Abstract
Plasmids have long been recognized as an important driver of DNA exchange and genetic innovation in prokaryotes. The success of plasmids has been attributed to their independent replication from the host's chromosome and their frequent self-transfer. It is thought that plasmids accumulate, rearrange and distribute nonessential genes, which may provide an advantage for host proliferation under selective conditions. In order to test this hypothesis independently of biases from culture selection, we study the plasmid metagenome from microbial communities in two activated sludge systems, one of which receives mostly household and the other chemical industry wastewater. We find that plasmids from activated sludge microbial communities carry among the largest proportion of unknown gene pools so far detected in metagenomic DNA, confirming their presumed role of DNA innovators. At a system level both plasmid metagenomes were dominated by functions associated with replication and transposition, and contained a wide variety of antibiotic and heavy metal resistances. Plasmid families were very different in the two metagenomes and grouped in deep-branching new families compared with known plasmid replicons. A number of abundant plasmid replicons could be completely assembled directly from the metagenome, providing insight in plasmid composition without culturing bias. Functionally, the two metagenomes strongly differed in several ways, including a greater abundance of genes for carbohydrate metabolism in the industrial and of general defense factors in the household activated sludge plasmid metagenome. This suggests that plasmids not only contribute to the adaptation of single individual prokaryotic species, but of the prokaryotic community as a whole under local selective conditions.
Keywords: metagenomic studies, mobilome
Introduction
Genome innovation and evolution in prokaryotes is essentially dependent on error-generating processes (Arber, 2000) and on influx of DNA from external sources (lateral gene transfer) (Ochman et al., 2000; Frost et al., 2005). The total contribution of noncognate DNA in prokaryotic genomes has been estimated from comparative genome sequencing to amount to 20% (Koonin and Wolf, 2008), whereas on average 75% of all known prokaryotic gene families may have been subject to lateral gene transfer at some point in evolutionary history (Kloesges et al., 2011). Mobile DNA elements have a significant role in the reshuffling of genetic material and its lateral distribution (Koonin and Wolf, 2008; Siefert, 2009). To a large extent plasmids, extrachromosomally replicating DNA in prokaryotes, are held responsible for lateral gene transfer and DNA reshuffling. This is because plasmids maintain themselves in the host cell independently, can frequently self-transfer or become mobilized by other plasmids and often accumulate transposable elements that mobilize genes onto plasmids (Thomas and Nielsen, 2005). Extensive pure culture evidence indicates that interspecies gene distribution by plasmids is one of the main causes for the rapid adaptation of prokaryotes, leading to antibiotic resistance formation (Mazel and Davies, 1999; Robicsek et al., 2006), distribution of virulence factors (Fondi et al., 2010) or metabolism of toxic compounds (Warren et al., 2004). However, most prokaryotic species thrive in natural communities and do not exist as pure laboratory culture (Zengler et al., 2002; Stepanauskas and Sieracki, 2007). Therefore, the full extent of plasmid DNA diversity, mobilization and adaptation potential at the prokaryotic community level is far from being understood, although several studies have suggested hidden reservoirs of mobile DNA (Cortez et al., 2009; Jones et al., 2010; Kav et al., 2012). In particular, the impact of local conditions imposed at the level of the microbial community on the types of gene functions carried on plasmids has never been studied without culturing bias. Here, we report the functional differences of collective gene information content of two plasmid pools in microbial communities using a culture-independent metagenomic approach. In contrast to now regular metagenomic approaches that analyze total DNA, metagenomic analysis of the mobilome requires specific separation and purification of, for example, closed circular supercoiled DNAs (CCSD) originating from, for example, plasmids. We optimize here methods to extract supercoiled DNAs from complex microbial communities. We focus specifically on activated sludge, an assembly of prokaryotes and small eukaryotes that spontaneously develops under the local operating conditions of wastewater treatment plants (WWTPs) (Daims et al., 2006). WWTPs arguably form the largest, open and noncontrolled mixed microbial cultures in human society. Because of its high species diversity and density of prokaryotic cells in flocs and biofilms, activated sludge is thought to be a hotspot for lateral gene transfer and, possibly, to favor the emergence of antibiotic-resistant pathogens (Szczepanowski et al., 2008). Hence, plasmids in the WWTP microbial communities are expected to be selected to provide necessary functions to their host, and such functions are expected to depend on the incoming wastewater composition and the WWTP operating conditions. To test this hypothesis we compared plasmid metagenomes from a municipal activated sludge WWTP (near Morges, Switzerland) receiving a mixture of household and hospital wastewaters, typically containing small amounts of pharmaceuticals, and from an industrial but similarly operated WWTP (near Visp, Switzerland) that treats 70% of incoming wastewater from chemical industry with up to ten times higher organic loads (Materials and methods). We describe for the first time in great depth plasmid replicons from two complex communities, recover a number of fully closed replicons and show substantial difference in plasmid gene content as a result of selective conditions at the community level.
