Abstract
Objective
To create an analytics platform for specifying and detecting clinical phenotypes and other derived variables in electronic health record (EHR) data for quality improvement investigations.
Materials and Methods
We have developed an architecture for an Analytic Information Warehouse (AIW). It supports transforming data represented in different physical schemas into a common data model, specifying derived variables in terms of the common model to enable their reuse, computing derived variables while enforcing invariants and ensuring correctness and consistency of data transformations, long-term curation of derived data, and export of derived data into standard analysis tools. It includes software that implements these features and a computing environment that enables secure high-performance access to and processing of large datasets extracted from EHRs.
Results
We have implemented and deployed the architecture in production locally. The software is available as open source. We have used it as part of hospital operations in a project to reduce rates of hospital readmission within 30 days. The project examined the association of over 100 derived variables representing disease and co-morbidity phenotypes with readmissions in five years of data from our institution’s clinical data warehouse and the UHC Clinical Database (CDB). The CDB contains administrative data from over 200 hospitals that are in academic medical centers or affiliated with such centers.
Discussion and Conclusion
A widely available platform for managing and detecting phenotypes in EHR data could accelerate the use of such data in quality improvement and comparative effectiveness studies.
Keywords: Healthcare analytics, Clinical data warehousing, Temporal abstraction, Quality improvement, Comparative effectiveness
1 INTRODUCTION1
Emerging changes in the United States’ healthcare delivery model [1] have led to renewed interest in data-driven methods for managing quality of care [2, 3]. Hospitals are evaluated using quality metrics [4] for mortality [5], length of stay [6], hospital readmissions within 30 days [7] and others [8]. Rankings are increasingly made public [9] and have begun to be used by insurers to penalize through lower reimbursement those that perform poorly [10, 11]. Hospitals need to track and understand their performance and determine whether interventions to improve have been successful.
Healthcare analytics [12] leverages clinical and administrative data in EHRs, and knowledge of clinical practice standards and guidelines, to manage metric-driven quality improvement. Goals include identifying inefficiencies in care delivery and deviations from practice standards, identifying opportunities for expanding services believed to enhance quality of care, efficient targeting of limited resources, and comparing risk-adjusted performance to peer institutions. Techniques [13, 14] include retrospectively identifying populations with high or low risk-adjusted metric scores that represent exemplars of high quality care and areas for improvement, respectively. Patient characteristics, or phenotypes, with strong associations with events of interest may help explain such scores. Phenotypes derived from EHR data also may be incorporated as variables into predictive models [15, 16] that prospectively compute and present patient-specific risk via clinical decision support. These analytic methods require mature EHR implementations.
EHRs with the required breadth of data increasingly are available at large hospital systems [17]. Institutions may make these data available through a clinical data warehouse, a relational database fed by an EHR’s transactional systems that supports efficient population queries [18–23]. National repositories of administrative and sometimes clinical data [24–26] provide institutions with access to comparative data, e.g. UHC’s CDB [27, 28]. These local and national datasets together represent a rich potential source of data for analytics, but they present substantial challenges in their use [29–33]. Diagnoses, co-morbidities and procedures typically are represented indirectly as billing codes [34]. Clinical data such as laboratory test results and medication histories may be recorded as local codes or in text, and they may require substantial clinical context to interpret. Efforts are underway to create publicly available libraries of phenotypes defined in terms of patterns in codes and discrete data [35–38] such as eMERGE [35]. eMERGE’s phenotypes are publicly available in document form [37]. Tools for translating these phenotypes into queries of local data warehouses and national repositories could make data-driven quality improvement more broadly practical.
To address this need, we have developed an architecture and implementation for healthcare analytics, the Analytic Information Warehouse (AIW). It supports specifying phenotypes in a database-agnostic manner as groups of administrative codes, classifications of numerical test results and vital signs, and frequency, sequential and other temporal patterns in coded and discrete data. This capability allows specifying phenotypes such as Patients with repeated elevated high blood pressure readings who have diagnosed hypertension and are being treated with a diuretic in terms of these data. The AIW extracts data and derived variables representing phenotypes of interest into delimited files suitable for statistical analysis and mining with standard tools. It alternatively imports data and found phenotypes into an i2b2 [19] project for query. I2b2 (Informatics for Integrating Biology and the Bedside) is a widely adopted clinical research data warehouse system. The AIW software is available as open source under the Apache 2 license [39] from http://aiw.sourceforge.net. The AIW’s development has been initiated and driven by an operational project at our institution to understand the causes of hospital readmissions locally and nationally. We show results from this effort, which included empirical descriptive analyses of hospital readmissions in local data and the UHC CDB.
2 BACKGROUND AND SIGNIFICANCE
Clinical data warehouses and national repositories have heterogeneous structure, semantics and analytics features. Their structure ranges in complexity from single star schemas [40] to more complex dimensional modeling [41] approaches with overlapping star schemas. They typically represent EHR data with the same codes and concepts as source systems, thus interpretation of their data presents the same challenges as with data from EHRs in general. Business intelligence tools [42] may support computing derived variables and metrics from data warehouses using proprietary mechanisms for inclusion in reports. Commercial analytics platforms may provide similar metric calculation capability with commonly used metrics preconfigured. They also may support integrating with EHR data national prescription and claims databases and administrative codes from affiliated hospitals and practices. The relatively standard semantic representation of administrative data in clinical systems enables such platforms. It also has enabled representing clinical phenotypes as variables derived from administrative codes in epidemiological studies [43]. Limited adoption of analogous standards for clinical data representation likely is in part responsible for limited availability and application of scores and variables computed from such data [44–47].
Addressing the challenges of computing phenotypes and risk scores from EHR extracts has become a requirement of multi-center gene-based disease management studies [48]. eMERGE’s phenotypes, which target such studies, employ multiple strategies for overcoming these challenges including categorizing administrative codes, classifying numerical test results, computing frequency, sequential and other temporal patterns, and selecting alternative phenotype definitions depending on data availability [49]. These strategies appear to be general purpose, thus we expect them to be applicable in quality improvement. SHARPn [36] aims to provide software for automating computation of eMERGE phenotypes cross-institutionally, but it primarily has targeted research use cases, and its software’s performance in quality improvement has not been tested.
Detecting temporal patterns and relationships in EHR data presents substantial challenges [50]. The SQL standard [51] has only limited temporal features [52, 53] despite creation of temporal extensions [54, 55] and attempts to add such extensions to the standard [50]. Separate development efforts have implemented temporal query layers on top of existing relational databases [56–68]. These systems typically implement a custom query language or user interface, translate queries into SQL that is appropriate for the underlying database to retrieve “raw” data, and process the retrieved data for temporal pattern finding. Some systems leverage a temporal abstraction ontology [69, 70] for specifying categorizations, classifications and temporal patterns of interest. These systems compute such abstractions as time intervals during which they are found in a patient’s data. The abstractions supported by these systems are similar to the derived variables representing phenotypes above, though using temporal abstraction systems for such phenotype detection has never been reported.
Leveraging phenotypes in quality improvement analytics is inherently experimental. This contrasts with the critical nature of healthcare operations, yet analytics and operations must work together to realize the goals of data-driven quality improvement. Software engineers, analysts and researchers need a sandbox with high performance hardware and software. This sandbox should allow secure retrieval and phenotyping of fully identified large EHR extracts from clinical data warehouses and national datasets. This work is expected to involve lengthy database queries, yet rapid iteration is needed in generating extracts in response to changing requirements. This rapidity is inconsistent with the time-intensive testing required in production clinical databases for new long-running queries. Thus, the sandbox should have a regularly updated data warehouse clone that is a snapshot of the production warehouse but is otherwise independent of the operational environment. It also should support the staging of national datasets such as the UHC CDB. Information produced in the sandbox may support implementing changes in clinical operations and practice as described above. The analytics and operational environments ideally engage in a feedback loop.
The AIW is a healthcare analytics sandbox. Its software substantially extends a temporal abstraction system, PROTEMPA [64, 65], to support data retrieval and phenotype detection in enterprise analytics, a capability that was not previously supported by PROTEMPA’s architecture. Areas of extension include flexible configuration to allow direct access by the software to enterprise data warehouses; a configurable mechanism for transforming data from its source system representation into a common data model; support for specifying abstractions in terms of data as represented in the common model to allow their reuse across datasets; support for leveraging associations between data elements in the common model in abstraction computation; efficient processing of large databases that maintains provenance of how abstractions were computed – the AIW can scale to tens of millions of encounters, corresponding to a multiple orders of magnitude increase in supported data volume as compared with PROTEMPA; and configurable support for importing processed data and found phenotypes into existing analysis, mining and query tools. The main contributions of this work are this software’s extended architecture and open source implementation. While quality improvement analytics has been the driving clinical problem for AIW development, AIW is also deployed in a variety of comparative effectiveness [71, 72] and other ongoing studies that use EHR data including the Minority Health GRID project, which studies genetics associated with hypertension in African American populations.
3 MATERIALS AND METHODS
The AIW data retrieval and phenotype detection software is a framework that programs utilize by calling defined Application Programming Interfaces (APIs). The steps for using the framework are: 1) specify a model of relational database information called a virtual data model (VDM) that is database-agnostic, mappings between the VDM and a database of interest, and database connection information; 2) specify phenotypes of interest as abstractions in a temporal abstraction ontology; 3) specify the rows and the column variables of the delimited file output’s grid in terms of abstractions and raw data elements; and 4) execute a data processing run. The output specification contains the names of selected abstractions and raw data needed to compute the column variables, thus it drives what data are retrieved and which of the abstractions in the ontology are computed. In general, a data modeler specifies the VDM once. An analyst specifies the mappings once with occasional modification as the underlying schema changes. A data modeler modifies the ontology as new or changed phenotypes are needed. An analyst specifies the contents of the delimited file output as the “query” for each processing run.
