Abstract
Proteins with β-sandwich and β-grasp topologies are resistant to mechanical unfolding as shown by single-molecule force spectroscopy studies. Their high mechanical stability has generally been associated with the mechanical clamp geometry present at the termini. However, there is also evidence for the importance of interactions other than the mechanical clamp in providing mechanical stability, which needs to be tested thoroughly. Here, we report the mechanical unfolding properties of ubiquitin-like proteins (SUMO1 and SUMO2) and their comparison with those of ubiquitin. Although ubiquitin and SUMOs have similar size and structural topology, they differ in their sequences and structural contacts, making them ideal candidates to understand the variations in the mechanical stability of a given protein topology. We observe a two-state unfolding pathway for SUMO1 and SUMO2, similar to that of ubiquitin. Nevertheless, the unfolding forces of SUMO1 (∼130 pN) and SUMO2 (∼120 pN) are lower than that of ubiquitin (∼190 pN) at a pulling speed of 400 nm/s, indicating their lower mechanical stability. The mechanical stabilities of SUMO proteins and ubiquitin are well correlated with the number of interresidue contacts present in their structures. From pulling speed-dependent mechanical unfolding experiments and Monte Carlo simulations, we find that the unfolding potential widths of SUMO1 (∼0.51 nm) and SUMO2 (∼0.33 nm) are much larger than that of ubiquitin (∼0.19 nm), indicating that SUMO1 is six times and SUMO2 is three times mechanically more flexible than ubiquitin. These findings might also be important in understanding the functional differences between ubiquitin and SUMOs.
Introduction
For more than a decade, it has been shown that pulling of polyproteins using single-molecule force spectroscopy (SMFS) to be very useful in characterizing the mechanical properties and the unfolding pathways of proteins (1–10). Proteins with β-sandwich topology or β-grasp topologies received much attention for their mechanical resistance against unfolding in single-molecule studies. In these studies, the mechanical stability has generally been attributed to the geometry in which the N- and C-terminal β-strands are parallel and directly connected by H-bonding (mechanical clamp). Mechanical clamp geometry is considered to provide resistance and rupture of it leads to unfolding as seen in single-molecule studies (11,12). On the other hand, it has been observed that the other interactions in the proteins, which are away from the mechanical clamp, are also important in providing mechanical resistance. In these studies, proteins with the same topology were used to provide support for the significance of interactions away from the mechanical clamp in providing the mechanical stability (13,14). Although, these previous studies provide vital evidence on the role of various interactions in proteins, the proteins were not of the same size to have a direct comparison between the mechanical stability and interactions. Here, we addressed this issue by comparing the mechanical properties of ubiquitin and small ubiquitin-related modifiers (SUMOs), which have the same topology, and more importantly, the same size.
SUMOs are members of the ubiquitin-like protein family that act as reversible posttranslational modifiers of proteins in eukaryotes (15–17). The mechanism of SUMO ligation to target proteins, which is called SUMOylation, is similar to that of ubiquitination. SUMOylation occurs through the formation of an isopeptide bond between the C-terminal Gly residue of SUMO protein and ε-amino group of Lys of the target protein through the action of E1, E2, and E3 enzymes in a manner similar to that of ubiquitination. Ubiquitin’s function is well defined in targeting proteins for degradation by tagging them with polyubiquitin. Furthermore, polyubiquitin chains are shown to resist mechanical unfolding in single-molecule experiments, and it has been associated with their function in proteasomal degradation (18). Unlike ubiquitin, the function of SUMOs is to modulate the function of the target protein. SUMO targets are involved in various cellular processes such as regulating mitochondrial function, chromatin structure, and signal transduction. In all these processes, SUMO proteins bind to other proteins and modify them (15,16,19,20).
In humans, there are four different SUMO proteins: SUMO1, SUMO2, SUMO3, and SUMO4. Although the sequences of SUMO2 and SUMO3 are >95% identical, they differ widely from that of SUMO1 with only ∼50% identity. Here, the sequence identity between two proteins is a pairwise identity between their amino acid sequences. The sequence identity of SUMO1 and SUMO2 with that of ubiquitin is below 20% (Fig. 1 A). Nevertheless, SUMO1, SUMO2, and SUMO3 share high structural homology (>95%) with ubiquitin (21–24). The structural homology between two protein structures was calculated using TopMatch protein structure alignment software (25,26). SUMOs have a characteristic long unstructured N-terminus that is absent in ubiquitin and other ubiquitin-like proteins (16,24). Structures of SUMOs consist of a classical β-grasp ubiquitin fold with four β-strands wrapped around a α-helix (Fig. 1 B). Despite their high structural similarity, SUMOs and ubiquitin are functionally divergent. This also raises an important question on what structural nuances and dynamics of these proteins make them functionally different. Furthermore, the terminal β-strands in the ubiquitin fold are directly connected to each other via backbone H-bonding, which acts as a mechanical clamp during unfolding as shown earlier by Carrion-Vazquez et al. (18). Based on the structural similarity between SUMOs and ubiquitin, their mechanical properties are expected to be similar, however, this remains to be tested experimentally. It must be noted that SUMO proteins are also found to undergo polymerization in a manner similar to polyubiquitination and it is not completely understood why such polySUMO proteins exist in vivo (27,28). The polymerization of SUMO proteins could be carried out in vitro as well (29). Hence, it would be important to study the biophysical properties of SUMOs and compare them with ubiquitin to gain further understanding on the functional diversity and the possible role of SUMO polymers in vivo.
