable genotypes on survival was modified by some important clinicopathological factors in Table 1. We found that only patients in the high-risk genotype group, but not the low-risk genotype group, showed substantially increased risk of death in the presence or absence of concomitant clinicopathological risk factors (e.g., thick tumor, involvement of lymph nodes, increased mitotic rate, advanced AJCC tumor stages, presence of tumor ulceration and tumor site in face and head and neck), except for the subgroups of thin tumor and without lymph node involvement (Figure 2).
Fig. 2.
Kaplan-Meier overall survival curves for patients with primary melanoma of 0–1 and 2–3 NER variant genotypes (i.e., rs28720291 GG, rs4150314 GG, rs2470458 AG+GG, and rs50871 AC+CC) by tumor related characters: (a) and (b) By tumor Breslow thickness (P < 0.0001); (c) and (d) By SLNB (P < 0.0001); (e) and (f) By mitotic rate (P < 0.0001); (g) and (h) By AJCC stages (P < 0.0001); (i) and (j). By ulceration (P < 0.0001); and (k) and (l). By primary tumor anatomic site (P < 0.0001).
DISCUSSION
In this relatively large melanoma patient cohort, we found that some variants of the NER genes (e.g., XPE rs28720291, ERCC5 rs4150314, XPC rs2470458 and ERCC2 rs50871) may independently or jointly modulate survival of melanoma patients. These genetic variants, in combination with clinicopathological factors, effectively predicted survival in subgroups of melanoma patients.
Previous studies demonstrated that some clinicopathological characteristics were associated with prognosis of melanoma patients, such as hair color, history of childhood freckling in the sun, tumor stage and ulceration status (Buettner et al. 2005). These results were also confirmed in the current analysis. However, we were interested in finding out some genetic variants of the NER genes that may play a role in modulating patient survival. This is because NER is an essential DNA repair mechanism that ensures genomic integrity. There is evidence that genomic instability increases not only in primary melanoma, compared with nevi, but also in metastases, compared with primary tumors (Chin et al. 2006). Hence, an increased NER capacity may reduce DNA mutations that may stimulate malignant progression and metastasis. More interestingly, previously reported two NER SNPs [ERCC5 p.Asp1104His (rs17655) and ERCC2 p.Lys751Gln (rs13181)] (Schrama et al. 2011) were replicated in our current analysis, because they are tagged by our selected tagging SNPs (ERCC5 rs4150314 and ERCC2 rs50871). However, we found that additional two NER SNPs (XPE rs28720291 and XPC rs2470458) also independently predicted the prognosis of melanoma. Therefore, our data further support the notion that genetic variants in the NER pathway may modulate clinical outcome of melanoma patients.
NER employs a relatively small number of essential repair proteins, including XPA, XPC, XPE/DDB1, ERCC1, ERCC2/XPD, ERCC3/XPB, ERCC4/XPF and ERCC5/XPG, to correct bulky DNA damage induced by chemical carcinogens or UV exposure. Briefly, repair of the damaged DNA strand includes making an incision at the 5′ and 3′ of the lesion, removing a 30-nucleotide section containing the damage, and ligating the gap by pairing DNA synthesis (Sancar 1995). XPC-hHR23B is a NER factor that detects DNA damage and recruits TFIIH to the damaged site (Yokoi et al. 2000); then proteins encoded by XPA-G genes, the ERCC1-hHR23B-RPA trimmers, and TFIIH are involved in the excision step (de Boer and Hoeijmakers 1999). Two helicase subunits of TFIIH (i.e., XPB and XPD), together with XPA, RPA, and XPG, form a 30 base-pair bubble around the lesion for damage verification and correct positioning of two endonucleases (i.e., XPG and XPF-ERCC1) before incision (Missura et al. 2001). The incisions made by XPG and XPF-ERCC1 are at the double- and single-stranded DNA border in the incision complex (Sijbers et al. 1996). Observation of the mobility of various NER proteins in living cells suggests that NER proceeds by the sequential assembly of individual factors involved, rather than through the action of a preassembled repairosome (Houtsmuller et al. 1999).