3.1 Relationship to Existing Software
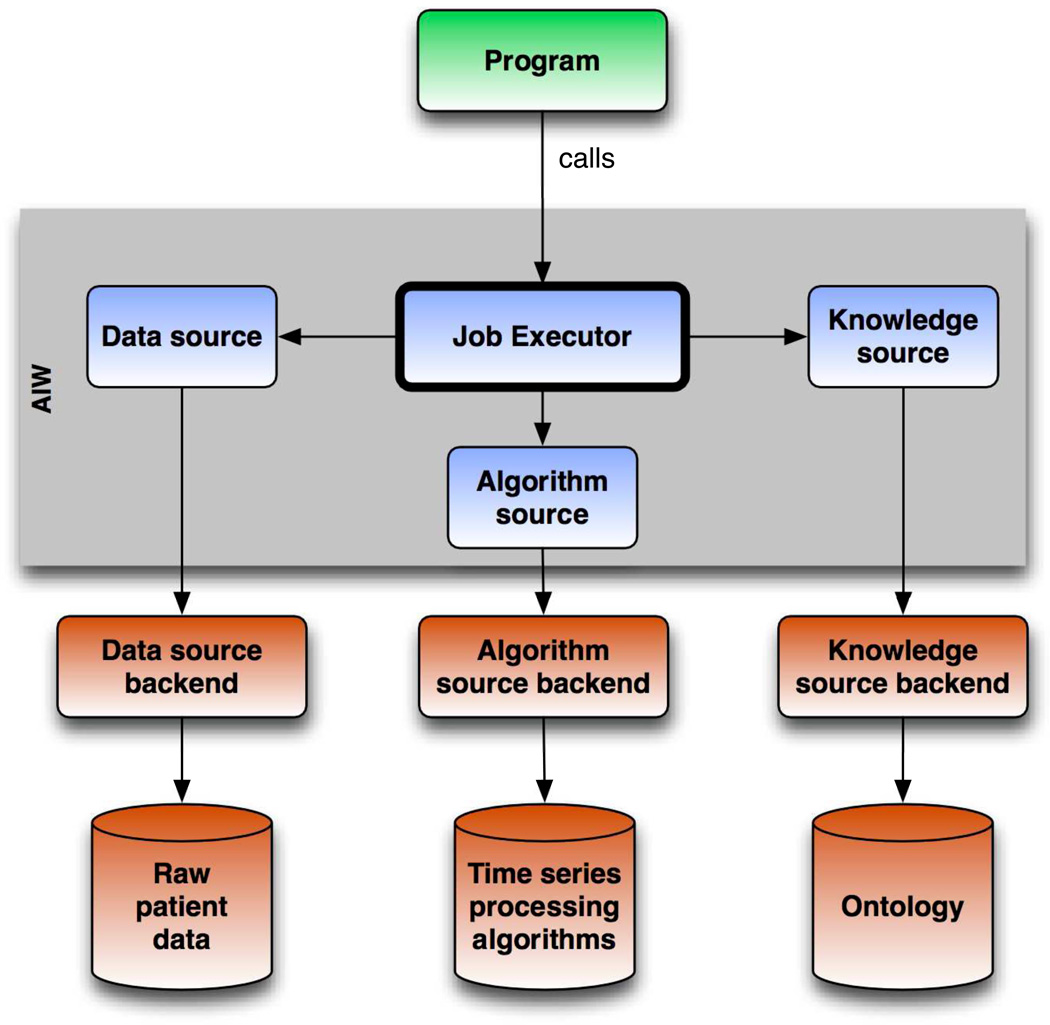
The AIW software includes substantial extensions of PROTEMPA as compared with that described in an earlier publication [64]. These extensions support the data volume, data and phenotype representation and sharing, and output format needs of clinical analytics. Shown in Figure 1, the AIW architecture specifies services providing relational database access (Data Source in Figure 1), definitions of data and abstractions (Knowledge Source), and implementations of algorithms that compute classifications of time series data (Algorithm Source). The Job Executor component (Figure 1) manages processing runs by orchestrating calls to the service providers to retrieve raw data and abstraction definitions, and compute frequency, sequential and other temporal patterns. Extensions as compared with PROTEMPA include 1) a Knowledge Source service provider that allows specifying non-temporal in addition to temporal abstractions in a temporal abstraction ontology, 2) a Data Source service provider that implements the VDM capability, and generates and executes SQL appropriate for databases of interest, 3) re-architected Data Source and Job Executor components that stream data from source systems into temporal abstraction processing, thus allowing tens of millions of hospital encounters to be processed efficiently on standard server hardware configurations and without the need for on-disk caching; and 4) a pluggable mechanism in the Job Executor called the Output Handler for implementing and configuring how PROTEMPA outputs computed intervals and raw data. We describe below an output plugin implementation that generates delimited files. An alternative plugin, a preliminary version of which is described elsewhere [73], loads raw data and computed intervals into an i2b2 project.
Figure 1.
AIW software architecture. The AIW software is a modular framework that extends the PROTEMPA temporal abstraction system for use in detecting clinical phenotypes in EHR data in quality improvement. The Job Executor controls data processing and is supported by the Data Source, Knowledge Source, and Algorithm Source services (blue boxes). Service provider implementations allow access to specific data or knowledge stores (red boxes). Arrows represent dependencies between components, not flow of information. A program (green box) calls the software through a defined API to retrieve data and detect phenotypes of interest.
3.2 Architectural Components
3.2.1 Temporal Abstraction Ontology
The temporal abstraction ontology contains raw data definitions and abstraction definitions. Raw data definitions are specified as instances of three classes: constant (atemporal data such as demographics), observation (data with a timestamp, a temporal granularity [74] and a value such as a vital sign), and event (data with a timestamp or time interval such as a diagnosis). Abstraction definitions are specified as instances of three classes. The low-level abstraction class allows specifying one or more sequential time-stamped observations with values that all meet a specified state threshold or have a slope meeting a trend threshold. The temporal pattern class allows specifying events, observations and/or other temporal abstractions with sequential, overlapping and/or co-occurrence temporal constraints on their order. Temporal patterns may be specified as having the same temporal extent as some or all of the data and/or intervals from which they are derived. The temporal slice class allows specifying the first, second, etc. interval of an event or observation on a timeline. The abstraction classes’ abstractedFrom relationship allows specifying the raw data and/or intervals from which an abstraction is computed. The inverseIsA relationship between instances of the same class allows specifying category abstractions. These relationships enable building up phenotypes that are computed from data in the source database.
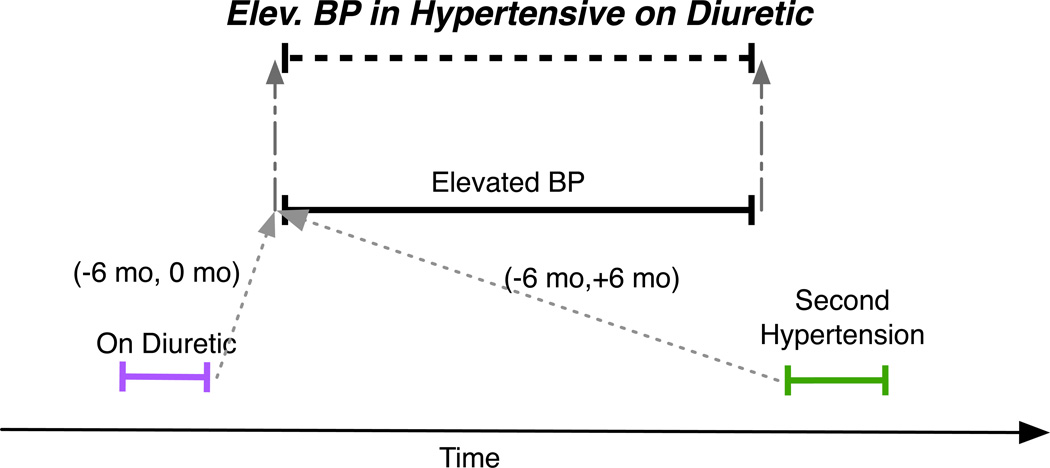
The hypothetical Elevated BP (blood pressure) in Hypertensive on Diuretic phenotype mentioned above can be expressed as a temporal abstraction using this ontology as depicted in Figure 2. Two low-level abstractions, High Systolic BP and High Diastolic BP, are specified as at least two sequential systolic and diastolic blood pressure observations greater than 140 and 90 mm Hg, respectively. A category, Elevated BP, contains High Systolic BP and High Diastolic BP, either of which suggests that the patient has high blood pressure. Two categories, Hypertension and On Diuretic, are specified as hypertension diagnosis codes (e.g., ICD9:401.1) and dispenses of diuretics such as hydrochlorothiazide (HCT Dispense), respectively. A temporal slice, Second Hypertension, is specified as the second Hypertension interval. Elevated BP in Hypertensive on Diuretic is a temporal pattern specified as Elevated BP with an On Diuretic interval within 6 months before the start of Elevated BP and Second Hypertension within 6 months of the start of Elevated BP. Elevated BP in Hypertensive on Diuretic intervals are assigned the same start and finish time as the Elevated BP interval from which they are derived.
Figure 2.
A hypothetical Elev. (elevated) BP (blood pressure) in Hypertensive on Diuretic phenotype is specified as a temporal pattern computed from blood pressure (Elevated BP), diagnosis (Second Hypertension) and medication dispense (On Diuretic) intervals. It has the same endpoints as the contributing Elevated BP interval (gray dashed arrows). Gray dotted arrows denote temporal relationships defined between endpoints of intervals and are labeled with minimum and maximum time constraints. For example, (−6 mo, 0 mo) indicates that the first time point must occur between 0 and 6 months before the second time point.
