Abstract
Highly variable regions called genomic islands are found in the genomes of marine picocyanobacteria, and have been predicted to be involved in niche adaptation and the ecological success of these microbes. These picocyanobacteria are typically highly sensitive to copper stress and thus, increased copper tolerance could confer a selective advantage under some conditions seen in the marine environment. Through targeted gene inactivation of genomic island genes that were known to be upregulated in response to copper stress in Synechococcus sp. strain CC9311, we found two genes (sync_1495 and sync_1217) conferred tolerance to both methyl viologen and copper stress in culture. The prevalence of one gene, sync_1495, was then investigated in natural samples, and had a predictable temporal variability in abundance at a coastal monitoring site with higher abundance in winter months. Together, this shows that genomic island genes can confer an adaptive advantage to specific stresses in marine Synechococcus, and may help structure their population diversity.
Keywords: marine picocyanobacteria, copper, oxidative stress, Synechococcus, horizontal gene transfer, genomic islands
Introduction
Marine picocyanobacteria including Synechococcus and its sister taxon Prochlorococcus, are a highly diverse group that are found throughout the world oceans, and contribute significantly to global primary production (Li, 1994; Richardson and Jackson 2007). Multiple Synechococcus clades or ‘species' have been identified using various phylogenetic marker genes, and many of these clades are found to coexist in a single location (Ferris and Palenik 1998; Rocap et al., 2002; Zwirglmaier et al., 2007). The physiological and ecological traits associated with this diversity are not completely understood. With the advent of genome sequencing and the ability to compare whole genomes of these related species, it was discovered that there are a large number of horizontally transferred genes only found in individual strains, and that these genes are concentrated in highly variable regions of the genome called ‘genomic islands' (Palenik et al., 2003; Coleman et al., 2006; Palenik et al., 2006). These regions appear to evolve quite rapidly via horizontal gene transfer as evidenced in part by a Synechococcus-enriched metagenome from coastal California waters that showed high recruitment to sequenced reference genomes, except in these island regions (Palenik et al., 2009).
Genomic islands have been found in a wide range of bacterial phyla, and in many cases carry genes known to be involved in niche differentiation, such as nitrogen fixation genes in the symbiotic Rhizobiaceae species (Sullivan and Ronson 1998). These acquisitions are considered by some to drive the evolution of bacterial species (Gogarten et al., 2002) and it was predicted that these islands might be a driving force behind niche differentiation in picocyanobacteria (Coleman et al., 2006). However, while some predictions can be made based on known gene function, these islands are characterized by their high number of novel genes of unknown function making it difficult to fully assess their ecological role (Hsiao et al., 2005).
Several studies of phosphate acquisition genes in Prochlorococcus have found evidence that certain phosphate nutrition-associated genes are only seen in ocean regions with low phosphate levels and can be found in many phylogenetically distinct clades, indicating that these genes are likely to be laterally transferred at a high rate dependent on environmental conditions (Martiny et al., 2006, 2009a). Given that many Prochlorococcus strains have small, streamlined genomes, acquisition and retention of new genetic material is most likely under negative selective pressure (Garcia-Fernandez et al., 2004), and island genes likely provide a positive selection advantage. Moreover, metatranscriptomic analyses have found that some of these variable genes in Prochlorococcus are very highly expressed and thus, may be important to cellular processes in specific strains, despite their low abundance in the metagenomic data sets (Frias-Lopez et al., 2008). Additionally, a recent study by Avrani et al. (2011) found that one island in Prochlorococcus is involved in phage-resistance. These studies have been key to our understanding of the role of these islands in marine picocyanobacterial ecology, but targeted gene inactivation of genomic island genes with a predicted function to prove they confer an adaptive response has not been done to our knowledge.
Marine picocyanobacteria are known to be the most sensitive of the major marine phytoplankton groups to copper stress (Brand et al., 1986) and given our knowledge of the temporal and spatial variability of copper in the marine environment (Coale and Bruland 1990; Moffett et al., 1997) it seems likely that tolerance to elevated copper levels would confer an ecological advantage. Indeed, copper has been suggested to affect marine picocyanobacterial distribution (Mann et al., 2002), and marine Synechococcus strains have been shown to produce strong extracellular binding ligands in response to excess copper (Moffett and Brand, 1996). Copper toxicity is thought to result from a number of different mechanisms including the production of hydroxyl radicals, the blocking of essential metal binding sites and inhibition of electron transport in photosystem II (Baron et al., 1995; Sunda and Huntsman, 1998; Pinto et al., 2003). Much of the characterized copper stress response can consequently overlap with the oxidative stress response. In addition, copper induces an osmotic shock-like response in Synechococcus (Stuart et al., 2009).
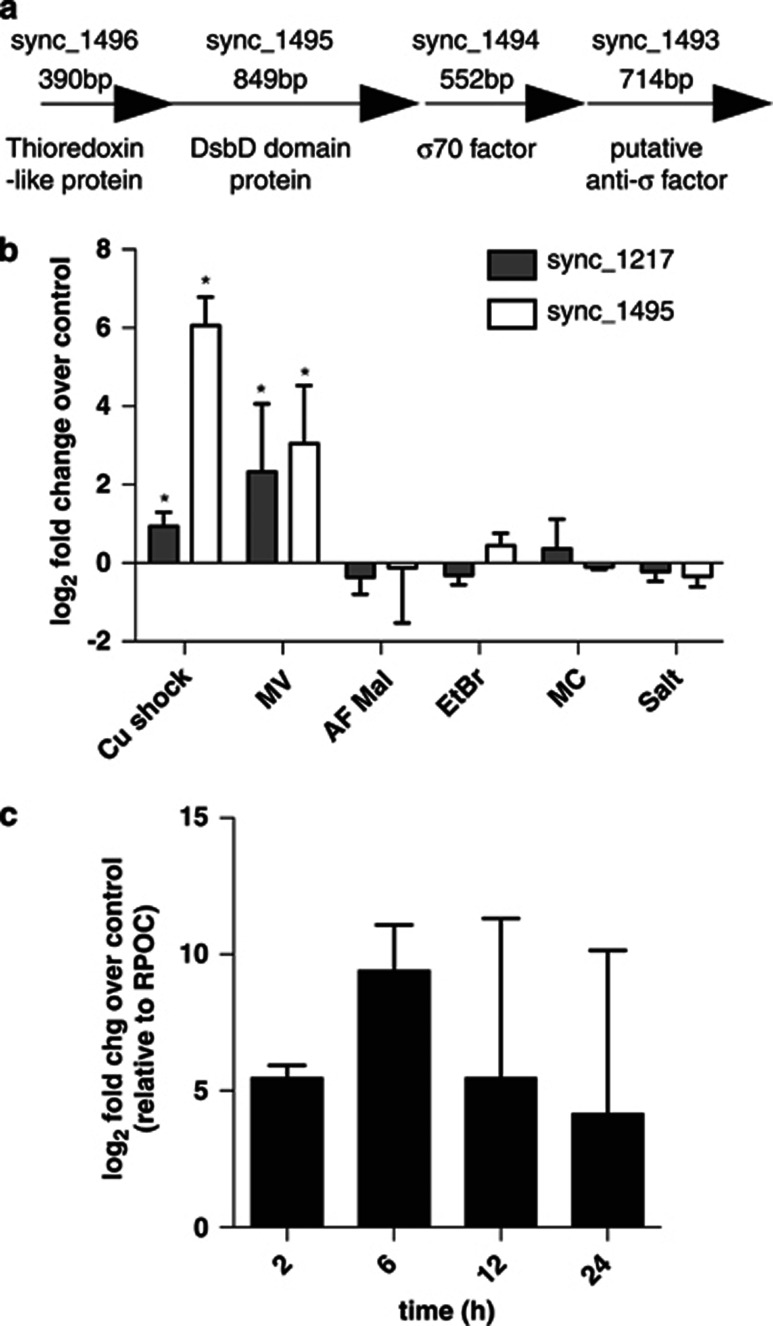
In a previous study done on the transcriptional response to copper stress, we found that the coastal Synechococcus sp. strain CC9311 (CC9311) had a response distinct from its open ocean cousin (Synechococcus sp. strain WH8102), in that a large percentage of the genes it upregulated came from putative horizontally transferred genes (Stuart et al., 2009). In particular, two of the most highly upregulated putative operons were from two genomic islands in this strain. The most highly expressed putative operon contains four genes that are predicted to be a thioredoxin-like protein (sync_1496), a cytochrome c biogenesis protein-like (sync_1495) an extracytoplasmic function sigma-70 factor (sync_1494) and a potential anti-sigma factor (sync_1493). It is on an 83 kb island, the largest island predicted in the genome, with 78 genes, many of which are metal-related (Dufresne et al., 2008).