Previous studies have extensively explored associations between NER and melanoma susceptibility, but few investigated the effect of NER on clinical outcomes of melanoma patients. In a study of 90 stage IV melanoma patients, an ERCC1-rs11615 SNP was found to be weakly associated with overall survival (Liu et al. 2005). In another study of 244 melanoma patients in Sweden, an ERCC2-rs13181 SNP was suggested to be a prognostic factor for melanoma progression (Kertat et al. 2008). More recently, both ERCC5-rs17655 and ERCC2-rs13181 SNPs were found to be independent prognostic factors in 742 melanoma patients (Schrama et al. 2010). However, none of the above SNPs were replicated or in linkage with the four positive SNPs (i.e., XPE-rs28720291, ERCC5-rs4150314, XPC-rs2470458 and ERCC2-rs50871) investigated in the present study. Several possible reasons may explain the discrepancies: first, the studies by Liu et al. and Kertat et al. were small sample-sized (90 cases and 244 cases, respectively), which could lead to chance findings or miss some SNPs with mild effects due to a limited study power; second, the majority of melanoma patients in these two studies had late-stage tumors (stages III/IV), while a large proportion of patients in our analysis and the study by Schrama et al. had early-stage tumors (866/1044 and 652/742, respectively).
Despite these discrepancies, there was some consistency among these published studies and ours. If excluding the most small-sized study by Liu et al., it appears there are increasing levels of significance as well as increasing number of genes as the study patient population size increases, and the significant SNPs/genes identified in previous small-sized study could be confirmed by later larger-sized studies (ERCC2 in 244 patients, ERCC2 and ERCC5 in 742 patients, and ERCC2, ERCC5, XPC and XPE in 1042 patients). Since SNPs may alter the related gene’s function, our analysis, together with previous studies, suggested that the four genes, ERCC2/XPD, ERCC5/XPG, XPC and XPE/ERCC3, may play important roles in modulating melanoma patients’ survival.
Although the effect of a single SNP on cancer risk or clinical outcomes, if any, may be limited, the combined effect of several SNPs in the same or different genes could be more significant. In the present analysis, we were also interested in whether there was an additive/synergistic effect in the association of the four SNPs (XPE-rs28720291, ERCC5-rs4150314, XPC-rs2470458 and ERCC2-rs50871) with melanoma survival. Indeed, patients with two or three unfavorable genotypes showed dramatically increased risk of death, compared with those with none or one unfavorable genotype. This is biologically plausible, because multiple variants may be more likely to have a substantial joint effect on the DNA repair capacity phenotype.
Through stratified analyses, we found that the genotype-survival association was most pronounced in the presence of clinicopathological risk factors, suggesting that suboptimal repair of DNA damage induced externally (UV-exposure) or internally (free radicals from metabolism) could aggregate the existing genomic instability of a fast-growing melanoma, promoting melanoma development and progression in the high-risk populations. Since these high-risk patients comprised 20.0% (198 out of 1042) of all the study subjects, our analysis identified a significant proportion of melanoma patients (such as those with unfavorable genotypes) that may require close clinical surveillance or alternative treatment to improve their survival.
The current analysis has some limitations. First, since we used a tagging SNP approach, we were not able to explore the mechanism by which the studied genetic polymorphisms influence melanoma patients’ survival. Although the four identified SNPs and their tagged SNPs (LD ≥0.8) may have potential biological functions as predicted by software tools (http://snpinfo.niehs.nih.gov/snpfunc.htm), none of them have been reported or investigated as functional SNPs in the literature. Only four SNPs tagged by XPC-rs2470458 were found to be associated with risk of bladder cancer (rs2228000, rs2470352, and rs2470458) and lung cancer (rs2229090) in previous association studies (Shen et al. 2005; Stern et al. 2009). Further functional studies of these SNPs are required. Second, there were only 52 deaths out of 1042 patients at our last follow-up at a median of nearly 3 years. Therefore, the current study is, to a large extent, an interim survival analysis. We will report updated results after we have a longer follow-up time. Third, we did not adjust for multiple tests, simply because this was an exploratory study with a limited study power. We plan to confirm current findings in our ongoing prospective expansion studies in more stringent conditions with a larger study population. Fourth, since treatment of melanoma, advanced melanoma in particular, has not been standardized, patients included in the current analysis who developed advanced melanoma may have received a wide variety of systemic therapies, often sequentially. The systemic therapies available for the cohort of patients included in our analysis would only have been expected to be modestly effective in a minority of melanoma patients (the study period ended in 2008; vemurafenib was approved by the FDA for the treatment of advanced melanoma in 2011). Because of the variety of treatments administered (often multiple types of treatments to the same patient) and their very modest anticipated effect on overall survival, we did not evaluate the potential role of these therapies in the outcomes of the patients, or their potential relationship to the polymorphisms identified. While evaluation of the association between the polymorphisms investigated and response to a variety of melanoma systemic therapies is important, such an evaluation is beyond the scope of the current analysis.
In summary, we identified four SNPs of the NER genes (i.e., XPE rs28720291, ERCC5 rs4150314, XPC rs2470458 and ERCC2 rs50871) that may have independent or joint effects on survival of melanoma patients. These findings, once validated in future prospective studies with large sample sizes and better study designs, will provide some promising guidance for clinical management and tailored or personalized therapeutics in treating melanoma patients.