3.2.2 Abstraction Mechanisms
For each abstraction class, there is a corresponding processing mechanism that the Query Executor uses for computing named intervals that meet an abstraction’s specified criteria. Another processing mechanism computes category intervals based on the presence of the inverseIsA relationships described above. Additional processing mechanisms make other inferences such as concatenating adjacent or closely spaced intervals of the same abstraction to form a single longer interval. See [64, 65] for further details. An example of an Elevated BP in Hypertensive on Diuretic interval is shown in Figure 3. The phenotypes we use in our quality improvement efforts are specified and computed in terms of these abstraction classes and mechanisms. Table 1 shows selected abstraction definitions representing exclusion criteria or populations found to have elevated readmission rates in our analyses (see Results).
Figure 3.
An example of the hypothetical Elev. (elevated) BP (blood pressure) in Hypertensive on Diuretic abstraction. It is computed from blood pressure (Elevated BP abstraction), diagnosis (Hypertension abstraction) and medication dispense (On Diuretic abstraction) intervals. See Methods for details. Each interval has a label with its name and the abstraction mechanism that computed it in parentheses.
Table 1.
Descriptions of the abstractions used in the reported readmissions analyses and the abstraction mechanisms used to compute them. These abstractions are represented and stored in computable form in the temporal abstraction ontology for use in data processing.
| Name | Definition | Abstraction mechanism |
Name | Definition | Abstraction mechanism |
|---|---|---|---|---|---|
| Encounter with subsequent 30-day readmission | A hospital encounter that is followed within 30 days of discharge by the start of another hospital encounter | Temporal pattern | Fourth encounter with subsequent 30-day readmission | The fourth Encounter with subsequent 30-day readmission across all encounters for a patient | Temporal slice |
| Frequent-flier encounter | An encounter after the patient’s Fourth encounter with subsequent 30-day readmission | Temporal pattern | Second readmit | The second Encounter with subsequent 30-day readmission across all encounters for a patient | Temporal slice |
| Myocardial infarction (MI) | Discharge ICD-9 code in 410.* | Categorization | Second MI | The second Myocardial infarction across all encounters for a patient. | Temporal slice |
| Diabetes | Discharge ICD-9 code in 250.* or 648.0* | Categorization | Uncontrolled diabetes | 1) Discharge ICD-9 code in 250.×2, 250.×3, 707.1; -or- 2) Hemoglobin A1c (HbA1c) test result > 9% |
1) Categorization 2) Low-level abstraction |
| Surgical procedure | ICD-9 procedure codes 01-86.99 | Categorization | Heart failure | Heart failure from diagnosis codes –or-Heart failure from BNP | Categorization |
| Heart failure from BNP | B-type natriuretic peptide (BNP) test result 100–300 pg/ml = suggest heart failure is present; 300–600 = mild heart failure; 600–900 = moderate heart failure; or > 900 = severe heart failure | Low-level abstraction | Heart failure from diagnosis codes | Discharge ICD-9 code in 402.01, 402.11, 402.91, 404.01, 404.03, 404.11, 404.13, 404.91, 404.93 or 428.* | Categorization |
| Encounter in last 90 days | Encounter that ends within 90 days of the start of another encounter | Temporal pattern | Encounter in last 180 days | Encounter that ends within 180 days of the start of another encounter | Temporal pattern |
| Chronic kidney disease (CKD) | Discharge ICD-9 code in 581.*, 582.*or 585.* | Categorization | End-stage renal disease (ESRD) | Discharge ICD-9 code 285.21 or 585.6 | Categorization |
| Chemotherapy encounter | Discharge ICD-9 code in V58.1* | Categorization | Radiation therapy encounter | Discharge ICD-9 code V58.0 | Categorization |
| Chemotherapy 180 days before surgery | Chemotherapy encounter that ends within 180 days before a surgical procedure | Temporal pattern | Chemotherapy 365 days before surgery | Chemotherapy encounter that ends within 365 days before a surgical procedure | Temporal pattern |
| Obesity | 1) Discharge ICD-9 code 278.00 or 278.01; -or- 2) Body mass index (BMI) > 30 |
1) Categorization 2) Low-level abstraction |
Stroke | Discharge ICD-9 code in: 430.*, 431.*, 432.9*, 433.01, 433.11, 433.21, 433.31, 433.81, 433.91, 434.00, 434.01, 434.10, 434.90, 434.91, 435.*, or 436.* | Categorization |
| Pressure ulcer | Discharge ICD-9 code 707.0 or 707.2 | Categorization | Methicillin-resistant staph aureus (MRSA) | Discharge ICD-9 code 041.12 or 038.12 | Categorization |
| Sickle cell anemia | Discharge ICD-9 code in 282.6* | Categorization | Sickle cell crisis | Discharge ICD-9 code 282.62 or 282.64 | Categorization |
| Chronic obstructive pulmonary disease | Discharge ICD-9 code in 491.20, 491.21, 491.22, 492.8, 493.20, 493.21, 493.22, 494.0, 494.1, 495.* or 496.* | Categorization | Cancer | Discharge ICD-9 code in 140–208, 209.0, 209.1, 209.2, 209.3, 225.*, 227.3, 227.4, 227.9, 228.02, 228.1, 230.*, 231.*, 232.*, 233.*, 234.*, 236.0, 237.*, 238.4, 238.6, 238.7, 239.6, 239.7, 259.2, 259.8, 273.2, 273.3, 285.22, 288.3, 289.83, 289.89, 511.81, 789.51, 795.06, 795.16, V58.0, V58.1*or V10.* | Categorization |
| Metastasis | Discharge ICD-9 code in 196.*, 197.*, or 198.* | Categorization | Pulmonary hypertension | Discharge ICD-9 code 416.0, 416.1, 416.8 or 416.9 | Categorization |
Asterisks represent ranges of codes (e.g., 428. means 428.0, 428.1, 428.2., …). Italicized text represents an abstraction definition specified elsewhere in the table. Abstraction definitions with temporal features include specifications of frequency over a patient’s entire medical record and temporal patterns relative to the encounter of interest. The abstraction mechanisms are described in Methods. All discharge ICD-9 codes may be primary or secondary unless otherwise specified.
3.2.3 Virtual Data Model and Mappings
The abstractions in the ontology are linked to a VDM, the common model, to support retrieval of raw data from a database of interest. A data modeler specifies a VDM as a Unified Modeling Language (UML) [75] diagram. VDMs support a subset of UML with classes, attributes and associations. Attributes may have a generic type (e.g., string, number) or an enumerated value set. See Figure 4 for an example. Mappings associate via naming convention each VDM class with one or more instances from the ontology that represent the same data. Each raw data definition in the ontology is named as the name of a VDM class concatenated to the value of the class attribute representing the code or concept of interest. For example, in the Elevated BP in Hypertensive on Diuretic example, the ontology would contain an instance VitalSign:SystolicBloodPressure that maps to a VDM class VitalSign with a code attribute that has a value set containing the value SystolicBloodPressure.
Figure 4.
UML class diagram of the readmissions virtual data model. There may be multiple PatientDetails records per Patient to represent changes in address, name, marital status and organ donor status over a lifetime. While birthdate, gender, race and ethnicity should not change, they may be recorded incorrectly or differently over time with no way to validate which is correct, thus we represent them in PatientDetails too. PatientDetails are associated with Encounters to represent the name, address and demographic information at that time. Encounters represent the admit and discharge date (start and finish attributes), the database’s unique identifier for encounters (encounterId attribute), the age of the patient in years at the time of the encounter (age attribute), the location of the encounter down to unit level (healthcareEntity, organization, hospitalPavilion and unit attributes), the National Uniform Billing Committee UB- 04 discharge status code of the encounter (dischargeDisposition attribute), insurance type (insuranceType attribute) and APR-DRG (All Patient Refined Diagnosis Related Group) risk of mortality and severity values. The ICD9:Diagnoses class’ position attribute represents whether a diagnosis is primary or secondary. Laboratory test result values (LAB:LabTest) have attributes for reference range, units of measure and interpretation (high, normal, low, critical). The Nominal VDM data type is a string data type. VDM data types with a name ending in Value are value sets. The AbsoluteTime VDM data type represents a timestamp with its granularity in parentheses.
A data analyst links the VDM and the physical schema of a database of interest by specifying a second set of mappings that allows generation of SQL select statements. These mappings are specified in a Data Source service provider (Figure 1). For each VDM class, the mappings specify a “core” lookup table in the source database where the class is represented in the physical schema. The mappings specify joins if needed to associate a VDM class with attribute values and a unique identifier. Plugins allow altering generated SQL according to the dialects of different database systems. An example of a mapping between a VDM and a physical schema is shown in Figure 5. In addition to these structural transformations, the mappings also allow specifying semantic transforms of coding systems and value sets in the database of interest to those in the VDM. These are specified as delimited files with one line per transformation. The first column represents the code or value in the physical schema, and the second column represents the corresponding code or value in the VDM. The name of the file indicates the VDM class and/or class attribute to which the transformations apply.
Figure 5.
UML class diagram showing how the UHC Clinical Database (CDB) schema maps into the readmissions virtual data model (VDM). The subset of the CDB that was mapped is shown on the left in green, and the relevant subset of the readmissions VDM is shown on the right in blue. Purple arrows show how primary keys in the CDB map to VDM unique identifier attributes (UID). Red arrows show how other attributes in the CDB map to VDM attributes. The Nominal VDM data type is a string data type. VDM data types with a name ending in Value are value sets. The AbsoluteTime VDM data type represents a timestamp with its granularity in parentheses.