Bacterial copper resistance has been best characterized in Escherichia coli (for review see (Rensing and Grass 2003)) and the gene sync_1495 has predicted transmembrane segments and a conserved domain, DsbD, that has been characterized in E. coli. In E. coli, DsbD is a inner membrane-bound protein that transfers electrons to DsbC, a disulfide isomerase that repairs nonnative disulfide bonds in the periplasm (Rietsch et al., 1997) and DsbE, a thioredoxin protein essential to cytochrome c maturation (Stirnimann et al., 2005). The DsbD knockout is extremely copper sensitive, which is not surprising, since copper as a redox metal catalyzes disulfide bonds quite readily (Katzen and Beckwith, 2003), and the entire pathway is thought to be involved in copper resistance and potentially other oxidative stressors (Hiniker et al., 2005). Indeed, in Synechocystis PCC6803 the orthologous operon (sll0685-sll0688) is transcriptionally upregulated in response to hydrogen peroxide stress, osmotic stress and DCMU ((3-(3,4-dichlorophenyl)-1,1-dimethylurea)) (Singh et al., 2010). The best available evidence thus, predicts that the sync_1496–93 putative operon in CC9311 might use a similar mechanism of disulfide repair to confer increased copper and oxidative stress tolerance.
The second most upregulated putative operon from the CC9311 copper stress studies includes an ABC transporter (sync_1214–1215), a HlyD secretion protein (sync_1216) and a 6.5 kb RTX toxin domain protein (sync_1217). This operon is on a much smaller 14 kb island with only eight genes, of which the four described above were induced in response to copper (Dufresne et al., 2008; Stuart et al., 2009). RTX toxin-domain proteins have a range of functions from enzymatic toxins to protective S-layer proteins, but most are secreted (Linhartova et al., 2010). An S-layer-localized RTX toxin domain protein in Synechocystis PCC6803 (sll1951) was found to be important in resistance to heavy metals, antibiotics and osmotic stress suggesting a role as general barrier against these stressors (Sakiyama et al., 2011). Although not an ortholog, we predicted that the sync_1214–1217 operon may secrete the RTX toxin-domain protein sync_1217, given the ABC transporter that is part of the putative operon, to function as an outer membrane barrier.
Using bioinformatic searches, we found that close homologs of these two copper-regulated operons were not abundant in marine metagenomic data sets, which was expected given the low recruitment to these genomic island regions in metagenomic data sets shown in Palenik et al. (2009). By using targeted gene inactivations of predicted copper tolerance genes, as well as investigation of the temporal and spatial distribution of one of these genes in the marine environment with quantitative PCR(qPCR), we have sought to better understand the functional and ecological role of genomic island genes in Synechococcus, using copper and oxidative stress tolerance in the coastal strain CC9311 as a model.
Materials and methods
Cell culture and growth assays
1 L batch cultures of CC9311, sync1495- and sync1217- were grown in chelexed synthetic ocean water on stir plates as previously described in Stuart et al. (2009). For mutant cultures, spectinomycin and streptomycin were both added at 2 μg ml−1 concentration at the time of inoculation, to maintain the insertion. For growth assays, cultures were grown to early log phase, and 50 ml were aliquoted in triplicate into acid-washed glass flasks and grown with shaking at 23 °C. Cultures were given 24 h in these conditions to adjust to the transfer then CuEDTA was added in triplicate at 10 μℳ, 4 μℳ, 1 μℳ, 0.4 μℳ for the copper stress, while sterile milli-Q water (Millipore, Billerica, MA, USA) was added to the control. Using the MINEQL algorithm (Westall et al., 1976), we calculated that this corresponds to equilibrium free copper levels equivalent to pCu 10.1, pCu 10.5, pCu 11.1 and pCu 11.5. For methyl viologen stress, methyl viologen (Sigma, St. Louis, MO, USA) was added at 500 nℳ, 100 nℳ, 50 nℳ and 5 nℳ. For AlexaFluor-488 C5 maleimide (Life Technologies, Carlsbad, CA, USA) treatment it was added at 1 μℳ. Flow cytometry samples were collected daily for up to 7 days and were fixed and analyzed as described in Stuart et al., 2009.
RNA extraction and quantitative reverse transcription-PCR
Cells were harvested and RNA extracted as described in Stuart et al., 2009. Briefly, cells were resuspended in Trizol Reagent (Life Technologies, Carlsbad, CA, USA), heated, extracted according to the manufacturers specifications, and then purified and concentrated on a glass fiber column. The following modifications were made to the original procedure: the 65 °C incubation was for 30 min instead of 60 min, a Zymo PCR inhibitor removal kit (Zymo, Irvine, CA, USA) was used to purify RNA instead of the Qiagen RNeasy kit (Qiagen, Venlo, Netherlands). Semiquantitative PCR was performed as described in (Stuart et al., 2009). Briefly, complimentary DNA was generated using a commercially available kit and a set amount of total starting RNA. Control reactions with no reverse transcriptase were also included. QPCR reactions included both a negative control and the complimentary DNA control as well as a 5 point standard curve. The following changes to Stuart, 2009 protocol were made: complimentary DNA was generated using 250 ng of total RNA using Ambion's Retroscript kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's specifications. QPCR was performed on a Qiagen RotorGene-Q with Promega GoTaq qPCR mastermix according to the manufacturer's specifications. Rpoc1 was used as a housekeeping gene, because its expression was high and did not change significantly in any of the expression microarrays datasets available for strain CC9311 or strain WH8102 (Stuart et al., 2009; Tai et al., 2009; Tetu et al., 2009).