Figure 1.
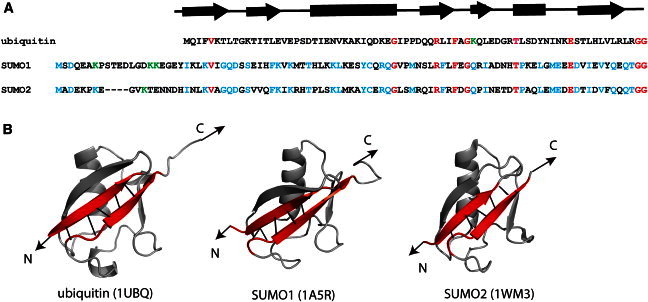
(A) The amino acid sequences of ubiquitin, SUMO1, and SUMO2. The residues that are identical in all three proteins are highlighted in red. In SUMO1 and SUMO2 there exists a long structureless sequence (∼20 residues) at N-terminus, which is absent in ubiquitin. The sequence identity between SUMO1/ubiquitin, SUMO2/ubiquitin pairs is ∼16%. The sequence identity between the SUMO1/SUMO2 pair is high (∼50%) and the residues identical in both of them are highlighted in blue. The C-terminal diglycine sequence is preserved in all of them. Ubiquitin forms Lys-48-C-linked polyubiquitin, whereas SUMO2 forms Lys-11-C-linked polymer chains. SUMO1 forms polymer chains in vitro through Lys-7, Lys-16, and Lys-17, which are highlighted in green. The secondary structural elements of ubiquitin are indicated directly above the sequence. (B) Structures of SUMO1, SUMO2, and ubiquitin. PDB IDs are given in parenthesis. The N-terminus structureless regions in SUMO1 and SUMO2 are not shown. In all the structures, the terminal β-strands (shown in red) are parallel to each other and connected by five H-bonds between the backbones (black lines). The arrows indicate the pulling direction used in the mechanical unfolding experiments.
Here, we report the mechanical properties of SUMO1 and SUMO2 measured in constant velocity pulling experiments using SMFS and compare them with those of ubiquitin. We find that SUMO1 and SUMO2 unfold at a much lower force than that of ubiquitin despite having similar structure. Furthermore, we have also varied pulling speed, used Monte Carlo simulations, and the Bell-like model to obtain the details of their unfolding energy landscapes. We find differences in their underlying energy landscapes, in terms of the distance to the transition state from the native state.
Materials and Methods
Polyprotein engineering
The genes of human SUMO1 and human SUMO2 were modified to insert the restriction sites of BamHI at the 5′ end and BglII, KpnI at the 3′ end. The genes were then cloned into pQE80L vector between BamHI and KpnI sites. The genes of (SUMO1)8 and (SUMO2)8 were constructed by an iterative cloning method as described by Carrion-Vazquez et al. (2). The octameric proteins, (SUMO1)8 and (SUMO2)8, were overexpressed in the BLR DE3 strain of Escherichia coli by inducing with 1 mM IPTG (isopropylthio-β-D-1-thiogalactopyranoside) for 6 h after the OD600 of the cell culture has reached 0.6. The harvested cells were suspended in phosphate buffered saline (PBS) (pH 7.4) containing 1 mM DTT (dithiothreitol) and 0.1 mM PMSF (phenylmethanesulfonyl fluoride). The cells were lysed by sonication, centrifuged, and the supernatant was applied to a column of Ni-NTA-coated agarose beads. The beads were washed with PBS containing 20 mM imidazole and the proteins were eluted with PBS containing 250 mM imidazole at pH 7.4. They were further purified by size exclusion chromatography using a superdex200 column (Amersham Biosciences). The purity of proteins was checked on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. All purified proteins were stored at 4°C. Polyubiquitin was expressed and purified using protocols described earlier by Carrion-Vazquez et al. (18).
SMFS
Single-molecule pulling experiments were performed on a custom-built atomic force microscope as described elsewhere (10). Approximately, 5 μl of polyprotein sample (∼2 μM) was added to ∼50 μl of PBS (pH 7.4) on a gold-coated glass coverslip. Gold-coated reflective cantilevers with a silicon-nitride tip with spring constants ∼35 pN/nm were purchased from Veeco, Singapore. Calibration of cantilevers was done using the equipartition theorem before each pulling experiment (30). All the experiments were performed at room temperature. All force-extension (FX) traces were collected at a sampling rate of 5 kHz.
Data analysis
In the data analysis, force peaks in FX traces were fitted to a worm-like chain (WLC) model of polymer elasticity (31) using Eq. 1
| (1) |
where p, Lc denotes the persistence length and contour length, respectively, kB is Boltzmann’s constant and T is absolute temperature. The persistence length was varied in the range of 0.3–0.8 nm with an average value of ∼0.5 nm. This range of persistence length for the WLC model was found to best fit the FX traces.