MATERIALS AND METHODS
Study Populations
Patients were accrued for an ongoing, hospital-based, case–control study of epidemiologic and genetic risk factors for melanoma. A total of 1042 histologically-confirmed patients with melanoma in situ and stage I to stage IV were enrolled between January, 2000 and September, 2008. Patients were enrolled into the study regardless of age, sex or disease stage. On entry into the study, each patient had a personal interview to elicit lifestyle factors, using a standardized questionnaire. Each patient also had a 30-mL sample of blood drawn for various biomarker studies, including genotyping. All patients were enrolled and diagnosed with staging system defined by the AJCC at The University of Texas M. D. Anderson Cancer Center, Houston, TX. Specifically, melanoma patients with melanoma in situ, stage I/II (primary tumor without evidence of regional or distant metastasis at diagnosis), stage III (locoregional disease, including in transit, satellite, and/or regional lymph node metastasis at diagnosis), and stage IV (distant metastasis at diagnosis) were classified according to the AJCC melanoma staging system (Balch et al., 2009). For patients with stage I/II disease, staging elements included Breslow primary tumor thickness, presence or absence of primary tumor ulceration, and mitotic rate (i.e., number of mitoses per square millimeter using dermal hotspot approach). All patients gave a written informed consent, and the protocol was approved by the M. D. Anderson Cancer Center Institutional Review Board. Patients were evaluated, staged, treated and followed using the standard guidelines, including the use of sentinel lymph node biopsy for high-risk primary melanoma (Gershenwald and Ross, 2011). Patients with high-risk local-regional, and those with recurrent and metastatic melanoma, received a variety of protocol-based and off-protocol systemic therapies, based on standard guidelines, physician recommendations, and patient preferences. The study protocol and informed consent were in compliance with Declaration of Helsinki Principles.
Polymorphism selection and genotyping
Genomic DNA was extracted from the buffy coat fraction of each blood sample by using a Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. DNA purity and concentrations were determined by spectrophotometric measurement of absorbance at 260 and 280 nm by a UV spectrophotometer (Nano Drop Technologies, Inc., Wilmington, DE). These 1042 patients complete set of selected SNPs was genotyped using the Illumina HumanOmni1-Quad_v1-0_B array and was called using the BeadStudio algorithm, at the John Hopkins University Center for Inherited Disease Research (CIDR). In this analysis, we selected the available 74 tagging SNPs in 8 core NER genes, including XPA (rs1800975 and rs2808667), XPC (rs1350344, rs2227999, rs2228000, rs2228001, rs2470458, rs2607772, rs2733533, rs2733537, rs3731062, rs3731125, rs3731127, rs3731146, rs3731149, and rs3731151), XPE (rs2230356, rs4939513, and rs28720291), ERCC1 (rs11615, rs1007616, rs2298881, and rs3212955), ERCC2 (rs13181, rs50871, rs171140, rs238406, rs238416, rs1052555, rs1618536, rs1799786, rs1799787, and rs1799793), ERCC3 (rs1566823, rs1803541, rs4150403, rs4150436, rs4150496, rs4150523, rs4662718, and rs9282675), ERCC4 (rs254942, rs1799801, rs1800067, rs1800124, rs2276464, rs2276465, rs2276466, rs3136146, rs3136166, rs3136187, rs3136189, rs3136195, rs3743538, and rs16963255), and ERCC5 (rs17655, rs751402, rs873601, rs1047768, rs1047769, rs2227869, rs2296147, rs2296148, rs4150260, rs4150275, rs4150314, rs4150330, rs4150339, rs4150342, rs4150355, rs4150383, rs4771436, rs8002276, and rs11069498). Any SNP with a call rate lower than 95% was excluded from further analysis.
Statistical Analysis
We used the Cox proportional hazards regression model to evaluate the effect of genotypes and clinicopathological variables on overall survival (OS), calculated as hazard ratios (HRs) with their corresponding 95% confidence intervals (CIs). We performed a stepwise conditional logistic regression analysis to explore the best model to predict the survival outcome. The survival time was calculated from the first day of diagnosis until the date of event or the last-known follow-up. All HRs were adjusted for age, sex, tumor stage, tumor Breslow thickness, ulceration of tumor, tumor cell mitotic rate, involvement of lymph node, and primary tumor anatomic site. Kaplan-Meier analysis was used to evaluate the effect of clinicopathological and genetic variables on the cumulative probability of overall survival. All reported P values were two-sided, and P < 0.05 was considered to indicate statistical significance. All analyses were performed using SAS software (version 9.1; SAS Institute, Cary, NC).