Insertional inactivation of sync_1495 and sync_1217
Gene inactivations were conducted as described in Brahamsha, 1996 (Brahamsha 1996a, 1996b). Synechococcus sp. strain sync1495- and sync1217-, in which the genomic gene sync_1495 or sync_1217 is interrupted by a duplication, and insertion of a nonreplicating plasmid was constructed as follows. First, the omega cassette conferring spectinomycin/streptomycin resistance was cut from the pDW9 vector (Golden and Wiest 1988) using BamHI and cloned into the BamHI site of pBR322 (New England Biolabs, Ipswich, MA, USA) resulting in plasmid pBR322_Ω. A 399 bp segment of sync_1495 that is completely internal to the gene was amplified from CC9311 genomic DNA using primers 1495mutfwd and 1495mutrvs (Supplementary Table S1). A 500 bp segment of sync_1217 that is completely internal to the gene was also amplified from CC9311 genomic DNA using primers 1217mutfwd and 1217mutrvs. The amplified fragments were ligated separately into pCR2.1-TOPO (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's specifications resulting in plasmids TOPO_1495 and TOPO_1217, respectively, and insertion of the correct fragment was verified by sequencing. The 407 bp EcoRI fragment from TOPO_1495 was ligated into the EcoRI site of pBR322_Ω resulting in plasmid pBR322_Ω_1495. The 507 bp EcoRI fragment from TOPO_1217 was ligated into the EcoRI site of pBR322_Ω resulting in plasmid pBR322_ Ω _1217. Both plasmids were then separately introduced via conjugation into Synechococcus sp. strain CC9311 as described in Brahamsha, 1996 (Brahamsha, 1996b), with the following modifications: SN pour plates contained 0.4% (wt/vol) SeaPlaque agar (Lonza, Koeln, Germany) and 2 μg ml−1 each of spectinomycin and streptomycin. Exconjugants were screened by PCR to verify that no wild-type copy of the gene remained and that insertion had occurred at the predicted region (Supplementary Figure S1 and Supplementary Table S1).
CAMERA search, environmental clone library construction and qPCR
Bioinformatic searches for sync_1495 homologs were performed on the CAMERA website (Sun et al., 2011) by first performing a tBLASTn (with an e-value cutoff of 10−6) of both the ‘All metagenomic open reading frames' and the ‘All metagenomic 454 Reads' databases and combined the hits. A reciprocal BLASTx using this combined list of hits was then performed to the non redundant database to assess the similiarity of the hits to sync_1495. Sequences that did not hit sync_1495 as a top hit were excluded.
To assess the abundance of this gene, primers to sync_1495 were designed with several considerations in mind. First, primers should amplify only close homologs of sync_1495, including all Clade I Synechococcus members with the gene or potentially very recent horizontally transferred versions, and excluding other DsbD genes. At the nucleotide level this gene is almost unique in the non redundant database, with no close BLASTn hits, which made it simple to exclude known DsbD genes. At the time of primer design, we did not have other Clade I versions of the gene to align to so we chose a region within the predicted DsbD domain, which extends from 198 to 811 bp, and would be less likely to have divergence within the clade. We also wanted to amplify as large a region as possible given qPCR limitations (which optimally amplifies 50–150 bp) in order to cover a sufficient portion of the gene for TOPO-cloning verification. In consequence, the 254 bp amplified region chosen covers from 549−802 bp of the gene and the primers had no close BLASTn hits in the database (Supplementary Table S1). DNA from Tai et al., 2009 (Tai and Palenik 2009) was used first to generate a clone library from one sample (14 May 2006) using TOPO TA Cloning Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's specifications. Five clones were picked and sequenced using the forward primer included in the kit. QPCR was performed on a Stratagene Mx300P using Promega GoTaq qPCR master mix (Promega, Madison, WI, USA) according to the manufacturer's specifications. QPCR samples included DNA from monthly samples from 2005 (no February or August samples due to sample loss) and 2007 from Tai et al., 2009 (Tai and Palenik 2009) and DNA from depth samples collected in July, 2007 as described in Tai et al., 2011. QPCR primers used on above samples were for sync_1495 and rpoc Clade I (Supplementary Table S1).
Results
Expression of sync_1495 and sync_1217 in wild type in response to stress
As the response to copper stress often shares similarities to the oxidative stress response, we wanted to determine whether copper-responsive genes could be induced by other sources of oxidative stress. We also wanted to investigate the timing of upregulation of copper responsive genes to confirm their importance in copper stress response. The first two genes in putative operon sync_1496−1493 (Figure 1a) had very high induction in response to copper (6.3 and 6.1 log2 fold changes, respectively) more than double the log2 fold changes of other induced genes (Stuart et al., 2009), so we chose one, sync_1495, to further investigate the specificity of this response. Sync_1495 was chosen because it is the longer of the two genes, which makes it a more feasible target for eventual genetic inactivation, which is discussed below. In quantitative reverse transcription-PCR (QRTPCR) experiments sync_1495 was also significantly induced by methyl viologen (Figure 1b), which is a mild oxidant that subsequently reduces oxygen and generates superoxide anions (Imlay, 2008). The gene was not induced by AlexaFluor-488 C5 maleimide, which binds to surface free thiols, and in microarray experiments (S Tetu, D Johnson, K Phillippy, R Stuart, C Dupont, K Hassan, B Palenik and I Paulsen, unpublished data), the gene was also not significantly induced by ethidium bromide, mitomycin C, or salt shock (Figure 1b). Induction in response to copper stress was significant at 2 h and 6 h after treatment (one sample t-test of log2 fold changes, P-values of 0.0022 and 0.0103, respectively), but began to decrease by 12 h with more variation between the three biological replicates (Figure 1c).
Figure 1.