3.2.4 Output Handler
The delimited file output handler outputs retrieved data and found intervals (phenotypes) as variable values in a spreadsheet. A data analyst specifies output as a column delimiter, a grouping of data and intervals (e.g., encounter) representing the rows of the grid, and variables representing the columns of the grid. Each variable represents a raw data type or abstraction. Column variables may be defined as the patient id, the start or finish time or duration of intervals of a particular abstraction, a VDM attribute value, the time duration between successive intervals of an abstraction, the frequency with which specified data or intervals occurred or a Boolean value indicating whether a threshold on that frequency was satisfied, or aggregate values computed from numerical observations (last, first, min, max, average). Using the hypertension example above, output might be specified as one encounter per row and a column that contains True or False depending on whether the Elevated BP in Hypertensive on Diuretic abstraction was computed from data recorded during each encounter. See Table 2 for an example output file.
Table 2.
Sample delimited file output. There is one encounter per row. Columns represent variables recorded during the encounter or the patient, or variables representing abstractions found in those data (derived variables). Columns with gray background indicate derived variables specified as the existence or value of the abstraction named in the column header. See Table 1 for abstraction definitions.
| PtIda | EncIdb | LOSc | Discharge Dispositiond |
HeartFailure FromLastBNPe |
Chemo180Days BeforeSurgeryf |
Readmit within 30 dg |
Multiple Readmith |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 7 | HomeSelfCare | T | Yes | F | |
| 2 | 2 | 5 | HomeSelfCare | F | No | F | |
| 3 | 3 | 8 | HomeSelfCare | IndicateMild HeartFailure | F | Yes | F |
| 4 | 4 | 17 | OtherRehabFacility | NoHeartFailure | T | Yes | T |
| 4 | 5 | 19 | LongTermCare Hospital | F | Yes | T | |
| 4 | 6 | 3 | ShortTermHospital | F | Yes | T | |
| 4 | 7 | 6 | HospiceHome | F | Yes | T | |
| 5 | 8 | 3 | HospiceHome | SuggestHeart FailureIsPresent | F | No | F |
| 5 | 9 | 9 | SkilledNursingCare | SuggestHeart FailureIsPresent | F | Yes | F |
| 6 | 10 | 4 | HomeHealthService | T | No | F | |
| 7 | 11 | 7 | HomeSelfCare | F | Yes | F | |
| 8 | 12 | 5 | HomeHealthService | NoHeartFailure | T | No | F |
| 8 | 13 | 19 | HomeHealthService | NoHeartFailure | F | No | F |
| 9 | 14 | 3 | HomeSelfCare | IndicateSevere HeartFailure | F | No | T |
| 10 | 15 | 8 | HomeSelfCare | F | No | T | |
| 10 | 16 | 19 | SkilledNursingCare | IndicateMild HeartFailure | F | Yes | T |
| 11 | 17 | 10 | HomeSelfCare | NoHeartFailure | T | No | F |
| 11 | 18 | 11 | HomeSelfCare | F | Yes | F | |
| 11 | 19 | 22 | Expired | F | No | F |
The patient identifier.
The encounter identifier.
Length of stay in days.
The encounter’s National Uniform Billing Committee UB-04 discharge status code.
Value of the last Heart failure from BNP interval computed from this encounter, if any.
Whether the encounter had a Chemotherapy 180 days before surgery interval computed from it.
Whether the encounter had an Encounter with subsequent 30-day readmission interval computed from it.
Whether the patient had a Second readmit interval computed from the dataset.
3.2.5 Job Execution
An AIW processing job is executed with database connection information, a VDM (such as that shown in Figure 4) and mappings to the database (such as those in Figure 5), abstraction definitions represented in the temporal abstraction ontology (such as those in Table 1 and Figure 2) and an output specification (delimited file or i2b2 import). Inclusion and exclusion criteria, called Filters, may be specified on raw data as defined in the VDM for restricting the date range of interest or limiting retrieved data to those with specified attribute values. Processing flow is shown in Figure 6. The Job Executor traverses the temporal abstraction ontology’s inverseIsA and abstractedFrom relationships, starting from the abstractions specified in the column definitions of the delimited file output (such as those in Table 2). It follows these relationships to the definitions of the raw data from which the abstractions are derived. The raw data definitions’ names are passed into the Data Source, which maps them to a VDM and generates a SQL query. Retrieved result sets are transformed into the structure and semantics of the VDM and streamed into the Job Executor, which performs temporal abstraction finding. The retrieved data and found intervals are then output to a delimited file as the variable values specified in the output specification.
Figure 6.
Flow chart showing processing steps during execution of an AIW job. A user specifies the rows and columns of the delimited file output as described in the text. After starting the job, the AIW determines from this specification what data to retrieve from the source database and what abstractions to compute. It generates and executes SQL queries, and transforms their result sets into the form of the virtual data model (VDM). It computes abstractions as specified in the temporal abstraction ontology (Abstraction definitions in figure), and it generates output. CDW=clinical data warehouse.
4 RESULTS
The AIW software is implemented in Java. It represents the temporal abstraction ontology above in a Protégé [76, 77] knowledge base, and it accesses data and abstraction definitions in the ontology via Protégé’s Java APIs. AIW stores VDMs in a Protégé ontology representing the basic UML described above. We author data definitions and VDMs using Protégé’s ontology editor. Abstraction mechanisms are implemented as rules in the Drools inference engine (www.jboss.org/drools). The AIW environment contains a clone of our institution’s data warehouse and a copy of the UHC CDB. Our AIW deployment is described in Appendix A.
We constructed a VDM for analysis of readmissions, shown in Figure 4, and mappings from it to the schemas of our local data warehouse and the UHC CDB, shown in Figure 5. We specified the abstraction definitions from Table 1 in the temporal abstraction ontology. These represent phenotypes that the project’s clinical investigators believed might be associated with elevated readmission rates, or exclusion criteria for encounters (chemotherapy, radiotherapy, newborn and psychiatric encounters) believed to represent planned or not preventable readmissions. A screenshot of the Protégé user interface displaying the Chemotherapy 180 days before surgery abstraction is shown in Figure 7.
Figure 7.
Screenshot of the temporal abstraction ontology in Protégé, showing the Chemotherapy 180 days before surgery temporal pattern abstraction. The AbstractedFrom slot shows that the temporal pattern is composed of the V58.1 ICD-9 code group and the SurgicalProcedure category of ICD-9 codes (contains all surgical procedure codes). The WithRelations slot contains an instance of the Relation class specifying the temporal constraint between the procedure and chemotherapy encounter codes. The TemporalOffset slot contains an instance of the TemporalOffsets class that, together with the MaxGap, Concatenable and Solid slots, specify that intervals created of this abstraction should have the same temporal extent as the chemotherapy encounter code from which they are derived. The InDataSource slot is unchecked to indicate that this data element should not be searched for in the source database (because it is computed).
We specified output as one encounter per row with columns containing patient id and demographics, admit and discharge dates and discharge status codes, MS-DRG codes and UHC product lines (groups of MS-DRG codes by clinical specialty and subspecialty, see Appendix B for selected product line definitions), and ICD-9 diagnosis and procedure codes. Additional columns specified co-morbidity counts and Boolean variables representing at least n instances of a specified temporal pattern. Other columns specified whether there was a subsequent hospital readmission within 30 days for any cause and diagnosis and other information from the readmit. For analyses using our local data warehouse, we additionally outputted aggregated laboratory test results, vital sign values and the existence of orders for medications from several drug classes.
While these analyses were performed as part of hospital operations, we obtained Institutional Review Board approval to publish them. A simple Java application invoked the AIW software separately for processing our local data warehouse and the UHC CDB. Filter criteria limited processing to hospital encounters between 2006 and 2011; verified, modified or complete vital sign observations; and new, changed or discontinued medication orders. The UHC CDB processing run required 2 GB of heap space and completed in 8 hours including database queries, temporal abstraction processing and output file generation. Local data warehouse processing required 8 GB of heap space and completed in 20 hours. The CDB output contained 17,982,679 encounters in 11,794,310 patients, and the local data warehouse output contained 238,996 encounters in 149,514 patients. While the UHC CDB contains information on a far larger number of patients, the local data warehouse is a much richer dataset that includes laboratory data, medication orders, patient demographic information and a rich set of radiology, pathology and surgical synoptics and free text notes. The output files were passed into Python scripts. The scripts removed encounters with planned or not preventable readmissions (defined above). They computed hospital readmission rates within 30 days (all-cause) for encounters with the phenotypes in Table 1 singly and in pairs. For the UHC extract, they also computed median readmission rates and interquartile range by UHC hospital (using the SciPy library, www.scipy.org).
We present results obtained from UHC CDB in this report; results from the local data warehouse are described elsewhere [78]. Median readmission rates in the UHC extract are shown in Table 3. Most of the shown phenotypes’ rates were elevated as compared with the overall rate (12%). The combination of uncontrolled diabetes and end-stage renal disease had a higher median readmission rate than either alone. Variability across hospitals was high for Sickle-cell anemia. Frequent fliers (>= 4 30-day readmissions, Table 1) had the highest median readmission rate and dominated all other phenotypes. Other analyses are described in Appendix C.
Table 3.