Spontaneous unfolding rate constant (ku0) and distance to the unfolding transition state (Δxu)
The spontaneous unfolding rate constant (ku0) and the distance to the unfolding transition state (Δxu) were calculated using two different methods: Monte Carlo simulations and by fitting the experimental data to the Bell-like model. The ku0, Δxu were calculated using Monte Carlo simulations as described by Oberhauser et al. (32). In brief, the dependence of unfolding force on pulling speed was modeled assuming a two-state Markov process (1,33). Initially, all the domains in a polyprotein chain were assumed to be folded (Nf) and were pulled at a given speed and the force was calculated using the WLC model as described previously. The probability of unfolding (Pu) of any of the domains is given by Pu = Nfk(F)Δt, where k(F) = ku0·eFΔxu/kBT. The unfolding event and force at which it occurred was recorded when the probability Pu, was greater than a random number between 0 and 1. The procedure was repeated until all the domains in the polyprotein chain unfolded and the unfolding force histograms were made. The values of ku0 and Δxu were varied such that the unfolding force histograms from simulations matched with those obtained experimentally at different pulling speeds.
We have also fitted the pulling-speed-dependent experimental unfolding forces, F(ν), to Bell-Evans-Ritchie approximation (34–37) and extracted the ku0, Δxu values. The following equation was used to fit the experimental values.
| (2) |
where v is the loading rate.
Transition state energy barrier and spring constant of the unfolding potential
The transition state energy barrier and spring constant of the unfolding potential were calculated using the method suggested by Dietz et al. (38). The transition state barrier height (ΔG#) was calculated using the Arrhenius equation ΔG# = −kBT × ln(ku0/kA), where kB is Boltzmann’s constant, T is temperature (= 298 K), ku0 is the rate constant for spontaneous unfolding and kA is the Arrhenius frequency factor. Here, kA is taken as 109s−1 (38,39). The unfolding potential is assumed to be harmonic and its spring constant (ks) is calculated using the equation ks = 2 ΔG#/(Δxu)2, where Δxu is the distance to the transition state or width of the unfolding potential.
Results and Discussion
SUMO1 and SUMO2 unfold at a lower mechanical force than that of ubiquitin
We performed pulling experiments on polyproteins (ubiquitin)9, (SUMO1)8, and (SUMO2)8 using SMFS at a pulling speed of 400 nm/s. The details of the protein engineering and the experimental technique are given in the Materials and Methods section. A typical FX trace of each polyprotein is shown in Fig. 2 A. For all three cases, the FX traces show a sawtooth pattern of force peaks indicating the sequential unfolding of individual protein units in the polyprotein. Furthermore, the spacing of ∼24 nm between adjacent peaks indicates that all three proteins have approximately the same number of amino acids contributing to the change in contour length upon unfolding. In these FX traces, the last force peak at >400 pN is due to the detachment of the protein either from the tip or from the substrate. For further analysis, we have taken all the FX traces that have at least four unfolding force peaks. The single-molecule FX traces were fitted to the WLC model (31) and the change in contour length of protein upon mechanical unfolding is ∼24 nm in all three cases. The histograms of the change in contour length upon unfolding for all three proteins are given in the Supporting Material (Fig. S1). However, their unfolding forces are different: ubiquitin (∼190 pN) unfolds at a higher mechanical force than SUMO1 (∼130 pN) and SUMO2 (∼120 pN) at a pulling speed of 400 nm/s. The unfolding force histograms are given in Fig. 2 B. Our results on ubiquitin are in agreement with those reported earlier (18,40). The results are given in Table 1.
Figure 2.
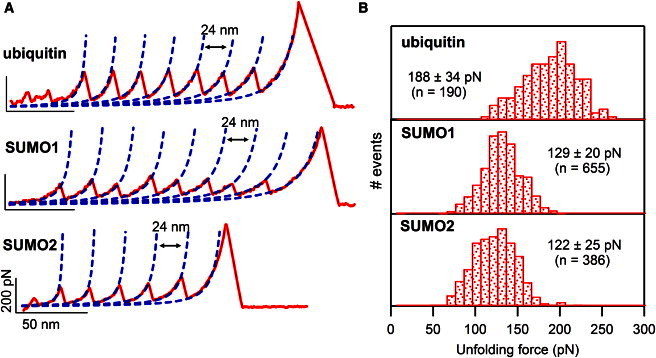
SUMOs are mechanically weaker compared to ubiquitin. (A) (top to bottom) Representative single-molecule force-extension (FX) traces of (ubiquitin)9, (SUMO1)8, and (SUMO2)8 at a pulling speed of 400 nm/s showing the sawtooth pattern with a contour length increment ∼24 nm. The traces were fit with the WLC model (dashed lines). The scale bars are 200 pN (vertical) and 50 nm (horizontal). (B) Unfolding force histograms (see Table 1 for more details).
Table 1.