Materials and methods
WWTP and sample description
Activated sludge was collected from the aeration basin of a municipal WWTP in the city of Morges, canton Vaud, Switzerland, (GPS: 46.5166N, 6.51014E) at an elevation of 270 m above sea level. This WWTP serves ca. 32 000 inhabitants and treats a mixture of wastewaters from households, a hospital (180 beds), agriculture (for example, vineyards), local enterprises, as well as runoff waters. The WWTP treats around 11 000 m3 sewage per day, with the following characteristics (averaged over the year 2007): biological oxygen demand=122 mg O2 l−1, chemical oxygen demand=378 mg O2 l−1, total organic carbon=144 mg C l−1. Concentrations of heavy metals in the sludge (27% dry matter content, averaged over four measurements in 2007) were as follows: Hg, 1.2 mg l−1; Mo, 4.8 mg l−1; Cd, 0.9 mg l−1; Co, 3.5 mg l−1; Ni 24.8 mg l−1; Cr, 66.5 mg l−1; Cu, 412.0 mg l−1; Pb, 39.0 mg l−1; Zn, 897.5 mg l−1 (Official Information Canton Vaud, Switzerland: http://www.vd.ch/fr/themes/environnement/eau/eaux-usees/controle-des-step/ files Bilan de l'exploitation des STEP (2007, pdf, 3.1 Mo), Bilan de l'exploitation des STEP du canton (2009, pdf, 5.0 Mo).
At the time of sampling the WWTP at Morges was in stable operation with a pH of between 7.0 and 7.2 in the activated sludge basin and a temperature of 14–16 °C. Three samples were taken: 3 and 6 May 2007, and 16 March 2009, each time at 0900 hours. Each time 10 l of raw sludge were taken, transported to the laboratory within 30 min and handled on ice or at +4 °C during the entire procedure (unless stated otherwise). The prokaryotic community in fresh sludge was profiled using fluorescent in situ hybridization (FISH) against 16S rRNA targeted probes covering the main phylogenetic groups. Hereto, freshly taken sludge (Morges 2007) was fixed with an equal volume of ethanol and shipped on ice for commercial analysis (Vermicon AG, München, Germany).
The second WWTP is located in Visp (Switzerland, GPS coordinates: 7.85940W, 46.30044N, elevation 658 m). It treats a mixture of household wastewater (on average 4990 m3 per day) and effluents of the chemical/pharmaceutical industry (LONZA, Visp, Switzerland, 8660 m3 per day). The treatment consists of mixing the influents, two sequential steps of aeration with activated sludge and a final sedimentation with partial sludge recycling (turnover rate is 100–150%). Temperature is maintained at 27–28 °C, pH at 7.6–7.9. Wastewater influent has a biological oxygen demand of 1.86 g O2 l−1, a chemical oxygen demand of 2.7 g O2 l−1 and total organic carbon of 920 mg l−1 (mainly acetate, but detailed composition not disclosed by the company), plus high ammonium loads. Activated sludge was collected on 19 February, 2009 2230 hours, and at the moment of sampling the WWTP was in the early stage of adaptation from a low to high load of chemical waste.