Hospital readmissions within 30 days (all-cause) by selected phenotype (see Table 1) or phenotype combination in the UHC CDB (2006–2011, all hospitals): readmission rate, and median readmission rate and interquartile range for UHC hospitals.
| Population | Number of encounters |
Number of readmissions |
Readmission rate (%) |
Median readmission rate by hospital (%) |
Interquartile range by hospital (%) |
|---|---|---|---|---|---|
| All patients | 15,699,788 | 1,969,858 | 13 | 12 | [10 – 13] |
| Chronic kidney diseasea | 1,706,325 | 385,090 | 23 | 21 | [19 – 23] |
| Heart failurea | 1,731,307 | 343,806 | 20 | 19 | [17 – 21] |
| Diabetesa | 3,149,083 | 542,760 | 17 | 16 | [15 – 18] |
| Cancera | 2,988,904 | 536,855 | 18 | 17 | [16 – 19] |
| Acute Myocardial Infarctiona | 410,335 | 57,310 | 14 | 14 | [12 – 16] |
| Sickle-cell anemiaa | 110,289 | 34,249 | 31 | 25 | [17 – 32] |
| Multiple myocardial infarction (MI) patientb | 195,263 | 69,471 | 36 | 36 | [33 – 40] |
| Metastasisa | 652,198 | 133,583 | 20 | 20 | [17 – 23] |
| End-stage renal diseasea | 663,230 | 178,222 | 27 | 24 | [22 – 28] |
| Uncontrolled diabetes mellitusa | 570,973 | 108,250 | 19 | 18 | [16 – 20] |
| Pressure ulcera | 293,757 | 62,848 | 21 | 20 | [17 – 23] |
| Uncontrolled diabetes mellitus and pressure ulcera | 30,667 | 7,170 | 23 | 22 | [18 – 26] |
| Uncontrolled diabetes mellitus and End-stage renal diseasea | 67,617 | 19,381 | 29 | 27 | [23 – 31] |
| Sickle cell crisisa | 81,049 | 27,849 | 34 | 27 | [19 – 36] |
| Methicillin-resistant Staph aureus (MRSA) infectiona | 107,119 | 18,525 | 17 | 17 | [13 – 19] |
| Stroke and MRSAa | 2,054 | 346 | 17 | 12 | [0 – 21] |
| Frequent-flier encountera | 455,550 | 202,470 | 44 | 43 | [39 – 45] |
| Chemotherapy 180 days before surgerya,c | 51,186 | 15,789 | 31 | 31 | [26 – 35] |
| Chemotherapy 365 days before surgerya,c | 60,507 | 18,305 | 30 | 30 | [26 – 34] |
| Encounter in last 180 daysa | 3,932,748 | 922,926 | 23 | 23 | [20 – 24] |
| Encounter in last 90 daysa | 3,150,364 | 792,867 | 25 | 24 | [21 – 26] |
| CKD and frequent-flier encounter a | 139,255 | 67,532 | 48 | 44 | [39 – 47] |
| Frequent-flier encounter in Multiple MI patienta | 25,193 | 12,092 | 48 | 44 | [38 – 50] |
| MI and MRSAa | 31,185 | 7,237 | 23 | 22 | [18 – 26] |
Row represents encounters from which the specified phenotype was computed.
Row represents all encounters in patients who express the specified phenotype.
Rates of readmission subsequent to the surgical encounter are reported.
5 DISCUSSION
We have created an open source healthcare analytics environment, the AIW, that was launched to address our institution’s need to understand the causes of hospital readmissions locally and nationally. The environment’s software (Figure 1) employs temporal abstraction to allow specifying phenotypes in terms of EHR data as categories of codes and concepts, classifications of numerical data, and temporal patterns and relationships. Phenotypes are specified in terms of a database-agnostic model (the VDM) that is mapped to a physical database schema, thus AIW phenotypes may be specified once and reused with multiple databases. We successfully tested these capabilities as part of an operational effort in analyzing hospital readmissions in our local data warehouse and the national UHC CDB. We expect such comparisons of local data to multi-institutional databases also to be of interest elsewhere. The AIW aims to support exploratory analyses of the causes of low metric scores and clinical events of interest such as readmissions. This capability is complementary to that of emerging commercial analytics platforms that have focused primarily on metric computation and data integration rather than exploratory data analysis (see Background). A platform such as AIW could process integrated data sets created by such systems.
The use of temporal abstraction in AIW has benefits beyond enabling temporal pattern recognition. It enables AIW to maintain data provenance. AIW currently records the data from which a derived interval was computed and metadata specifying what abstraction mechanism was used to compute it. The software maintains consistent unique identifiers for data values across processing runs, assuming the source database maintains such identifiers. It also records unique identifiers for the source of each data value (see Methods). We plan to support maintaining a persistent store of retrieved data, intervals and abstraction definitions that is updatable and allows maintaining history. The ontological storage of data definitions and abstraction definitions enables efficient and systematic testing of the impact of changes to a data or abstraction definition on derived information generated by AIW. We are creating a framework for running such tests [79].
The use of temporal abstraction also allows us to leverage clinical data in specifying phenotypes. Billing codes have the benefit of being widely available nationally in standardized form, and they enabled the UHC analyses above. However, they may not represent diagnoses at the desired level of detail, they may not accurately represent the temporal extent of disease, and they represent only conditions that were billed for. They also may only be recorded after a patient’s discharge, thus rendering them unavailable for decision support during the current hospital stay. The AIW mitigates these issues by allowing creation of custom categories of such codes, specification of heuristics for inferring clinical events from billing codes based on their temporal sequence relationships and frequencies (the temporal abstractions described above), and leveraging of clinical data values in combination with billing codes when clinical data is available. We have defined and utilized a substantially broader set of clinical and socio-cultural characteristics including information abstracted from geo-location of patient addresses coupled with census and American Community Survey (http://www.census.gov/acs/www/) data in the development of predictive models of readmission risk, described elsewhere [78].
Making temporal abstraction the basis for representing and computing phenotypes has several benefits for phenotype maintenance. Temporal abstraction enables sharing of phenotypes across sites with heterogeneous data warehouses and schemas. It forces the creation of phenotype definitions to adhere to a relatively small but flexible set of templates that we expect will speed development and reduce bugs. It enables sharing these templates and the corresponding mechanisms for computing phenotypes. In most data warehouse environments, defining and computing complex phenotypes involves implementing and deploying stored procedures that are run during the ETL cycle. An ontology organizes phenotype definitions in a far more searchable structure than attempting to organize hundreds of text files (or more) containing database-specific stored procedure and query code on a file system. AIW has greatly enhanced our ability to generate datasets for our readmissions analyses with consistent numerators and denominators because it makes repeating analyses with slight variations straightforward.
The AIW outputs data and expressed phenotypes as delimited files (Table 2). This capability allowed us to use off-the-shelf analysis software (SciPy) to compute readmission rates for disease, co-morbidity and other phenotypes (Table 1) singly and in combination (Table 3). These analyses and others described in Appendix C yielded “hot spots” [80] in readmission rates in our local and national datasets. To turn a hot spot into actionable information, typical follow up includes analyses of statistical significance, identification of changes if any in the affected population’s readmission rate over time, and collaborations with clinical care teams to investigate practice patterns. As part of these activities, we analyze AIW output in SAS (SAS Institute Inc., Cary, NC), R (www.r-project.org) and Excel (Microsoft Corp., Redmond, WA).
Similar phenotypes are needed in comparative effectiveness studies, and the AIW is being extended for use in these investigations (see above and [73]). The AIW’s capability for phenotype reuse could allow broad evaluations of phenotypes’ sensitivity and specificity. For phenotypes with known performance characteristics, our goal is for institutions to be able to apply such phenotypes without having to undertake substantial software development or configuration beyond an initial investment in specifying appropriate mappings from their local database(s) to a VDM. A mechanism for sharing AIW phenotypes, which is future work, will utilize emerging clinical phenotype repositories such as pheKB (http://phekb.org) that support collaborating on and publishing phenotype specifications in textual and ultimately computable form. As an interim step, computable representations of the phenotypes shown in Table 1 are provided in an ontology that is shipped with the open source distribution of the AIW software (see Introduction for access information). Open source tools such as AIW in combination with such repositories could provide a foundation for the comparative effectiveness community to build robust clinical phenotyping capabilities.
The AIW software’s predecessor, PROTEMPA, was evaluated in proof-of-concept form [64]. AIW extends PROTEMPA into a production-quality system that is deployed at our institution in support of clinical analytics. We created our own SQL generation solution rather than leverage something off-the-shelf [81] in order to tailor generated queries for AIW’s relatively limited set of database access patterns and to support streaming data from the database into the software. This allowed efficient retrieval of over ten million patients’ data in our analyses above. While we believe our solution supports the most common types of transforms needed for accessing typical star and dimensional modeling-based schemas, we expect new features to be required to support data warehouses and databases more broadly. While Protégé meets our current needs as an ontology server, AIW’s service provider-based architecture (Figure 1) will allow us to migrate to a more complete ontology server platform with high performance support for editing and retrieval of large subsets of ontologies, automated solutions to maintaining up-to-date versions of standard terminologies, and support for managing the ontology development, test and release lifecycle. The AIW’s support for outputting data as flat files enables it to fit into existing data analysis workflows. The common data model’s representation of significant associations between data enables the flat file output mechanism to support grouping data by those associations (e.g., data and phenotypes grouped by hospital encounter, by organization). Much work remains to extend the software for deployment elsewhere. Making AIW available as open source is expected to provide an avenue for enhancing the software further for adoption by other groups.
Clinical and operational studies require cleansing of EHR extracts to resolve inconsistencies, interpret missing values, identify obviously invalid data, and harmonize alternative representations of the same information [33, 82]. Data cleansing is laborious and could be automated substantially by software such as the AIW. The AIW’s data mapping capability supports harmonizing multiple data representations into a single VDM class, and it allows mapping null values to a specific interpretation. Value range and other validation checks are not yet implemented. Clinical information relevant in quality improvement is embedded in text reports, and while we have a capability planned to extract concepts from free-text, such support has not been implemented.