Mechanical properties of ubiquitin and SUMO proteins
| Protein | Change in contour length (nm)*† | Unfolding force (pN)‡ | Δxu(nm) | ku0(s−1) | ΔG# (kBT) | ks(N/m) | ||
|---|---|---|---|---|---|---|---|---|
| Ubiquitin | 23.9 ± 1.2 (n = 190) | 188 ± 2 | 0.19§ | 0.15 ± 0.01¶ | 8 × 10−3§ | 0.45 ± 0.12¶ | 25.5 | 5.8 |
| SUMO1 | 24.1 ± 0.9 (n = 655) | 129 ± 0.8 | 0.51§ | 0.51 ± 0.04¶ | 1.15 × 10−5§ | 1.8 × 10−4 ±1.6 × 10−4¶ | 32.1 | 1.0 |
| SUMO2 | 24.1 ± 1.3 (n = 386) | 122 ± 1 | 0.33§ | 0.28 ± 0.02¶ | 5 × 10−3§ | 0.32 ± 0.19¶ | 26.1 | 2.0 |
Mean ± SD.
The n in the parentheses is the number of events used in the data analysis.
Mean ± SE.
From Monte Carlo simulation.
From Bell-Evans-Ritchie approximation.
SUMO1 and SUMO2 have 97 and 93 residues, respectively (Fig. 1 A). SUMO proteins are known to have a long and highly flexible N-terminus, which is absent in ubiquitin (24). A contiguous stretch of 76 residues spanning to the C-terminus in SUMO1 and SUMO2, forms a structure similar to that of ubiquitin with >95% homology (Fig. 1 B). Based on the high similarity between the contour length change and the structures of SUMOs and ubiquitin, it can be envisaged that the structureless region at the N-terminus of SUMO1 and SUMO2 unravels without any discernible force peak in the FX trace and the contiguous stretch of 76 residues spanning to the C-terminus unravel in a cooperative manner resulting in a single force peak with a contour length change of 24 nm (Fig. 2 A). Indeed, we have analyzed the featureless spacer before the sawtooth pattern in FX traces for ubiquitin, SUMO1, and SUMO2 (Fig. S2). The spacer lengths are longer for SUMO1 and SUMO2 in comparison to ubiquitin, confirming that the structureless region at the N-terminus of SUMO1 and SUMO2 unravels without any mechanical resistance. As expected, the change in contour lengths of SUMO1 and SUMO2 are exactly identical to that of ubiquitin (Fig. 2, Fig. S1, and Table 1).
Role of noncovalent interactions on protein mechanical stability
In the case of ubiquitin, the origin of mechanical stability was attributed to the five H-bonds between the force-bearing parallel β-strands at the termini (18). Terminal β-strands of SUMO1 and SUMO2 are also connected by five H-bonds (Fig. 1 B). Despite the same pulling geometry and rupture of the same number of H-bonds between the force-bearing structures, the unfolding force is very low for SUMO1 and SUMO2.
Previously, the parallel geometry of the terminal β-strands and the number of H-bonds connecting them in proteins is hypothesized to be the main reason for their very high mechanical stability (11) and their rupture leads to the mechanical unfolding as shown for many β-sheet and α/β proteins like I27, GB1, etc. (41,42). Proteins of this structural geometry at the termini have been found to unfold at higher forces (>180 pN at a pulling speed of ∼400 nm/s). However, proteins that lack this structural geometry at the termini, such as barnase, barstar, and DHFR tend to be mechanically compliant and unfold at much lower unfolding forces (<100 pN at a pulling speed of ∼400 nm/s) (43–45). Based on these general observations, SUMO1 and SUMO2 are also expected to be as strong as ubiquitin. However, our experiments demonstrate that this is not the case and SUMO1 and SUMO2 are mechanically weaker compared to ubiquitin.
In previous experiments, Li et al. (46) have shown that I1 and I27, the 1st and the 27th domains of I-band of human cardiac titin, respectively, have different mechanical stabilities as well as different distances to transition states despite similar structures. The I1 has a much lower mechanical unfolding force (130 pN) than that of I27 (210 pN) at the same pulling speed. These differences in mechanical properties were attributed to the variation in the distribution of H-bonds between the two terminal β-strands, hydrophobic interactions, and salt bridges in the protein (46–48). From our experiments, we found out that the mechanical properties of SUMO1, SUMO2, and ubiquitin have similar features as that of I1 and I27. However, the distinctly different mechanical properties of SUMOs from that of ubiquitin cannot be due to the difference in the number of H-bonds joining the two terminal β-strands as this is the same (five) in all the cases (Fig. 1 B). Hence, it is very likely that the mechanical stability of SUMOs is not solely dependent on the H-bonding at the termini but is also influenced by other interactions within the protein. This issue was earlier highlighted by research groups of Clarke (14,44) and Radford (13,49) where they have used interresidue contact maps in interpreting their experimental findings. TNfn3, a fibronectin-like domain from tenascin, despite having the same topology as I27, unfolds at a much lower force (∼100 pN) and the resistance to mechanical unfolding of TNfn3 was attributed to interresidue interactions present in the protein core in addition to those between the termini (14). Similarly, while comparing protein L with ubiquitin, Brockwell et al. (13) suggested that hydrophobic interactions in the protein structure might play an important role in modulating the mechanical stability of protein L because it was found to unfold at a lower force (∼130 pN) than ubiquitin at a similar pulling speed despite having the same structural fold.