Sludge biomass purification
Sludge biomass was purified by centrifuging 5 l of sludge at 1000 g for 3 min, decanting and resuspending the biomass pellet in 1 l of ice-cold poly(beta-amino) esters (PBAE) buffer (PBAE buffer is 10 mℳ Na-phosphate, 10 mℳ Na-ascorbate, 5 mℳ EDTA, pH 7.0). Each of four 250 ml aliquots of the PBAE-biomass slurry were combined with a further 600 ml of PBAE buffer and homogenized in a kitchen blender (Bosch MMB-2000) for 15 min at minimal power setting. After that, coarse particles, cellular aggregates and eukaryotic cells were removed by low speed centrifugation for 6 min in 50 ml conical tubes using an Eppendorf swing-out rotor AA-4-62 at 800 r.p.m. (ca. 160 g). The resulting supernatants were harvested and pooled together, and the microbial biomass was collected by centrifugation at 6000 g for 6 min in a Sorvall SLA-3000 rotor. The pellets were resuspended in 500 ml of PBAE buffer supplemented with 95 U ml−1 of pectinase from Aspergillus aculeatus (Sigma-Aldrich Chemie, GmbH, Buchs, Switzerland, P2611) and 0.01% Zwittergent 3–14 (Merck (Schweiz) AG, Zug, Switzerland, 693017), and incubated on a rotary shaker at 180 r.p.m. and 25 °C for 30 min. Subsequently, the total volume was adjusted to 850 ml with ice-cold PBAE buffer, blended, centrifuged at low speed and harvested as before. Finally, the biomass was resuspended in 500 ml PBEA.
Metagenome plasmid DNA isolation
CCSD was isolated from the washed microbial sludge biomass by means of either (i) a modified alkaline lysis procedure (Birnboim and Doly, 1979)—for the Morges 2007 sample, or (ii) a hot alkali lysis method in combination with acid phenol–chloroform treatment (Kieser, 1984)—for the Morges 2009 sample. Although no method will be perfect to lyse all prokaryotic cells in a diverse mixture, such as microbial sludge biomass, visual and microscopic inspection indicated a cleared lysate with very few remaining cells. Subsequently, CCSD was separated from other forms of DNA using isopycnic density centrifugation in a classical CsCl-ethidium bromide (EB) gradient. For the modified Birnboim and Doly method, the washed microbial sludge biomass in PBAE was adjusted to a culture turbidity of 4 at 600 nm and divided into 200 ml portions in screw cap polypropylene tubes. Cellular biomass was collected by centrifugation at 4000 g for 6 min in a Sorvall GSA rotor, and resuspended in 20 ml GTED buffer (GTED is 50 mℳ glucose, 50 mℳ Tris-HCl, 10 mℳ EDTA at pH 8.0) supplemented with 2 mg ml−1 lysozyme and 40 mg ml−1 polyvinylpolypyrrolidone (Sigma-Aldrich). Mixtures were incubated at room temperature for 30 min on a rocking platform. Then 40 ml of the lysis solution (0.2 ℳ NaOH, 1% SDS, 100 mℳ EDTA) were added and mixed by five inversions, and after 4–5 min incubation the lysate was neutralized by addition of 30 ml of ice-cold 3 ℳ potassium acetate (pH 4.8). The mixture was further incubated on ice for 15 min and cleared by centrifugation at 20 000 g and 4 °C for 20 min. After filtering through Whatman No.1 paper, the DNA in the cleared lysate was precipitated for 30 min at room temperature by addition of 0.8 volume of 2-isopropanol. DNA was collected by centrifugation for 30 min at 20 000 g and 20 °C, washed with 75% ethanol, drained and dissolved (usually within 1 h at 37 °C) in 2 ml of a buffer containing 10 mℳ Tris-HCl at pH 8.0, 10 mℳ EDTA, 20 μg ml−1 RNAse A, 0.5 mg ml−1 Zwittergent 3–14 detergent (Calbiochem-Novabiochem AG, Läufelfingen, Switzerland, cat no.693017), and 1 mg ml−1 proteinase K (Roche, Basel, Switzerland). Finally, isogenous DNA preparations were pooled together.
For the hot alkali/acid phenol:chloroform extraction, 200 ml of cell suspension in PBAE buffer at a culture turbidity of 3.6 were harvested by centrifugation for 6 min at 4000 g, and resuspended in 104 ml of STED buffer (STED is 0.3 ℳ sucrose, 20 mℳ Tris-HCl, 25 mℳ EDTA at pH 8.0) supplemented with 2 mg ml−1 lysozyme. The suspension was incubated at room temperature for 30 min on a rocking platform. Then 52 ml of the lysis solution (0.3 ℳ NaOH, 3% SDS, 100 mℳ EDTA) were mixed in, and the mixture was incubated for 45 min at 65 °C with periodic gentle mixing. The resulting lysate was chilled on ice for 15 min and neutralized by thorough mixing with 20 ml of acid phenol:chloroform (1:1, v-v), and cleared by centrifugation at 10 000 g and 4 °C for 30 min. The supernatant was aspirated, filtered through Whatman No. 1 filter paper, and supplemented with NaCl to a final concentration of 250 mℳ. DNA was precipitated for 30 min at room temperature with 0.8 v/v of 2-isopropanol, recovered by centrifugation and dissolved as described for the method.