The AIW’s current implementation has met the needs of its original driving problem of enabling exploratory analyses of the causes of hospital readmissions. It has provided a maintainable and flexible mechanism for representing clinical phenotypes that are of interest to our stakeholders. It has enabled a highly repeatable process for generating datasets and analyses that ensures consistency of data transformations using two databases with markedly different schemas (local data warehouse and UHC CDB). The primary limitation of the current implementation is the need for software engineers and data modelers fluent in ontologies to configure the system. We are working with our local IT departments on how to extend the software to enable its use by a standard complement of data analysts as well as by technologysavvy clinical investigators. This work, supported by the CardioVascular Research Grid [83], will include development of web-based user interfaces for specifying phenotypes, configuring database access, configuring delimited output, and managing job execution. A preliminary release, called Eureka! Clinical Analytics, is available in demonstration form at https://eureka.cci.emory.edu and as an Amazon Machine Image (AMI) for deployment in the Amazon Elastic Compute Cloud (http://aws.amazon.com/ec2/). User interfaces for specifying temporal patterns and relationships are challenging to build [53, 61, 74, 84].
6 CONCLUSIONS
The AIW enables quality improvement studies to leverage phenotypes expressed in EHR data as categories of codes and concepts and as frequency, sequential and other temporal relationships. It supports processing and comparison of heterogeneous data sources through capabilities to specify phenotypes in database-agnostic form and transform retrieved data into that form. These features allowed a large-scale comparison of readmitted patients at our institution with those in a national dataset. The AIW’s temporal abstraction capability provides significant and needed flexibility in specifying phenotypes in quality improvement. Its data mapping capability may ultimately provide a mechanism for cross-institutional clinical phenotype reuse. The availability of software that allows straightforward deployment of phenotype repositories may substantially accelerate the application of EHR data in comparative and clinical effectiveness.
Highlights.
We propose a temporal abstraction-based methodology for healthcare analytics.
We implemented this methodology in software and deployed it in production.
We identified in EHR data clinical phenotypes associated with hospital readmissions.
Temporal abstraction is a scalable and flexible method for clinical phenotyping.
Clinical phenotyping helps leverage EHR data in quality improvement analyses.
ACKNOWLEDGMENTS
This work was supported in part by PHS Grant UL1 RR025008, KL2 RR025009 and TL1 RR025010 from the CTSA program, NIH, NCRR; NHLBI grant R24 HL085343; NIH/ARRA grant 325011.300001.80022; M01 RR-00039 from the GCRC program, NIH, NCRR; and Emory Healthcare.
ROLE OF THE FUNDING SOURCES
Other than the named authors, the funders of this work had no role in study design, collection, analysis or interpretation of data, writing this manuscript, or in the decision to submit the manuscript for publication.
APPENDICES
Appendix A AIW Operational Environment
The AIW operates in Health Insurance Portability and Accountability Act (HIPAA)- compliant network zones at our institution that are accessible only by virtual private network (VPN) or Citrix (Citrix Systems, Inc., Fort Lauderdale, FL) clients. Two Oracle 11g database servers contain a clone of the Emory Clinical Data Warehouse that is updated monthly, 5 years of data from the UHC CDB that is updated quarterly and several data marts. Three Linux application servers provide for running AIW software, statistical analysis, data mining, and data file archiving. One Windows (Microsoft Corp., Redmond, WA) server provides secure remote access to Windows applications.
We download UHC CDB data from www.uhc.edu as delimited files and bulk import the files into a database schema with a simple script. Our local data warehouse clone is updated using tools supplied by Oracle. We have created data source service providers for both with appropriate mappings to a VDM for analyses of readmissions (see Figure 5 for selected structural mappings from the UHC CDB to the readmissions VDM). We validate AIW-generated SQL and compare selected records in the result sets with the source database tables by manual inspection.
Appendix B
UHC Product Lines
Table A.1.
Definitions of selected UHC product lines are in the table below. For other product line definitions, see http://www.uhc.edu.
| MS-DRG Codes in Selected UHC Product Lines | |||
|---|---|---|---|
| Medicine General | Surgery General | Medical Oncology |
Gastroenterology |
| 075–079, 094–099, 152–153, 175–179, 186–206, 208, 294–295, 299–301, 304–305, 312–313, 383–384, 539–541, 551–552, 592–594, 600–605, 637–645, 682–685, 689–690, 698–700, 808–816, 862–869, 871–872, 915–918, 922–923, 947–951 | 239–241, 255–257, 264, 326–358, 405–425, 579–581, 584–585, 614–621, 625–630, 799–804, 853–858, 876, 901–903, 939–941, 981–983, 987–989 | 054–055, 180–182, 374–376, 435–437, 542–544, 597–599, 686–688, 834–849 | 368–373, 377–382, 385–395, 432–434, 438–446 |
Appendix C
Additional Readmissions Analyses of the UHC CDB (2006–2011, All Hospitals)
Of the 1,969,858 inpatient encounters that were followed by a 30-day readmission (Table 3), 238,542 had a National Uniform Billing Committee UB-04 discharge status code (www.nubc.org) representing a lower intensity of post-discharge care as compared with the discharge status code of the subsequent readmit (rows with gray background in Table A.2). For example, 140,109 encounters with a discharge status code of Home self-care were followed by a readmission within 30 days with a discharge status code of Home health service. The Home selfcare code indicates that the patient was sent home and was to receive no organized care prior to their scheduled clinic visit for follow-up. The Home health service code indicates that the patient was to be seen at home on some schedule by a nurse or other clinician that is part of an organized home health service agency. These readmissions represent a population that might have benefitted from more intensive follow-up following the initial encounter.
Table A.2.
Top 10 UB-04 discharge status code pairs for hospital encounters (Index encounter) with a subsequent all-cause readmission within 30 days (Readmit encounter) in the UHC CDB (2006–2011, all hospitals): overall count with percent. Pairs with gray background represent those for which the index encounter’s discharge code represents a lower intensity of post-discharge care than the readmit’s discharge code.
| UB-04 discharge status code description (code in parentheses) | Number of pairs |
Percent of pairs |
|
|---|---|---|---|
| Index encounter | Readmit encounter | ||
| Home self-care (01) | Home self-care (01) | 909,219 | 46.2 |
| Home health service (06) | Home health service (06) | 151,247 | 7.7 |
| Home self-care (01) | Home health service (06) | 140,109 | 7.1 |
| Nursing facility Medicare (03) | Nursing facility Medicare (03) | 119,585 | 6.1 |
| Home health service (06) | Home self-care (06) | 96,595 | 4.9 |
| Other rehab facility (62) | Home health service (06) | 44,924 | 2.3 |
| Other rehab facility (62) | Home self-care (01) | 40,501 | 2.1 |
| Home self-care (01) | Nursing facility Medicare (03) | 40,204 | 2.0 |
| Home health service (06) | Nursing facility Medicare (03) | 29,932 | 1.5 |
| Home self-care (01) | Expired in hospital not Medicare or CHAMPUS Hospice (20) | 28,297 | 1.4 |
| Grand total | 1,600,613 | 81.3 | |
We binned patients into percentile bands by total number of hospital days and calculated the readmission rate for each band, shown in Table A.3. Patients in the upper quartile accounted for 84% of readmissions, and the patients in the 95th percentile and above accounted for 44% of readmissions. This suggests that such “frequent stayers” should receive further study for readmissions reduction efforts.
Table A.3.
The relationship between cumulative hospital days and rate of readmission within 30 days (all-cause) in the UHC CDB (2006–2011, all hospitals). Patients were binned into percentile bands by their cumulative hospital days. Planned readmissions (defined in Results), newborn encounters and psychiatric encounters were included in this analysis. Readmissions are clustered in the upper quartile.
| Percentile band |
Hospital days |
Max length of stay |
Number of encounters |
Number of readmissions |
Readmission rate |
Percent of overall readmissions |
|---|---|---|---|---|---|---|
| 0–10 | 1,611,384 | 1 | 1,611,384 | 87 | 0.0% | 0.0% |
| 10–40 | 3,583,236 | 2 | 1,873,606 | 20,480 | 1.1% | 1.0% |
| 40–55 | 8,265,025 | 4 | 2,777,131 | 78,616 | 2.8% | 4.0% |
| 55–65 | 3,388,305 | 5 | 896,237 | 50,695 | 5.7% | 2.6% |
| 65–75 | 8,530,036 | 8 | 1,857,092 | 158,255 | 8.5% | 8.0% |
| 75–85 | 10,010,465 | 13 | 1,717,041 | 242,162 | 14.1% | 12.3% |
| 85–95 | 20,283,704 | 30 | 2,568,866 | 562,540 | 21.9% | 28.6% |
| 95–98 | 17,396,822 | 69 | 1,629,260 | 516,485 | 31.7% | 26.2% |
| 99–100 | 11,439,212 | 1,401 | 769,171 | 340,538 | 44.3% | 17.3% |
| Grand total | 84,508,189 | -- | 15,699,788 | 1,969,858 | -- | 100.0% |
We assessed the relative frequency with which encounters are assigned MS-DRG codes and UHC product lines (Appendix B) in frequently readmitted versus not frequently readmitted patients. The Medicine General product line accounted for a large patient volume in general (Figure A.1a). The frequency of encounters assigned to the Surgery General product line decreased as the number of readmits increased (Figure A.1a). For hospital encounters in the Surgery General product line, two-thirds of subsequent readmissions within 30 days were assigned to product lines other than Surgery General (data not shown). The Med Oncology product line exhibited the reverse trend with the number of encounters assigned to it increasing as the number of readmits increased (Figure A.1a). Within the Medicine General product line, encounters tended to be assigned the Red Blood Cell Disorders w/o MCC MS-DRG code (code 812), which includes Sickle-cell anemia (overall readmission rate shown in Table 3), with greater frequency in frequently readmitted patients than in not frequently readmitted patients (Figure A.1b).