Acknowledgments
We thank Margaret Lung and Jessica Fiske for assistance in recruiting the subjects; Yawei Qiao, Jianzhong He, and Kejing Xu for laboratory assistance; This study was supported by the National Institutes of Health, National Cancer Institute Grants R01 CA 100264 and CA 131274 (Q.W.) and P50 CA 093459 (E.A.G.), and in part by the National Institute of Environmental Health Sciences Grants R01 ES11740 (Q.W.) and P30 CA16672 (M.D. Anderson Cancer Center).
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, Qingyi Wei
Financial support: Christopher I. Amos, Jeffrey E. Lee, Elizabeth A. Grimm, Qingyi Wei
Administrative support: Christopher I. Amos, Jeffrey E. Lee, Elizabeth A. Grimm, Qingyi Wei
Provision of study materials or patients: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, Qingyi Wei
Collection and assembly of data: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Dakai Zhu, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, Qingyi Wei
Data analysis and interpretation: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Dakai Zhu, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, Qingyi Wei
Manuscript writing: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Dakai Zhu, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, Qingyi Wei
Final approval of manuscript: Chunying Li, Ming Yin, Li-E Wang, Christopher I. Amos, Dakai Zhu, Jeffrey E. Lee, Jeffrey E. Gershenwald, Elizabeth A. Grimm, and Qingyi Wei.
References
- Amos CI, Wang LE, Lee JE, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20:5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, et al. Final Version of 2009 AJCC Melanoma Staging and Classification. J Clin Oncol. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner PG, Leiter U, Eigentler TK, et al. Development of prognostic factors and survival in cutaneous melanoma over 25 years: An analysis of the Central Malignant Melanoma Registry of the German Dermatological Society. Cancer. 2005;103:616–624. doi: 10.1002/cncr.20816. [DOI] [PubMed] [Google Scholar]
- Chin L, Garraway LA, Fisher DE, et al. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81:127–137. doi: 10.1016/s0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- DiFronzo LA, Gupta RK, Essner R, et al. Enhanced humoral immune response correlates with improved disease-free and overall survival in American Joint Committee on Cancer stage II melanoma patients receiving adjuvant polyvalent vaccine. J Clin Oncol. 2002;20:3242–3248. doi: 10.1200/JCO.2002.01.065. [DOI] [PubMed] [Google Scholar]
- Gershenwald JE, Ross MI. Sentinel-lymph-node biopsy for cutaneous melanoma. N Engl J Med. 2011;364:1738–1745. doi: 10.1056/NEJMct1002967. [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB, Rademakers S, Nigg AL, et al. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Kertat K, Rosdahl I, Sun XF, et al. The Gln/Gln genotype of XPD codon 751 as a genetic marker for melanoma risk and Lys/Gln as an important predictor for melanoma progression: a case control study in the Swedish population. Oncol Rep. 2008;20:179–183. [PubMed] [Google Scholar]
- Li C, Hu Z, Liu Z, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–2532. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- Liu D, O’Day SJ, Yang D, et al. Impact of gene polymorphisms on clinical outcome for stage IV melanoma patients treated with biochemotherapy: an exploratory study. Clin Cancer Res. 2005;11:1237–1246. [PubMed] [Google Scholar]
- Missura M, Buterin T, Hindges R, et al. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. DNA repair in humans. Annu Rev Genet. 1995;29:69–105. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- Schrama D, Scherer D, Schneider M, et al. ERCC5 p.Asp1104His and ERCC2 p.Lys751Gln polymorphisms are independent prognostic factors for the clinical course of melanoma. J Invest Dermatol. 2011;131:1280–1290. doi: 10.1038/jid.2011.35. [DOI] [PubMed] [Google Scholar]
- Shen M, Berndt SI, Rothman N, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116:768–773. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Spatz A, Batist G, Eggermont AM. The biology behind prognostic factors of cutaneous melanoma. Curr Opin Oncol. 2010;22:163–168. doi: 10.1097/CCO.0b013e328337fe8f. [DOI] [PubMed] [Google Scholar]
- Stern MC, Lin J, Figueroa JD, et al. Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res. 2009;69:6857–6864. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Thaler AK, Kamenisch Y, Berneburg M. The role of ultraviolet radiation in melanomagenesis. Exp Dermatol. 2010;19:81–88. doi: 10.1111/j.1600-0625.2009.01025.x. [DOI] [PubMed] [Google Scholar]
- Woods JE, Soule EH, Creagan ET. Metastasis and death in patients with thin melanomas (less than 0.76 mm) Ann Surg. 1983;198:63–64. doi: 10.1097/00000658-198307000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Masutani C, Maekawa T, et al. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]