To understand the discrepancy in the mechanical stability of SUMO1 and SUMO2 with that of ubiquitin, we have adopted a similar strategy of analyzing interresidue contacts in these proteins as discussed previously. In Fig. 3 A, the interresidue side-chain contact maps of SUMO1 and SUMO2 in comparison to ubiquitin are shown. It clearly shows that the total number of contacts are different for SUMO1 (138 contacts), SUMO2 (137 contacts), and ubiquitin (154 contacts) as calculated using CM-view software (50). In this calculation, an interresidue contact was assumed to be present if the shortest distance between side-chain atoms of different residues is <5 Å. We have also varied the cutoff distance and found that ubiquitin always has a higher number of contacts than SUMO1 and SUMO2 (Fig. S3). We have also calculated the number of side-chain contacts between the N-terminal (ββα) and C-terminal (ββ) halves, which form the force bearing regions of each protein when pulled in the N-C direction (Fig. S4). The number of contacts between the halves also follow the same trend as the overall contacts: ubiquitin (45 contacts) > SUMO1 (37 contacts) > SUMO2 (34 contacts). We have dissected the total number of side-chain contacts into hydrophilic/H-bond, aromatic, and hydrophobic interactions using CSU software (51). A detailed classification of interactions for each protein is provided in the Supporting Material, Table S1. Nearly 65% interactions are hydrophobic and 35% hydrophilic/H-bond interactions for all three proteins with a couple of aromatic interactions in the case of SUMO1 and SUMO2. The difference in mechanical stability between SUMOs and ubiquitin does not seem to be due to the hydrophilic or hydrophobic interactions alone as both are higher in ubiquitin. Hence, it is possible that an overall lower number of contacts present in SUMOs might be the reason for its lower mechanical stability. Using CSU software, we have also estimated the contact surface area of these contacts and it follows the same trend as the number of contacts (Table S1). Contact maps were used earlier to explain the differences in the mechanical stabilities of protein L and ubiquitin (13). However, the sizes of protein L (62 residues) and ubiquitin (76 residues) are different, though their overall fold is the same (13). Here, we have compared proteins from the same subfamily, with similar structural fold, and more important, having the same number of amino acids. There is a good correlation between the number of contacts and the unfolding force for SUMO1, SUMO2, and ubiquitin (Fig. 3 B). The contact maps seem to explain the discrepancies in the mechanical stabilities. Furthermore, we also investigated the details of the unfolding energy landscape of these proteins by performing pulling speed-dependent mechanical unfolding experiments.
Figure 3.
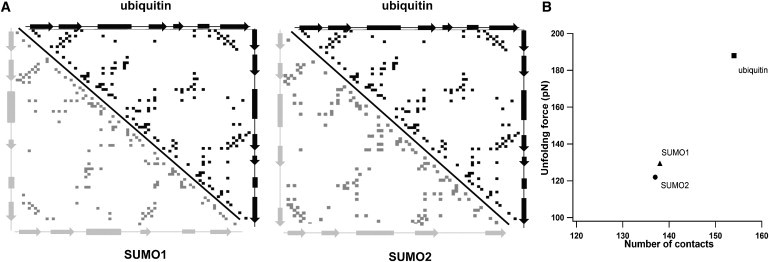
The number of interresidue contacts correlates with the unfolding force. (A) Contact maps of ubiquitin, SUMO1, and SUMO2. Ubiquitin paired with SUMO1 and SUMO2 separately for direct comparison. The definition of an interresidue contact is defined in the text. The total number of contacts in ubiquitin (154) are higher than SUMO1 (138) and SUMO2 (137), and this could be a reason for the lower mechanical stability of SUMO1 and SUMO2 compared to ubiquitin. (B) Plot of number of contacts versus unfolding forces for ubiquitin (■), SUMO1 (▴), and SUMO2 (●).
Unfolding energy landscapes of SUMO1, SUMO2, and ubiquitin
A semilogarithmic plot of unfolding force versus pulling speed for SUMO1, SUMO2, and ubiquitin is shown in Fig. 4. The speed-dependent unfolding force histograms are given in the Supporting Material (Fig. S5). The unfolding forces of SUMO1 and SUMO2 are consistently below ubiquitin throughout the pulling speed range (Fig. 4). Similar to Fig. 3 B, we have compared the number of contacts with unfolding forces at different pulling speeds (Fig. S6). For SUMOs and ubiquitin in general, the relationship that lower the number of contacts mean lower the mechanical stability seems to be valid in the entire pulling speed range (40–4000 nm/s). From this data, it can be said that the number of contacts could serve as an indicator of mechanical strength when comparisons have to be made within a class of structurally homologous proteins.
Figure 4.
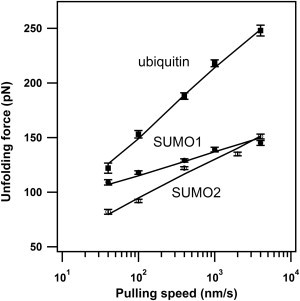
Pulling speed-dependent unfolding properties of ubiquitin, SUMO1, and SUMO2. Plot of unfolding force versus pulling speed. Error bars indicate SE. Solid lines are fits from Monte Carlo simulations. The results of simulations are given in Table 1.