Sync_1495 gene expression in response to stressors and over time. (a) is a schematic of the operon sync_1496−1493. (b) is gene expression in response to various stressors. Log2 fold change is calculated relative to a control treatment. Error bars represent one s.d. between three biological replicates. ‘Cu' is quantitative reverse transcription (RT)-PCR after 2 h of copper addition (pCu10.1, 10 μℳ CuEDTA), ‘MV' is qRT-PCR of methyl viologen treatment 2 h after addition (100 nℳ). ‘AF Mal' is qRT-PCR AlexaFluor-488 C5 maleimide 2 h after addition (1 μℳ). ‘EtBR', ‘MC' and ‘salt' are microarray expression data (S Tetu, D Johnson, K Phillippy, R Stuart, C Dupont, K Hassan, B Palenik and I Paulsen, unpublished data). (c) is gene expression of sync_1495 over 24 h after copper stress (pCu10.1, 10 μℳ CuEDTA).
In addition, sync_1217 was induced by methyl viologen with a 2.3 log2 fold change (±1.7), but not by ethidium bromide, mitomycin C, or salt shock (S Tetu, D Johnson, K Phillippy, R Stuart, C Dupont, K Hassan, B Palenik and I Paulsen, unpublished data). These results provide evidence that these acquired genes are responsive to methyl viologen stress and copper stress, but not other tested stressors. Methyl viologen and copper are similar as known inducers of oxidative stress-like responses.
Mutant sensitivity to methyl viologen and copper stress
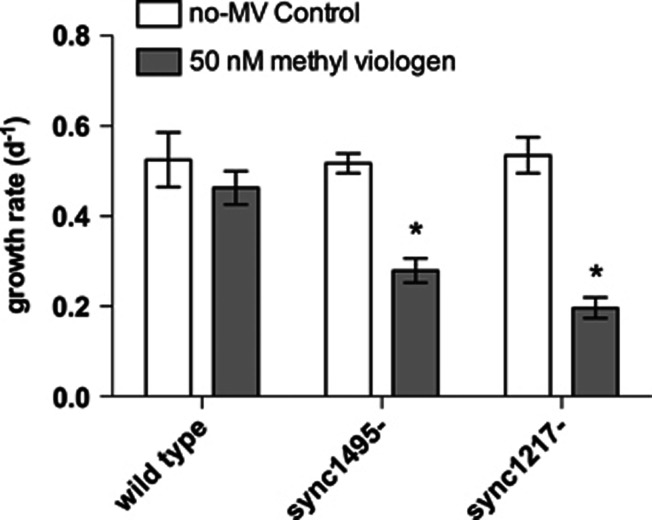
Insertional inactivations of sync_1495 (designated strain ‘sync1495-') and sync_1217 (designated strain ‘sync1217-') were validated using PCR (Supplementary Figure S1). Growth assays with methyl viologen additions showed that both sync1495- and sync1217- exhibited significant sensitivity to methyl viologen stress relative to the wild type. By 4 days after methyl viologen treatment at 50 nℳ, the wild type showed no difference in growth rate from the control treatment, whereas the mutants exhibited significantly slower growth rates (Figure 2, Supplementary Figure S2, Table 1).
Figure 2.
Synechococcus CC9311 wild type and mutant methyl viologen growth assays. Growth rates in both wild-type CC9311 (‘wt'), sync1495- and sync1217-. Cells per ml from flow cytometry counts were ln-transformed and a linear regression was performed on each. Bars represent linear regression slope, error bars represent one s.d. between three biological replicates. Star indicates significant difference from no-MV control treatment (one sample t-test, P<0.05).
Table 1. Growth rates and maximum yield after methyl viologen treatments.
| Treatmenta | Wild typeb | Sync1495-b | Sync1217-b |
|---|---|---|---|
| 0 (no-MV control) | 0.5251±0.060 (3.023±0.23) | 0.5170±0.022 (8.366±0.518) | 0.5353±0.040 (12.193±1.726) |
| 50 nM | 0.4627±0.037 (1.303±0.131) | 0.2791±0.027 (1.972±0.468) | 0.1964±0.023 (1.369±0.154) |
| 5 nℳ | 0.5558±0.0439 (2.806±0.696) | 0.4199±0.026 (6.065±0.753) | 0.4218±0.039 (5.682±1.037) |
Concentration methyl viologen (MV) added.
Growth rate (d−1)±1 s.d. and in parentheses maximum yield cells per ml (*10^8)±1 s.d.; bold indicates significant difference from no-MV control treatment (P<0.05, one sample t-test).
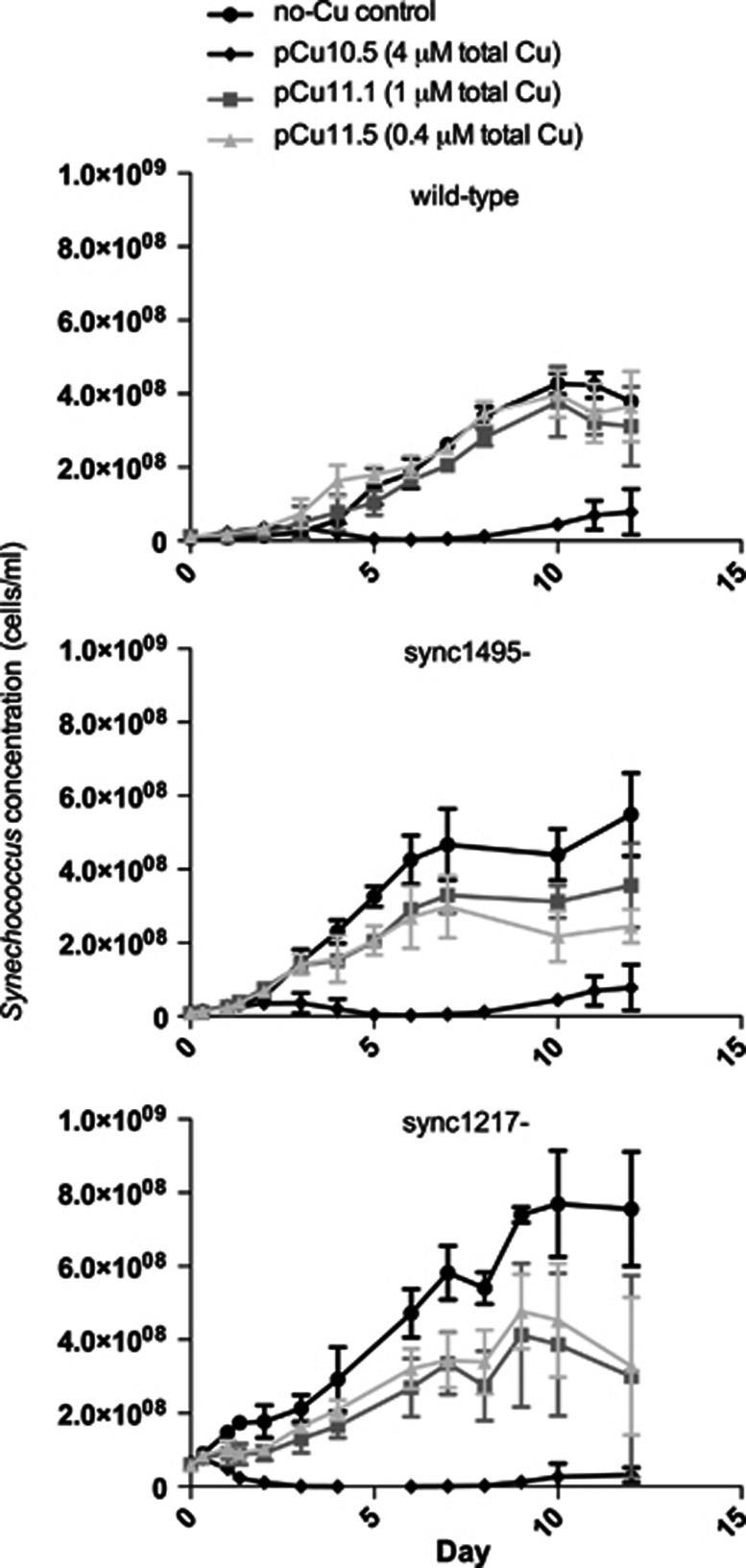
The response to copper stress in both mutants was slightly more complicated, as copper is both a micronutrient and a toxicant so growth stimulation is possible with copper additions. This can be seen in the wild-type growth assays, where in the first several days the lowest copper addition led to higher cell numbers than the control treatment, although the final maximum yields between these two treatments was not significantly different (Figure 3).