For the isopycnic density centrifugation, 7.7 g CsCl were dissolved in 7.0 g of DNA solution, supplemented with 0.7 ml of 10 mg ml−1 EB and cleaned from precipitate by a 20 min centrifugation at 6000 g and 20 °C, and filtration through glass wool. The resulting DNA-CsCl-EB solution was then centrifuged in 16 × 60 mm Optiseal tubes (Beckman, 361623) in a Kontron TFT 80.13 fixed angle rotor at 40 000 r.p.m. and 20 °C for 48 h. Both the lower (CCSD) and upper (linear and nicked circular DNA) bands seen under long wave ultraviolet light were harvested by tube puncture. DNAs were extracted four times with water-saturated 1-butanol, and precipitated using ethanol. The two samples taken from the Morges WWTP on 3 and 6 May 2007 yielded 262 μg and 150 μg of CCSD, respectively. The Morges 2009 sample produced 90 μg of CCS DNA. The Visp 2009 sample produced 80 μg CCS DNA.
Library construction, sequencing and assembly
Libraries were prepared and sequenced at the United States Department of Energy Joint Genome Institute (US DOE JGI) as part of the Community Sequencing Program 2007 (project CSP 776886). For the sample Morges 2007, three clone libraries were constructed in the vectors pUC18c (2–4 kb insert size, libraries FPCF and FOTH) and pMCL200 (8–10 kb insert size, library FXNX) according to JGI standard protocols. Sanger clones were sequenced on an ABI PRISM 3730 capillary DNA sequencer (Applied Biosystems, Carlsbad, CA, USA) according to the JGI standard protocols (www.jgi.doe.gov), screened for the presence of vector, and quality trimmed with LUCY version 1.19p (Chou and Holmes, 2001) with quality cutoff set to 15. This yielded 73 225 reads totaling 56 mega base pairs (Mbp) of sequence. CCSD from sample Morges 2009 was sequenced at JGI using a Genome Sequencer FLX System (454 Life Sciences, http://www.454.com/) (Margulies et al., 2005) with long-read GS FLX Titanium chemistry according to the manufacturer protocol, generating 180 548 reads totaling 48.7 Mbp of sequence. DNA sample Visp 2009 was sequenced at JGI using both pyro and Sanger sequencing approach (as described above for Morges 2007/2009 samples) to yield 238 059 trimmed 454 reads plus 12 065 trimmed Sanger reads from 8–10 kb insert library, altogether totaling 97 Mb. All reads from the sample Visp 2009 were assembled at JGI by Newbler, automatically annotated by the IMG/M pipeline and deposited to IMG/M under taxon object ID 2035918001.
Quality trimmed sequences were used to generate three assemblies: (i) Morges 2007 trimmed Sanger reads were assembled at JGI with PGA (Paracel Genome Assembler 2.6.2, Paracel, Pasadena, CA, USA), (ii) Morges 2009 454 reads were assembled using Newbler (assembler version 2.0.0-Post-Release-9 May 2008 (Roche/454)), and (iii) pooled reads from both samples were de novo assembled with MIRA 3 using the ‘accurate' option and disabling the ‘uniform read distribution' (Chevreux et al., 1999). The MIRA assembly of all three data sets yielded 13 099 contigs totaling 11 612 275 bp. One-third of all contigs were covered on average at least fourfold, comprising a total sequence space of 3 895 880 bp over 1324 contigs with an average length of 2942 bp and a maximum length of 39 565 bp. About half of those contigs was even covered eightfold, comprising a total sequence space 1 744 501 bp over 330 contigs with an average length of 5286 bp and a maximum length of 39 565 bp. A restricted subset of high quality major contigs longer than 1 kb and covered by sequencing more than five times was chosen as a representative nonredundant data set named ‘MIRA CONTIGS' (649 contigs covering 2 205 428 bp in total). A total of 40 contigs was closed as circular replicons and verified by PCR.