In Fig. 4, the slopes of the experimental data are different for the three proteins indicating the differences in their unfolding potentials. To extract the unfolding energy landscape parameters, we have performed Monte Carlo simulations assuming a two-state unfolding process where the native state and unfolded states are separated by a single transition state barrier (32). The simulated data along with the experimental data are shown in Fig. 4. We found out that the distance to the unfolding transition state (Δxu) to be 0.19 nm for ubiquitin, 0.51 nm for SUMO1, and 0.33 nm for SUMO2. Our measured Δxu for ubiquitin is comparable to the earlier reported values, 0.225 nm from constant velocity measurements by Carrion-Vazquez et al. (18) and 0.17 nm from constant force measurements by Schlierf et al. (52). The calculated spontaneous unfolding rates (ku0) for three proteins are given in Table 1. To independently estimate Δxu and ku0, we have used Bell-Evans-Ritchie approximation as described previously (34–36). The estimates from this model are also given in Table 1. The Δxu values from this model are in excellent agreement with that of Monte Carlo simulations. The ku0 values from this model differ from Monte Carlo simulations by about an order of magnitude. Both Monte Carlo simulations and the Bell-like model give more accurate estimates of Δxu than ku0. In fact, the Δxu values from both the simulation and the model estimate that the unfolding potential of ubiquitin is much narrower than SUMO1 and SUMO2. These results can be explained in the form of an energy landscape where the activation energies are comparable, but the distance to the transition state (Δxu) is longer for SUMO1 and SUMO2 than ubiquitin and hence a lower unfolding force applied over a longer distance can decrease the barrier for SUMO1 and SUMO2 during the mechanical unfolding. A schematic of the unfolding energy landscapes of SUMO1, SUMO2 and ubiquitin describing the experimental and simulation results is shown in Fig. 5.
Figure 5.
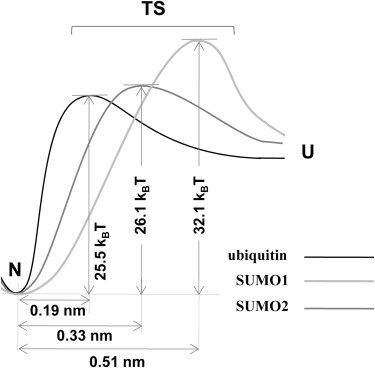
A schematic of the unfolding energy landscape. The unfolding potential width Δxu (or the distance to the transition state (TS)) of ubiquitin is much shorter than SUMO1 and SUMO2. See text and Table 1 for more details.
As seen in the previous section, the number of contacts is directly correlated with mechanical stability, whereas it is not correlated with ku0 and Δxu. Mechanical stability of a protein is both due to the unfolding barrier (or ku0) and Δxu, but either parameter alone might not explain it. Mechanical resistance can be correlated to an unfolding barrier for proteins that have the same Δxu and vice versa. However, in the case of SUMO1, SUMO2, and ubiquitin, both ku0 and Δxu are varying, and hence mechanical resistance cannot be directly correlated to ku0 (or Δxu) alone. Similarly, it is possible that the number of contacts that impart stability to a protein might not be correlated independently to the unfolding barrier or Δxu.
Furthermore, the Δxu is the magnitude of deformation along the pulling direction that occurs in protein to reach the transition state before crossing the unfolding energy barrier and it is usually taken as a measure of protein deformation response (38). We have estimated the stiffness of ubiquitin, SUMO1, and SUMO2 from their potential width (Δxu) and the energy barrier as suggested by Dietz et al. (38). We have used the Δxu and ku0 values obtained from Monte Carlo simulations in estimating the protein stiffness. We found out that ubiquitin (5.8 N/m) has a much larger spring constant for the unfolding potential (ks) than SUMO1 (1.0 N/m) and SUMO2 (2.0 N/m), which was calculated from the curvature of the potential well that precedes the unfolding energy barrier. This suggests that SUMO1 is six times and SUMO2 is three times more flexible than ubiquitin during mechanical unfolding. Between SUMO1 and SUMO2, SUMO2 is more similar to ubiquitin based on the unfolding energy landscape parameters—potential width, barrier height, and stiffness (Fig. 5 and Table 1). This is despite SUMO2 being less similar to ubiquitin (14% sequence identity) than SUMO1 (46% sequence identity) (Fig. 1). This could be explained by comparing the structures of SUMOs and ubiquitin, as done by Ding et al. (23), where it was shown that the root mean-square deviation value of average core structure of SUMO1 and SUMO3 pair is 2.2 Å, whereas the root mean-square deviation value for SUMO3, ubiquitin pair is 1.4 Å, which means the backbone core structure of SUMO3 is closer to ubiquitin than SUMO1. SUMO3 can represent SUMO2 as there is 97% sequence identity between them and the residues that are not common lie only in the unstructured N-terminus of SUMO2. On the basis of this comparison, it can be said that the backbone core structure of SUMO2 is more similar to ubiquitin than SUMO1. Hence, it is expected that the SUMO2/ubiquitin pair will have similar unfolding properties than the SUMO1/ubiquitin pair. Indeed, this is in concurrence with our observations (Fig. 5).