Figure 3.
Synechococcus CC9311 wild type and mutant copper growth assays. Wild-type CC9311 (‘wt'), sync1495- and sync1217- cells per ml from flow cytometry counts. Error bars represent one s.d. between three biological replicates. Cells/ml do not take into account mean cell size, which was significantly smaller in sync1217- (see Supplementary Figure S3).
This partially positive effect of providing some copper may have complicated the growth rate data, and we were not able to demonstrate a statistically significant difference in mutant growth rates relative to wild-type controls (Figure 3, Table 2). However, both mutants were sensitive to copper stress relative to the wild type as evidenced by the differences in maximum cell yield (Figure 3, Table 2). In the wild type the growth rate and the maximum yield for pCu11.5 (Cu2+ ion concentration of 10−11.5 ℳ; 0.25 μℳ CuEDTA, lowest copper addition) was not significantly different from the no-Cu control. By contrast, both mutants showed significantly lower maximum yields in the copper treated cultures (Figure 3, Table 2). Additionally, for the next highest copper level, pCu11.1, the wild type showed no difference in either growth rates or maximum yield from the control, whereas sync1495- and sync1217- had significantly lower maximum cell yields (Table 2).
Table 2. Growth rates and maximum yield after copper treatments.
| Treatmenta | Wild typeb | Sync1495-b | Sync1217-b |
|---|---|---|---|
| 0 (no-Cu control) | 0.6656±0.044 (4.269±0.288) | 0.7023±0.039 (5.781±0.765) | 0.3012±0.028 (8.447±0.870) |
| pCu10.1 | NG (0.195±0.008) | NG (0.206±0.020) | NG (0.642±0.057) |
| pCu10.5 | NG (0.447±0.006) | NG (0.406±0.107) | NG (0.762±0.0186) |
| pCu11.1 | 0.4816±0.079 (3.783±0.951) | 0.6253±0.057 (3.565±1.145) | 0.2305±0.026 (4.129±1.954) |
| pCu11.5 | 0.6108±0.051 (3.983±0.633) | 0.6278±0.069 (2.985±0.851) | 0.2633±0.017 (4.767±1.010) |
Concentration copper added.
Growth rate (d-1)±1 s.d. and in parentheses maximum yield cells per ml (*10^8)±1 s.d.; bold indicates significant difference from no-Cu control treatment (P<0.05, one sample t-test); ‘NG' indicates no significant increase in cells per ml over experiment.
Under no-copper conditions sync1217- had much higher cell yields than the wild type, however, the mean cell size of sync1217- over the course of the experiment was significantly smaller than the wild type, indicating that although cell number may be higher, total biomass may be similar between the two (Supplementary Figure S3). Sync1495- also had higher maximum cell yields than wild type, although not as high as sync1217-, and given that it did not have a smaller average cell size, it may be that sync1495- may actually have a higher biomass yield than wild type in the no-copper conditions, indicating that there may be a tradeoff in fitness associated with this operon. Conversely, it is also possible that the spectinomycin/streptomycin resistance cassette provides a slight growth advantage to the mutants.
Downstream gene expression in sync1495-
As insertional inactivation can sometimes lead to inactivation of downstream genes, we looked at the expression of downstream genes in the sync_1495 putative operon (Figure 1a). QRTPCR of the genes surrounding sync_1495 in the wild type, revealed that the upstream gene and two downstream genes were highly upregulated in response to 2 h of copper stress (Supplementary Figure S4a). In the mutant, transcripts of these two downstream genes were detectable, but there was no induction of these genes under copper stress relative to the control (Supplementary Figure S4b).
Temporal and spatial distribution of sync_1495 gene in the marine environment
A BLAST-based search was performed to find homologs of sync_1495 in marine metagenomic data sets using the CAMERA website (Sun et al., 2011), and in accordance with our predictions, only five hits were found to closely match sync_1495 (Supplementary Table S2).
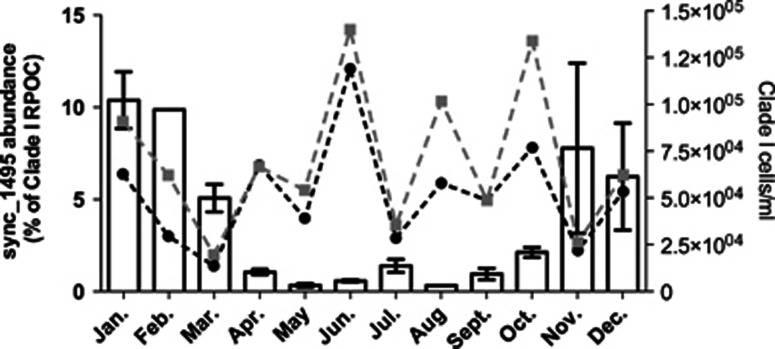
Primers were designed to amplify 254 bp of the 849 bp gene, sync_1495, that match the genome sequence exactly without introduced degeneracy (Supplementary Table S1). The primers were tested for specificity to sync_1495 in one DNA sample extracted from water collected at the Scripps Institution of Oceanography pier by generating a clone library from, which five clones were sequenced. All five clones aligned very closely to sync_1495 with 98.0−100% identity at the nucleotide level (Supplementary Figure S5). There were single nucleotide polymorphisms detected at residues 26, 68, 70, 85, 169, 178, 209 and 226, which resulted in amino acid substitutions in single nucleotide polymorphisms 26, 68 and 209 (Supplementary Figure S5). Using quantitative reverse transcription-PCR, the above primers along with Clade I-specific rpoc1 primers from Tai et al., 2009 (Tai and Palenik, 2009) were then used on monthly DNA samples from 2005 and 2007 from surface waters off the Scripps Institution of Oceanography pier. Sync_1495 was more abundant in the winter months and decreased to almost undetectable levels in the summer, relative to total Clade I Synechococcus abundance (Figure 4). This trend is not correlated to either total Synechococcus numbers or Clade I Synechococcus abundance (Figure 4, Supplementary Figure S6). We found that abundance of sync_1494, the gene directly downstream of sync_1495, was also higher in the winter months (Supplementary Figure S7), indicating that this trend holds for other genes in this putative operon. Sync_1494 was also detected using perfect primers (Supplementary Table S1) and compared with Clade I-specific rpoc1 abundance.