Figure A.1.
The percentage of hospital encounters in patients with exactly 1, 2, 3, etc. readmissions that were assigned to selected product lines (a) or MS-DRG codes (b) in the UHC CDB (2006–2011, all hospitals). These plots distinguish types of encounters that tend to occur in patients with few readmissions versus many readmissions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: AIW – Analytic Information Warehouse; UHC – University HealthSystem Consortium; CDB – UHC Clinical Database; CDW – Emory Clinical Data Warehouse.
DISCLOSURE STATEMENT
Dr. Saltz is on the Scientific Advisory Board of a company called Appistry.
Contributor Information
Andrew R. Post, Email: arpost@emory.edu.
Tahsin Kurc, Email: tkurc@emory.edu.
Sharath Cholleti, Email: sharath.cholleti@emory.edu.
Jingjing Gao, Email: jingjing.gao@emory.edu.
Xia Lin, Email: xia.lin@emory.edu.
William Bornstein, Email: wbornst@emory.edu.
Dedra Cantrell, Email: dedra.cantrell@emoryhealthcare.org.
David Levine, Email: levine@uhc.edu.
Sam Hohmann, Email: hohmann@uhc.edu.
Joel H. Saltz, Email: jhsaltz@emory.edu.
REFERENCES
- 1.Kocher R, Emanuel EJ, DeParle NA. The Affordable Care Act and the future of clinical medicine: the opportunities and challenges. Ann Intern Med. 2010;153:536–539. doi: 10.7326/0003-4819-153-8-201010190-00274. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal D. Implementation of the federal health information technology initiative. N Engl J Med. 2011;365:2426–2431. doi: 10.1056/NEJMsr1112158. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal D. Wiring the health system--origins and provisions of a new federal program. N Engl J Med. 2011;365:2323–2329. doi: 10.1056/NEJMsr1110507. [DOI] [PubMed] [Google Scholar]
- 4.National Quality Measures Clearinghouse | Tutorial on Quality Measures. Agency for Healthcare Research and Quality. [(accessed 2012 Apr 3)]; doi: 10.1080/15360280802537332. http://www.qualitymeasures.ahrq.gov/tutorial/index.aspx. [DOI] [PubMed]
- 5.Shahian DM, Wolf RE, Iezzoni LI, Kirle L, Normand SL. Variability in the measurement of hospital-wide mortality rates. N Engl J Med. 2010;363:2530–2539. doi: 10.1056/NEJMsa1006396. [DOI] [PubMed] [Google Scholar]
- 6.Bueno H, Ross JS, Wang Y, Chen J, Vidan MT, Normand SL, et al. Trends in length of stay and short-term outcomes among Medicare patients hospitalized for heart failure, 1993–2006. JAMA. 2010;303:2141–2147. doi: 10.1001/jama.2010.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kansagara D, Englander H, Salanitro A, Kagen D, Theobald C, Freeman M, et al. Risk prediction models for hospital readmission: a systematic review. JAMA. 2011;306:1688–1698. doi: 10.1001/jama.2011.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giordano LA, Elliott MN, Goldstein E, Lehrman WG, Spencer PA. Development, implementation, and public reporting of the HCAHPS survey. Med Care Res Rev. 2010;67:27–37. doi: 10.1177/1077558709341065. [DOI] [PubMed] [Google Scholar]
- 9.Fung CH, Lim YW, Mattke S, Damberg C, Shekelle PG. Systematic review: the evidence that publishing patient care performance data improves quality of care. Ann Intern Med. 2008;148:111–123. doi: 10.7326/0003-4819-148-2-200801150-00006. [DOI] [PubMed] [Google Scholar]
- 10.Lindenauer PK, Remus D, Roman S, Rothberg MB, Benjamin EM, Ma A, et al. Public reporting and pay for performance in hospital quality improvement. N Engl J Med. 2007;356:486–496. doi: 10.1056/NEJMsa064964. [DOI] [PubMed] [Google Scholar]
- 11.Medicare program; hospital inpatient prospective payment systems for acute care hospitals and the long-term care hospital prospective payment system and fiscal year 2013 rates; hospitals' resident caps for graduate medical education payment purposes; quality reporting requirements for specific providers and for ambulatory surgical centers. Final rule. Federal register. 2012;77:53257–53750. [PubMed] [Google Scholar]
- 12.Mouttham A, Peyton L, Kuziemsky C. Leveraging Performance Analytics to Improve Integration of Care. Proceedings of the 3rd Workshop on Software Engineering in Health Care. 2011:56–62.
- 13.Gregor S. The Nature of Theory in Information Systems. MIS Quarterly. 2006;30:611–642. [Google Scholar]
- 14.Brown DE. Introduction to data mining for medical informatics. Clin Lab Med. 2008;28:9–35. doi: 10.1016/j.cll.2007.10.008. v. [DOI] [PubMed] [Google Scholar]
- 15.Shmueli G, Koppius O. Predictive Analytics in Information Systems Research. MIS Quarterly. 2011;35:553–572. [Google Scholar]
- 16.Wharam JF, Weiner JP. The promise and peril of healthcare forecasting. Am J Manag Care. 2012;18:e82–e85. [PubMed] [Google Scholar]
- 17.Jha AK, DesRoches CM, Campbell EG, Donelan K, Rao SR, Ferris TG, et al. Use of Electronic Health Records in U.S. Hospitals. N Engl J Med. 2009;360:1628–1638. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 18.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE--An integrated standards-based translational research informatics platform; AMIA Annu Symp Proc; 2009. pp. 391–395. [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17:131–135. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyman JA, Scully K, Harrison JH., Jr. The development of health care data warehouses to support data mining. Clin Lab Med. 2008;28:55–71. doi: 10.1016/j.cll.2007.10.003. vi. [DOI] [PubMed] [Google Scholar]
- 22.Kamal J, Liu J, Ostrander M, Santangelo J, Dyta R, Rogers P, et al. Information warehouse - a comprehensive informatics platform for business, clinical, and research applications; AMIA Annu Symp Proc; 2010. pp. 452–456. [PMC free article] [PubMed] [Google Scholar]
- 23.Wade TD, Hum RC, Murphy JR. A Dimensional Bus model for integrating clinical and research data. J Am Med Inform Assoc. 2011;18(Suppl 1):i96–i102. doi: 10.1136/amiajnl-2011-000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khuri SF. The NSQIP: a new frontier in surgery. Surgery. 2005;138:837–843. doi: 10.1016/j.surg.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Lipscomb J, Gillespie TW. State-level cancer quality assessment and research: building and sustaining the data infrastructure. Cancer J. 2011;17:246–256. doi: 10.1097/PPO.0b013e3182296422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brindis RG, Fitzgerald S, Anderson HV, Shaw RE, Weintraub WS, Williams JF. The American College of Cardiology-National Cardiovascular Data Registry (ACC-NCDR): building a national clinical data repository. J Am Coll Cardiol. 2001;37:2240–2245. doi: 10.1016/s0735-1097(01)01372-9. [DOI] [PubMed] [Google Scholar]
- 27.About UHC. UHC. [(accessed 2012 May 3)];2012 http://www.uhc.edu/12443.htm. [Google Scholar]
- 28.Clinical Data Base/Resource Manager. UHC. [(accessed 2012 May 3)];2012 http://www.uhc.edu/11536.htm. [Google Scholar]
- 29.Chan KS, Fowles JB, Weiner JP. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev. 2010;67:503–527. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 30.Kahn MG, Ranade D. The impact of electronic medical records data sources on an adverse drug event quality measure. J Am Med Inform Assoc. 2010;17:185–191. doi: 10.1136/jamia.2009.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benin AL, Fenick A, Herrin J, Vitkauskas G, Chen J, Brandt C. How good are the data? Feasible approach to validation of metrics of quality derived from an outpatient electronic health record. Am J Med Qual. 2011;26:441–451. doi: 10.1177/1062860611403136. [DOI] [PubMed] [Google Scholar]
- 32.Parsons A, McCullough C, Wang J, Shih S. Validity of electronic health record-derived quality measurement for performance monitoring. J Am Med Inform Assoc. 2012 doi: 10.1136/amiajnl-2011-000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hripcsak G, Knirsch C, Zhou L, Wilcox A, Melton G. Bias associated with mining electronic health records. J Biomed Discov Collab. 2011;6:48–52. doi: 10.5210/disco.v6i0.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Serv Res. 2005;40:1620–1639. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathak J, Wang J, Kashyap S, Basford M, Li R, Masys DR, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled terminologies: the eMERGE Network experience. J Am Med Inform Assoc. 2011;18:376–386. doi: 10.1136/amiajnl-2010-000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea S, Pathak J, Savova G, Oniki TA, Westberg L, Beebe CE, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: The SHARPn project. J Biomed Inform. 2012 doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.eMERGE. eMERGE Network Phenotype Library. eMERGE. [(accessed 2012 Apr 3)]; http://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Library_of_Phenotype_Algorithms. [Google Scholar]
- 38.Pathak J, Pan H, Wang J, Kashyap S, Schad PA, Hamilton CM, et al. Evaluating Phenotypic Data Elements for Genetics and Epidemiological Research: Experiences from the eMERGE and PhenX Network Projects. AMIA Summits Transl Sci Proc. 2011:41–45. [PMC free article] [PubMed] [Google Scholar]
- 39.Apache License, Version 2.0. [(accessed 2012 Apr 3)];The Apache Software Foundation. 2004 http://www.apache.org/licenses/LICENSE-2.0.html. [Google Scholar]