Biological significance of mechanical properties of SUMOs
Our finding that SUMO1 and SUMO2 are mechanically weaker and flexible when compared to ubiquitin, might have implications for their function. In addition, the polyproteins (SUMO1)8 and (SUMO2)8 studied here are analogous to multichain SUMO polymers found in vitro and in vivo. SUMO2 and SUMO3 are known to form multimers via an internal consensus site (Val-Lys-Thr-Glu) present at the N-terminus with the C-terminal Gly and can form Lys-11-C-linked chains (53,54). This internal consensus site for SUMOylation is absent in SUMO1. Interestingly, SUMO1 is found to form polymers linked via Lys-7, Lys-16, and Lys-17 in vitro (54,55). SUMO1 by itself does not form polymers in vivo but participates as a chain terminator in mixed SUMOylation with SUMO2 and SUMO3. However, in all SUMOs, the polymerization sites are located within the structureless N-terminal extensions that are absent in ubiquitin. These polySUMOs are equivalent to polyproteins (SUMO1)8 and (SUMO2)8 used in the current study, as SUMO proteins are linked at the N- and C-termini in a head-to-tail manner via peptide bonds. Interestingly, the mechanical stability of polySUMOs and N-C-linked polyubiquitin is higher than the physiologically relevant Lys-48-C-linked polyubiquitin (18). The differences in the unfolding forces are due to the anisotropic nature of the force, because these proteins are pulled along different directions. However, more studies would be required to understand if the higher mechanical resistance of polySUMOs has any role in proteasomal degradation given the recent findings on the cross talk between polySUMOs and polyubiquitin (15,56).
Conclusion
In summary, we have constructed octameric polyproteins of SUMO1 and SUMO2, and measured their mechanical properties. Both SUMO1 and SUMO2 have lower unfolding forces than ubiquitin. Although, SUMO1, SUMO2, and ubiquitin have the same overall topology and the structural arrangement at the termini (terminal β-strands, which are H-bonded to each other), their structures have a different number of contacts. The measured mechanical stabilities are in correlation with their interresidue contacts. Our study further supports the hypothesis that the interresidue contacts, in addition to mechanical clamp, are important in providing proteins with mechanical resistance and stability. Our study is also important because it used proteins not only from the same structural family but also of the same size. Furthermore, the unfolding energy landscape parameters obtained from experiments, simulations, and theoretical models suggest that SUMO1 and SUMO2 are mechanically more flexible than ubiquitin.
Acknowledgments
The authors thank Prof. R. V. Hosur for SUMO1 clone, Prof. Julio Fernandez for polyubiquitin plasmid, Prof. Hsiu-Ming Shih for SUMO2 clone, and TIFR for the financial support.
Supporting Material
References
- 1.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 2.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 4.Crampton N., Brockwell D.J. Unravelling the design principles for single protein mechanical strength. Curr. Opin. Struct. Biol. 2010;20:508–517. doi: 10.1016/j.sbi.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Li H., Cao Y. Protein mechanics: from single molecules to functional biomaterials. Acc. Chem. Res. 2010;43:1331–1341. doi: 10.1021/ar100057a. [DOI] [PubMed] [Google Scholar]
- 6.Puchner E.M., Gaub H.E. Force and function: probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 2009;19:605–614. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Galera-Prat A., Gómez-Sicilia A., Carrión-Vázquez M. Understanding biology by stretching proteins: recent progress. Curr. Opin. Struct. Biol. 2010;20:63–69. doi: 10.1016/j.sbi.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ainavarapu S.R., Wiita A.P., Fernandez J.M. A single-molecule assay to directly identify solvent-accessible disulfide bonds and probe their effect on protein folding. J. Am. Chem. Soc. 2008;130:436–437. doi: 10.1021/ja077851s. [DOI] [PubMed] [Google Scholar]
- 9.Bornschlögl T., Rief M. Single-molecule protein unfolding and refolding using atomic force microscopy. Methods Mol. Biol. 2011;783:233–250. doi: 10.1007/978-1-61779-282-3_13. [DOI] [PubMed] [Google Scholar]
- 10.Aggarwal V., Kulothungan S.R., Ainavarapu S.R. Ligand-modulated parallel mechanical unfolding pathways of maltose-binding proteins. J. Biol. Chem. 2011;286:28056–28065. doi: 10.1074/jbc.M111.249045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eom K., Li P.C., Rodin G.J. Relationship between the mechanical properties and topology of cross-linked polymer molecules: parallel strands maximize the strength of model polymers and protein domains. J. Phys. Chem. B. 2003;107:8730–8733. [Google Scholar]
- 12.Hoffmann T., Dougan L. Single molecule force spectroscopy using polyproteins. Chem. Soc. Rev. 2012;41:4781–4796. doi: 10.1039/c2cs35033e. [DOI] [PubMed] [Google Scholar]
- 13.Brockwell D.J., Beddard G.S., Radford S.E. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 2005;89:506–519. doi: 10.1529/biophysj.105.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng S.P., Rounsevell R.W., Clarke J. Mechanical unfolding of TNfn3: the unfolding pathway of a fnIII domain probed by protein engineering, AFM and MD simulation. J. Mol. Biol. 2005;350:776–789. doi: 10.1016/j.jmb.2005.04.070. [DOI] [PubMed] [Google Scholar]