Figure 4.
Temporal distribution of sync_1495 gene over 2 years off the Scripps Institution of Oceanography pier. Bars show qPCR of sync_1495 as a percentage of total Clade I Synechococcus abundance (as determined by Clade I rpoc1 abundance) in surface samples from 2005 and 2007. Error bars represent +/− one s.e.m. between 2 replicate years (2005 and 2007). For Feb. and Aug. months no data was available for 2005 and only 2007 value is shown. Lines show Synechococcus abundance data from Tai et al., 2009; black line shows Clade I total abundance from qPCR and gray line shows total Synechococcus numbers from flow cytometry.
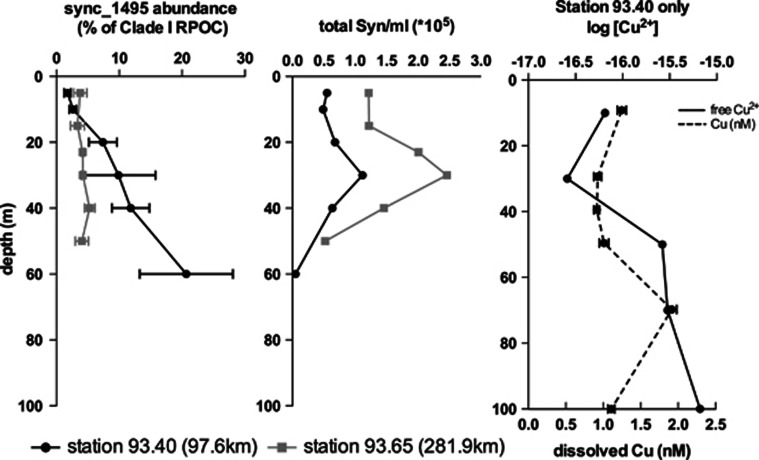
Additionally, from two sites off the California coast, DNA from several depths was obtained in July of 2007. Site 93.40 (32.51304°N, 118.21386°W), which was closer to the coast, showed an increase in sync_1495 abundance with depth. Site 93.65 (31.6776°N, 119.9025°W), which was further from shore, did not show an increase of the gene with depth in the top 100 m, while copper measurements were not available for this station (Figure 5). Along this transect, total dissolved copper ranged between 0.9 and 2.0 nℳ, and did not seem to increase consistently with distance from shore and only slightly increased with depth to 250 m (K. Buck and K. Barbeau unpublished data). Free copper and total copper generally have been shown to increase with depth (Coale and Bruland 1990; Moffett and Dupont 2007), however, marine Synechococcus is known to produce strong copper binding ligands in response to excess copper (Moffett and Brand 1996) and could influence free copper levels. At 60 m at site 93.40 the sharp decrease in total Synechococcus numbers (Tai et al., 2011) coincides well with a clear increase in Cu2+ concentration as found using CLE-ACSV (Figure 5, Supplementary Figure S8) (Buck and Bruland 2005; Buck et al., 2010)), and the largest positive difference between total dissolved copper and free Cu2+ occurs at the depth (30 m) with maximal Synechococcus numbers. Accordingly, we would predict that site 93.65, which has two to four times higher abundance of Synechococcus, might have lower free copper levels due to high levels of organic ligand.
Figure 5.
Spatial distribution of sync_1495 gene from two stations in the Southern California Bight. Numbers in parentheses indicate distance from shore. Left vertical profile shows qPCR of sync_1495 as a percentage of total Clade I Synechococcus abundance (as determined by Clade I rpoc1 abundance). Error bars represent one s.d. between three technical replicates. Middle profile shows Synechococcus counts from flow cytometry taken from Tai et al., 2011. Right profile shows both total dissolved copper concentration (dashed line) and free copper (-log[Cu2+]) concentrations (solid line). -log[Cu2+] measurements from copper speciation using CLE-ACSV at 25 μℳ SA (Buck et al., 2010).
Discussion
Highly variable genomic regions in picocyanobacteria have been implicated in picocyanobacterial adaptation to various ecological niches by conferring advantage, be it nutrient acquisition or some other stressor (Coleman et al., 2006; Martiny et al., 2009a). Identifying and characterizing an advantage can best be done in culture by knocking out these genes, and once this has been done it permits further analysis of environmental data to better understand the ecological relevance of these genes. The present data show that both sync_1495 and sync_1217 confer methyl viologen and copper tolerance to CC9311, suggesting that both genes have a role in an oxidative stress-like response, as both copper and methyl viologen can act as electron oxidants. Both are upregulated in the wild type in response to methyl viologen and copper, but not other stressors, again helping to confirm this role. The increase in the expression of sync_1495 is sustained over at least 6 h after copper stress in wild type, indicating it may be necessary for more than just early shock response.
As might be expected for genes that are not part of the Synechococcus core genome, the stress tolerance is manifested as a decrease in growth rate in response to methyl viologen in the mutants at levels that elicited no effect in the wild type. In response to copper the effect is slightly more complex. However, although the growth rate differences are not as large as for the methyl viologen treatments, comparison of the maximum cell yields clearly indicate that both mutants are impaired by copper levels that have no effect on wild-type cell yields.
Interestingly, the closest homologs to both these operons are found in estuarine cyanobacteria (Synechococcus WH5701 gene locus WH5701_04100 and Cyanobium PCC7001 gene locus CPCC7001_837), suggesting the possibility of gene flow between coastal marine and estuarine populations of cyanobacteria. On the basis of GC content, the genes in these two estuarine cyanobacteria do not themselves appear to have been laterally acquired. Indeed, the levels of copper, and other heavy metals, can be much higher in estuarine environments due to both natural and anthropogenic effects (for review of San Franscisco Bay see (Buck et al., 2007)), and that these groups would have extra copper resistance mechanisms is logical.