- 40.Murphy SN, Morgan MM, Barnett GO, Chueh HC. Optimizing healthcare research data warehouse design through past COSTAR query analysis; Proc AMIA Symp; 1999. pp. 892–896. [PMC free article] [PubMed] [Google Scholar]
- 41.Kimball R, Ross M. The Data Warehouse Toolkit: The Complete Guide to Dimensional Modeling. 2nd ed. New York: Wiley Computer Publishing; 2002. [Google Scholar]
- 42.Ferranti JM, Langman MK, Tanaka D, McCall J, Ahmad A. Bridging the gap: leveraging business intelligence tools in support of patient safety and financial effectiveness. J Am Med Inform Assoc. 2010;17:136–143. doi: 10.1136/jamia.2009.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zekry D, Loures Valle BH, Graf C, Michel JP, Gold G, Krause KH, et al. Prospective comparison of 6 comorbidity indices as predictors of 1-year post-hospital discharge institutionalization, readmission, and mortality in elderly individuals. J Am Med Dir Assoc. 2012;13:272–278. doi: 10.1016/j.jamda.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, et al. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100:1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- 45.Levy WC, Mozaffarian D, Linker DT, Sutradhar SC, Anker SD, Cropp AB, et al. The Seattle Heart Failure Model: prediction of survival in heart failure. Circulation. 2006;113:1424–1433. doi: 10.1161/CIRCULATIONAHA.105.584102. [DOI] [PubMed] [Google Scholar]
- 46.Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D'Agostino RB., Sr. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med. 2007;167:1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 47.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 48.Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 49.Conway M, Berg RL, Carrell D, Denny JC, Kho AN, Kullo IJ, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms; AMIA Annu Symp Proc; 2011. pp. 274–283. [PMC free article] [PubMed] [Google Scholar]
- 50.Adlassnig KP, Combi C, Das AK, Keravnou ET, Pozzi G. Temporal representation and reasoning in medicine: research directions and challenges. Artif Intell Med. 2006;38:101–113. doi: 10.1016/j.artmed.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Elmasri R, Navathe SB. Fundamentals of Database Systems. 3rd ed. New York: Addison- Wesley; 2000. [Google Scholar]
- 52.SQL:2011 (ISO/IEC 9075-1:2011) International Organization for Standardization (ISO) 2011 http://www.iso.org/iso/search.htm?qt=9075&searchSubmit=Search&sort=rel&type=simple&published=true (accessed 2012 Apr 3) [Google Scholar]
- 53.Plaisant C, Lam S, Shneiderman B, Smith MS, Roseman D, Marchand G, et al. Searching electronic health records for temporal patterns in patient histories: a case study with Microsoft Amalga; AMIA Annu Symp Proc; 2008. pp. 601–605. [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connor MJ, Tu SW, Musen MA. The Chronus II Temporal Database Mediator; Proc AMIA Symp; 2002. pp. 567–571. [PMC free article] [PubMed] [Google Scholar]
- 55.Das AK, Musen MA. SYNCHRONUS: A Reusable Software Module for Temporal Integration; Proc AMIA Symp; 2002. pp. 195–199. [PMC free article] [PubMed] [Google Scholar]
- 56.Nigrin DJ, Kohane IS. Temporal expressiveness in querying a time-stamp--based clinical database. J Am Med Inform Assoc. 2000;7:152–163. doi: 10.1136/jamia.2000.0070152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dorda W, Gall W, Duftschmid G. Clinical data retrieval: 25 years of temporal query management at the University of Vienna Medical School. Methods Inf Med. 2002;41:89–97. [PubMed] [Google Scholar]
- 58.Stacey M, McGregor C. Temporal abstraction in intelligent clinical data analysis: A survey. Artif Intell Med. 2007;39:1–24. doi: 10.1016/j.artmed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Martins SB, Shahar Y, Goren-Bar D, Galperin M, Kaizer H, Basso LV, et al. Evaluation of an architecture for intelligent query and exploration of time-oriented clinical data. Artif Intell Med. 2008;43:17–34. doi: 10.1016/j.artmed.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.German E, Leibowitz A, Shahar Y. An architecture for linking medical decision-support applications to clinical databases and its evaluation. J Biomed Inform. 2009;42:203–218. doi: 10.1016/j.jbi.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 61.Klimov D, Shahar Y, Taieb-Maimon M. Intelligent visualization and exploration of time-oriented data of multiple patients. Artif Intell Med. 2010;49:11–31. doi: 10.1016/j.artmed.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 62.O'Connor MJ, Shankar RD, Parrish DB, Das AK. Knowledge-level querying of temporal patterns in clinical research systems. Stud Health Technol Inform. 2007;129:311–315. [PubMed] [Google Scholar]
- 63.O'Connor MJ, Shankar RD, Parrish DB, Das AK. Knowledge-data integration for temporal reasoning in a clinical trial system. Int J Med Inform. 2009;78(Suppl 1):S77–S85. doi: 10.1016/j.ijmedinf.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 64.Post AR, Harrison JH., Jr. PROTEMPA: A Method for Specifying and Identifying Temporal Sequences in Retrospective Data for Patient Selection. J Am Med Inform Assoc. 2007;14:674–683. doi: 10.1197/jamia.M2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Post AR, Sovarel AN, Harrison JH. Abstraction-based temporal data retrieval for a Clinical Data Repository; AMIA Annu Symp Proc; 2007. pp. 603–607. [PMC free article] [PubMed] [Google Scholar]
- 66.Huser V, Narus SP, Rocha RA. Evaluation of a flowchart-based EHR query system: a case study of RetroGuide. J Biomed Inform. 2010;43:41–50. doi: 10.1016/j.jbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Combi C, Keravnou-Papailiou E, Shahar Y. Temporal Information Systems in Medicine. New York: Springer; 2010. [Google Scholar]
- 68.Combi C, Pozzi G, Rossato R. Querying temporal clinical databases on granular trends. J Biomed Inform. 2012;45:273–291. doi: 10.1016/j.jbi.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Shahar Y. A framework for knowledge-based temporal abstraction. Artif Intell. 1997;90:79–133. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 70.O'Connor MJ, Das AK. A lightweight model for representing and reasoning with temporal information in biomedical ontologies; Proceedings of the Third International Conference on Health Informatics; 2010. pp. 90–97. [Google Scholar]
- 71.Bergun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform. 2008:91–101. [PMC free article] [PubMed] [Google Scholar]
- 72.Sox HC, Greenfield S. Comparative effectiveness research: a report from the Institute of Medicine. Ann Intern Med. 2009;151:203–205. doi: 10.7326/0003-4819-151-3-200908040-00125. [DOI] [PubMed] [Google Scholar]
- 73.Post A, Kurc T, Overcash M, Cantrell D, Morris T, Eckerson K, et al. A Temporal Abstraction-based Extract, Transform and Load Process for Creating Registry Databases for Research. AMIA Summits Transl Sci Proc. 2011:46–50. [PMC free article] [PubMed] [Google Scholar]
- 74.Combi C, Oliboni B. Visually defining and querying consistent multi-granular clinical temporal abstractions. Artif Intell Med. 2012;54:75–101. doi: 10.1016/j.artmed.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 75.Singh Y, Sood M. Model Driven Architecture: A Perspective; IEEE International Advance Computing Conference; 2009. pp. 1644–1652. [Google Scholar]
- 76.Gennari JH, Musen MA, Fergerson RW, Grosso WE, Crubezy M, Eriksson H, et al. The evolution of Protege: an environment for knowledge-based systems development. International Journal of Human-Computer Studies. 2003;58:89–123. [Google Scholar]
- 77.Stanford Medical Informatics. The Protege Ontology Editor and Knowledge Acquisition System. [(accessed 2012 December 11)];2012 http://protege.stanford.edu/ [Google Scholar]
- 78.Cholleti S, Post A, Gao J, Lin X, Bornstein W, Cantrell D, et al. Leveraging Derived Data Elements in Data Analytic Models for Understanding and Predicting Hospital Readmissions; Proc AMIA Annu Fall Symp; 2012. pp. 103–111. [PMC free article] [PubMed] [Google Scholar]
- 79.Kim M, Cobb J, Harrold MJ, Kurc T, Orso A, Saltz J, et al. Efficient Regression Testing of Ontology-Driven Systems; Proceedings of the 2012 International Symposium on Software Testing and Analysis; 2012. pp. 320–330. [Google Scholar]
- 80.Gawande A. The Hot Spotters. [(accessed 2012 May 2)];The New Yorker. 2011 http://www.newyorker.com/reporting/2011/01/24/110124fa_fact_gawande. [Google Scholar]
- 81.Hibernate -- JBoss Community. [(accessed 2012 Apr 9)]; http://www.hibernate.org/M. [Google Scholar]
- 82.Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc. 1997;4:342–355. doi: 10.1136/jamia.1997.0040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winslow RL, Saltz J, Foster I, Carr JJ, Ge Y, Miller MI, et al. The CardioVascular Research (CVRG) Grid; Proceedings of the AMIA Summit on Translational Bioinformatics; 2011. pp. 77–81. [Google Scholar]
- 84.O'Connor MJ, Bingen M, Richards A, Tu SW, Das AK. Web-based Exploration of Temporal Data in Biomedicine; Proceedings of the 7th International Conference on Web Information Systems and Technologies; 2011. pp. 352–359. [Google Scholar]