- 15.Hay R.T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 17.Vertegaal A.C. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrion-Vazquez M., Li H., Fernandez J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 19.Müller S., Hoege C., Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 20.Melchior F. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 21.Song J., Zhang Z., Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 22.Huang W.C., Ko T.P., Wang A.H. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 2004;271:4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 23.Ding H., Xu Y., Shi Y. Solution structure of human SUMO-3 C47S and its binding surface for Ubc9. Biochemistry. 2005;44:2790–2799. doi: 10.1021/bi0477586. [DOI] [PubMed] [Google Scholar]
- 24.Bayer P., Arndt A., Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 25.Sippl M.J., Wiederstein M. Detection of spatial correlations in protein structures and molecular complexes. Structure. 2012;20:718–728. doi: 10.1016/j.str.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sippl M.J., Wiederstein M. A note on difficult structure alignment problems. Bioinformatics. 2008;24:426–427. doi: 10.1093/bioinformatics/btm622. [DOI] [PubMed] [Google Scholar]
- 27.Vertegaal A.C. SUMO chains: polymeric signals. Biochem. Soc. Trans. 2010;38:46–49. doi: 10.1042/BST0380046. [DOI] [PubMed] [Google Scholar]
- 28.Vertegaal A.C. Small ubiquitin-related modifiers in chains. Biochem. Soc. Trans. 2007;35:1422–1423. doi: 10.1042/BST0351422. [DOI] [PubMed] [Google Scholar]
- 29.Matic I., van Hagen M., Vertegaal A.C. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florin E.L., Rief M., Gaub H.E. Sensing specific molecular-interactions with the atomic-force microscope. Biosens. Bioelectron. 1995;10:895–901. [Google Scholar]
- 31.Bustamante C., Marko J.F., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 32.Oberhauser A.F., Marszalek P.E., Fernandez J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 33.Rief M., Fernandez J.M., Gaub H.E. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 1998;81:4764–4767. [Google Scholar]
- 34.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 35.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bustamante C., Chemla Y.R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 37.Li M.S., Kouza M. Dependence of protein mechanical unfolding pathways on pulling speeds. J. Chem. Phys. 2009;130:145102-1–145102-7. doi: 10.1063/1.3106761. [DOI] [PubMed] [Google Scholar]
- 38.Dietz H., Berkemeier F., Rief M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bieri O., Wirz J., Kiefhaber T. The speed limit for protein folding measured by triplet-triplet energy transfer. Proc. Natl. Acad. Sci. USA. 1999;96:9597–9601. doi: 10.1073/pnas.96.17.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chyan C.L., Lin F.C., Yang G. Reversible mechanical unfolding of single ubiquitin molecules. Biophys. J. 2004;87:3995–4006. doi: 10.1529/biophysj.104.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marszalek P.E., Lu H., Fernandez J.M. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y., Lam C., Li H. Nonmechanical protein can have significant mechanical stability. Angew. Chem. Int. Ed. Engl. 2006;45:642–645. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- 43.Ainavarapu S.R., Li L., Fernandez J.M. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Best R.B., Li B., Clarke J. Can non-mechanical proteins withstand force? Stretching barnase by atomic force microscopy and molecular dynamics simulation. Biophys. J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma D., Feng G., Li H. Stabilization provided by neighboring strands is critical for the mechanical stability of proteins. Biophys. J. 2008;95:3935–3942. doi: 10.1529/biophysj.108.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Fernandez J.M. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J. Mol. Biol. 2003;334:75–86. doi: 10.1016/j.jmb.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 47.Gao M., Wilmanns M., Schulten K. Steered molecular dynamics studies of titin I1 domain unfolding. Biophys. J. 2002;83:3435–3445. doi: 10.1016/S0006-3495(02)75343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayans O., Wuerges J., Wilmanns M. Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure. 2001;9:331–340. doi: 10.1016/s0969-2126(01)00591-3. [DOI] [PubMed] [Google Scholar]
- 49.Sadler D.P., Petrik E., Brockwell D.J. Identification of a mechanical rheostat in the hydrophobic core of protein L. J. Mol. Biol. 2009;393:237–248. doi: 10.1016/j.jmb.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vehlow C., Stehr H., Lappe M. CMView: interactive contact map visualization and analysis. Bioinformatics. 2011;27:1573–1574. doi: 10.1093/bioinformatics/btr163. [DOI] [PubMed] [Google Scholar]
- 51.Sobolev V., Sorokine A., Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 52.Schlierf M., Li H., Fernandez J.M. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geoffroy M.C., Hay R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 54.van der Veen A.G., Ploegh H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 55.Pedrioli P.G., Raught B., Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 56.Tatham M.H., Geoffroy M.C., Hay R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