Although the exact mechanisms of action of each of these genes has yet to be confirmed, we can make informed predictions based on both their phenotypes and what we know of homologous genes. The cellular localization of sync_1217 in strain CC9311 is not known, but we have some initial indications that a large protein of this size (the largest predicted protein in the genome) can be found in the outer membrane (R. Stuart, B. Brahamsha, unpublished data). A recent review of RTX domain genes surveyed all available genomes, and cyanobacteria tend to have a relatively high number of these genes most of which are of unknown function (Linhartova et al., 2010). Three that have been characterized in cyanobacteria are involved in motility including SwmA from Synechococcus WH8102 (swimming), oscillin from Phormidium uncinatum (gliding) and a hemolysin-like protein in Synechocystis PCC6803 that is involved in motility through an unknown mechanism, but CC9311 is nonmotile. In E.coli, a Type I secretion system secretes HlyA, an RTX hemolysin protein and the operon, HlyCABD, includes an ABC transporter (HlyB), an membrane fusion protein (HlyD) that is involved in the folding of HlyA and a toxin activation protein (HlyC). The sync_1217 putative operon does include both an ABC transporter and a HlyD homolog, which strongly suggests that sync_1217 is secreted. The S-layer RTX protein that has been best characterized is RsaA in Caulobacter crescentus that is thought to help form a selective porosity barrier that may prevent predation (Linhartova et al., 2010), and it could be that sync_1217 has a similar function, acting as an outer membrane barrier to toxins.
Although sync1495- was also sensitive to copper and methyl viologen, it did not have a smaller average cell size than wild type and indeed had higher maximum cell yields than wild type in some cases. This indicates that the activity of the proteins in the operon may have a cost in terms of cell growth under control conditions even, while it provides an advantage under copper and oxidative stress conditions. Copper induction of the downstream genes, the sigma factor and putative anti-sigma factor (sync_1494 and sync_1493) were also inactivated in this mutant, although the upstream thioredoxin-like protein (sync_1496) was still copper inducible. As noted in the introduction sync_1495 has a domain similar to DsbD in E.coli, which is involved in disulfide bond isomerization. Additionally, the DsbD superfamily is present in many bacterial groups and the DsbD homolog in Caulobacter crescentus, ScsB, acts on a different set of substrates from E.coli, including a novel cell envelope reductive pathway providing electrons to a thioredoxin-like protein, which then reduces a novel peroxiredoxin that acts in cell envelope to directly scavenge peroxides (Cho et al., 2012). This suggests that this superfamily may be involved in several mechanisms of reduction in the cell envelope including disulfide isomerization as in E.coli, but also potentially directly scavenging of reactive oxygen species produced by copper or methyl viologen in the periplasm.
On the basis of qPCR, sync_1495 was relatively more abundant within the clade I species in the winter months in 2005 and 2007 off the Scripps Institution of Oceanography pier. The primers used were not degenerate and matched the CC9311 version of sync_1495 and although we know little about the rate of transfer of these genomic island genes, unless it is very rapid, it is likely that these primers only amplify versions of this gene that are in strains closely related to CC9311, that is, the clade I species. Our data thus show that the strain(s) that includes sync_1495 is under a different selective pressure than the clade I species as a whole. The surface waters off the pier in the winter months are characterized by lower water temperature, a higher degree of mixing and more runoff from rains. The latter two of these could lead to episodic pulses of higher copper in surface waters, and although we have no direct measurements of seasonal copper at this location, it is well-known that runoff does tend increase copper levels in San Diego Bay, and the rainy winter months have significantly higher copper levels (Blake et al., 2004; Buck et al., 2007). Additionally, it could be that copper is also upwelled seasonally (Takesue and van Geen 2002).
At one site in the Southern California Bight the gene also appeared to increase with depth, but more samples are needed to explain the depth distribution pattern seen as the two sites sampled were quite different. In Tai et al. (2011) distribution of several subclades from clades I and IV were quantified at these sites and it was found that at site 93.40 (and other coastal stations) the main clade I groups increased with depth, whereas the dominant clade IV group decreased with depth. In contrast, at site 93.65 most of the groups increased with depth and thus, at depth these sites had quite different relative abundances of the various groups. Moreover, at site 93.40 where sync_1495 was found to increase with depth, Cu2+ did increase between 30 m and 70 m. Although copper measurements were not available for site 93.65 it did have much higher abundances of Synechococcus, which could mean lower free copper levels.
Although it is possible that copper is driving these trends in sync_1495 abundance, it is also possible that some other variable that increases in the winter months and causes increased envelope or oxidative stress is the driver of this trend. Increased runoff, a deeper mixed layer and decreased temperature can lead to an increase in many environmental stressors (that is, anthropogenic toxins, fecal bacteria (Dwight et al., 2002)) and an increase in dynamic conditions. So while copper might not be the driver of this trend, the more important point is that a genomic island gene that is known to confer stress tolerance in cultures is more abundant in the winter months, when many different environmental stressors are increasing, irrespective of seasonal variability in the main group, in which the gene has been found.
Most intraclade (intraspecies) strain variability in Synechococcus appears to lie within genomic islands (Palenik et al., 2009) and accordingly it is likely that the island(s) carried by the CC9311 strain and closely related strains are the drivers of the seasonal variability in relative abundance seen in this one gene, sync_1495, involved in copper/oxidative stress response. In Prochlorococcus, macronutrient levels in the ocean are predicted to drive genomic content and ecotype formation, potentially through lateral gene transfer (Coleman et al., 2006; Martiny et al., 2006, 2009a, 2009b). Although copper and oxidative stress response genes may be less recognizable than nutrient acquisition genes, our data predicts that this stress response differentiation can also influence genomic content and may be an additional mechanism driving population structure in picocyanobacteria. A collection of strains with different genomic islands including ones conferring tolerance to these stresses could contribute to the overall fitness of Synechococcus in dynamic coastal conditions.
Acknowledgments
This work was supported by the National Science Foundation (grants NSF: MCB0744334 and California Current Ecosystem – Long Term Ecological Research (CCE-LTER) program). We would like thank V. Tai for providing the environmental DNA samples. We also want to thank K. Buck and K. Barbeau for copper measurements and S Tetu, D Johnson, K Phillippy, C Dupont, K Hassan and I Paulsen for sharing unpublished data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474:604–608. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- Baron M, Arellano JB, Gorge JL. Copper and photosystem-II - A controversial relationship. Physiol Plant. 1995;94:174–180. [Google Scholar]
- Blake AC, Chadwick DB, Zirino A, Rivera-Duarte I. Spatial and temporal variations in copper speciation in San Diego Bay. Estuaries. 2004;27:437–447. [Google Scholar]
- Brahamsha B. A genetic manipulation system for oceanic cyanobacteria of the genus. Synechococcus. Appl Environ Microbiol. 1996a;62:1747–1751. doi: 10.1128/aem.62.5.1747-1751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium. Synechococcus.Proc Natl Acad Sci USA. 1996b;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LE, Sunda WG, Guillard RRL. Reduction of marine-phytoplankton reproduction rates by copper and cadmium. J Exp Mar Bio Ecol. 1986;96:225–250. [Google Scholar]
- Buck K, Ross J, Russell Flegal A, Bruland K. A review of total dissolved copper and its chemical speciation in San Francisco Bay, California. Environ Res. 2007;105:5–24. doi: 10.1016/j.envres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Buck KN, Bruland KW. Copper speciation in San Francisco Bay: a novel approach using multiple analytical windows. Mar Chem. 2005;96:185–198. [Google Scholar]
- Buck KN, Selph KE, Barbeau KA. Iron-binding ligand production and copper speciation in an incubation experiment of Antarctic Peninsula shelf waters from the Bransfield Strait, Southern Ocean. Mar Chem. 2010;122:148–159. [Google Scholar]
- Cho S-H, Parsonage D, Thurston C, Dutton RJ, Poole LB, Collet J-F, et al. 2012A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope Mbio 3piie00291–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coale KH, Bruland KW. Spatial and temporal variability in copper complexation in the North Pacific. Deep-Sea Res Part A-Oceanog Res. 1990;37:317–336. [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;95:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight RH, Semenza JC, Baker DB, Olson BH. Association of urban runoff with coastal water quality in Orange County, California. Water Environ Res. 2002;74:82–90. doi: 10.2175/106143002x139776. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez JM, de Marsac NT, Diez J. Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol Mol Biol Rev. 2004;68:630–638. doi: 10.1128/MMBR.68.4.630-638.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Golden JW, Wiest DR. Genome rearrangement and nitrogen-fixation in Anabaena blocked by inactivation of xisA gene. Science. 1988;242:1421–1423. doi: 10.1126/science.3144039. [DOI] [PubMed] [Google Scholar]
- Hiniker A, Collet JF, Bardwell JCA. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- Hsiao WWL, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FSL. Evidence of a large novel gene pool associated with prokaryotic genomic islands. Plos Genet. 2005;1:540–550. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F, Beckwith J. Role and location of the unusual redox-active cysteines in the hydrophobic domain of the transmembrane electron transporter DsbD. Proc Natl Acad Sci USA. 2003;100:10471–10476. doi: 10.1073/pnas.1334136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WKW. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton - measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, et al. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EL, Ahlgren N, Moffett JW, Chisholm SW. Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol Oceanogr. 2002;47:976–988. [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009a;11:1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009b;106:10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JW, Brand LE. Production of strong, extracellular Cu chelators by marine cyanobacteria in response to Cu stress. Limnol Oceanogr. 1996;41:388–395. [Google Scholar]
- Moffett JW, Brand LE, Croot PL, Barbeau KA. Cu speciation and cyanobacterial distribution in harbors subject to anthropogenic Cu inputs. Limnol Oceanogr. 1997;42:789–799. [Google Scholar]
- Moffett JW, Dupont C. Cu complexation by organic ligands in the sub-arctic NW Pacific and Bering Sea. Deep-Sea Res Part I-Oceanog Res. 2007;54:586–595. [Google Scholar]
- Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, et al. The genome of a motile marine. Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- Palenik B, Ren QH, Dupont CL, Myers GS, Heidelberg JF, Badger JH, et al. Genome sequence of Synechococcus CC9311: Insights into adaptation to a coastal environment. Proc Natl Acad Sci USA. 2006;103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Ren Q, Tai V, Paulsen IT. Coastal Synechococcus metagenome reveals major roles for horizontal gene transfer and plasmids in population diversity. Environ Microbiol. 2009;11:349–359. doi: 10.1111/j.1462-2920.2008.01772.x. [DOI] [PubMed] [Google Scholar]
- Pinto E, Sigaud-Kutner TCS, Leitao MAS, Okamoto OK, Morse D, Colepicolo P. Heavy metal-induced oxidative stress in algae. J Phycol. 2003;39:1008–1018. [Google Scholar]
- Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Richardson TL, Jackson GA. Small phytoplankton and carbon export from the surface ocean. Science. 2007;315:838–840. doi: 10.1126/science.1133471. [DOI] [PubMed] [Google Scholar]
- Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama T, Araie H, Suzuki I, Shiraiwa Y. Functions of a hemolysin-like protein in the cyanobacterium Synechocystis sp PCC 6803. Arch Microbiol. 2011;193:565–571. doi: 10.1007/s00203-011-0700-2. [DOI] [PubMed] [Google Scholar]
- Singh AK, Elvitigala T, Cameron JC, Ghosh BK, Bhattacharyya-Pakrasi M, Pakrasi HB. Integrative analysis of large scale expression profiles reveals core transcriptional response and coordination between multiple cellular processes in a cyanobacterium. BMC Syst Biol. 2010;4:105. doi: 10.1186/1752-0509-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann CU, Rozhkova A, Grauschopf U, Grutter MG, Glockshuber R, Capitani G. Structural basis and kinetics of DsbD-dependent cytochrome c maturation. Structure. 2005;13:985–993. doi: 10.1016/j.str.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Stuart RK, Dupont CL, Johnson DA, Paulsen IT, Palenik B. Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response relative to those of open-ocean strains. Appl Environ Microbiol. 2009;75:5047–5057. doi: 10.1128/AEM.00271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:51. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunda WG, Huntsman SA. Interactive effects of external manganese, the toxic metals copper and zinc, and light in controlling cellular manganese and growth in a coastal diatom. Limnol Oceanogr. 1998;43:1467–1475. [Google Scholar]
- Tai V, Palenik B. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. Isme J. 2009;3:903–915. doi: 10.1038/ismej.2009.35. [DOI] [PubMed] [Google Scholar]
- Tai V, Paulsen IT, Phillippy K, Johnson DA, Palenik B. Whole-genome microarray analyses of Synechococcus-Vibrio interactions. Environ Microbiol. 2009;11:2698–2709. doi: 10.1111/j.1462-2920.2009.01997.x. [DOI] [PubMed] [Google Scholar]
- Tai V, Burton RS, Palenik B. Temporal and spatial distributions of marine Synechococcus in the Southern California Bight assessed by hybridization to bead-arrays. Mar Ecol-Progr Ser. 2011;426:133–147. [Google Scholar]
- Takesue RK, van Geen A. Nearshore circulation during upwelling inferred from the distribution of dissolved cadmium off the Oregon coast. Limnol Oceanogr. 2002;47:176–185. [Google Scholar]
- Tetu SG, Brahamsha B, Johnson DA, Tai V, Phillippy K, Palenik B, et al. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. 2009. [DOI] [PubMed]
- Westall JC, Zachary JL, Morel FMM. MINEQL - General algorithm for computation of chemical-equilibrium in aqueous systems. Abstracts of Papers of the American Chemical Society. 1976;172:8–8. [Google Scholar]
- Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EMS, Zubkov MV, Scanlan DJ. Basin-scale distribution patterns lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.