Annotation and comparative analysis
Contigs and singletons yielded by lucyPGA (sample ‘MORGES-2007-PGA'), or Newbler (sample ‘VISP 2009') and a non redundant set of contigs from Morges 2007/2009 assembly (‘MIRA CONTIGS') were annotated using the JGI microbial genomics/metagenomics pipeline (IMG/M), curated using an expert review system (Markowitz et al., 2008, 2009) and were deposited under the IMG/M database taxon object ID 2013843001, ID 2035918001 and ID 2209111023, respectively. In addition, the following data sets were functionally annotated using the MG-RAST (Meta Genome Rapid Annotation using Subsystem Technology; v3.1) server at the Argonne National Library (http://metagenomics.nmpdr.org) and more specifically by using the subsystem-based annotation within the SEED database (Meyer et al., 2008): ‘MIRA CONTIGS', ‘MORGES2007-SANGER' (MG-RAST Acc. Nr. 4464065.3); ‘MORGES2009-454' (MG-RAST Acc. Nr. 4474065.3); ‘VISP2009-SANGER (MG-RAST Acc. Nr. 4464108.3); VISP2009-454 (MG-RAST Acc. Nr. 4464109.3); VISP2009-SANGER/454'. By default we used an e-value cutoff filter of 0.00001.
To establish the possible phylogenetic origin of the dominant and known fraction of the WWTP mobile gene pool encoding DNA, polypeptide sequences were called on ‘MIRA CONTIGS' using MetaGene (Noguchi et al., 2006) in the CAMERA environment (release 2.0.6.2). Sequences were queried against ‘All prokaryotic proteins (P)' database using BLASTP (Altschul et al., 1997) with CAMERA default parameters and an e-value cutoff of 0.01. Finally, hierarchical taxonomy mapping was done in MEGAN (Huson et al., 2007) with the following last common ancestor parameters: min support=1, min score=35, top percent=10.
As an estimate of the proportions of functionally characterized genes in different data sets, we used the value of ‘protein coding genes with function prediction percentage' as defined by IMG/M annotation. To calculate abundances of cluster of orthologous groups (COG) categories in different individual metagenomic or collective data sets, we summed up hit counts to each COG category and divided by the total number of COG assignments in a given data set. All data sets were hereto annotated uniformly in the IMG/M pipeline. To search for protein family (Pfam) conserved domains using profile hidden Markov models, we implemented the RAMMCAP pipeline (Li, 2009) either as a standalone application or via the CAMERA web portal for the following data sets: ‘MIRA CONTIGS', ‘MORGES2007-SANGER' ‘MORGES2009-454' ‘VISP2009-SANGER; VISP2009-454; VISP2009-SANGER/454' ‘PLASMIDS NCBI' and the ‘WWTP NC' metagenome. Pfam conserved replicon domains were searched using the IMG/M annotation pipeline in the data sets Morges 2007 (PGA assembly, ID 2013843001) and Visp 2009 (Newbler assembly, ID 2035918001).
Plasmid metagenome data sets were further compared with a variety of other metagenomes, all available and annotated uniformly in IMG/M on 29 September 2011 (Supplementary Table S3), and one artificially assembled metagenome from existing plasmids in IMG/M 'PLASMIDS-IMG' (Supplementary Table S4). As reference for activated sludge whole-metagenome in MG-RAST we used pyrosequencing reads from the North Carolina WWTP metagenome, named ‘WWTP NC' (Sanapareddy et al., 2009) and from the Aalborg WWTP EBPR (MG-RAST, Acc. Nr. 4463936.3). The alternative comparative plasmid data set in MG-RAST named ‘Plasmids-NCBI' was compiled from the list of plasmids from the domain Bacteria in the size range 1–150 kb (1878 plasmids totaling 55 029 614 bp, available from NCBI database as on 24 May 2011, see Supplementary Table S10), and was annotated by RAMMCAP and MG-RAST.
The 40 closed replicons were automatically annotated using the GenDB expert system (Meyer et al., 2003). Thirteen representatives of those were manually curated, exported and displayed using DNAplotter (Carver et al., 2009). Eight replicons were submitted to GenBank and are available under accession numbers JX194159-JX194161 and JX202560-JX202564.
Comparisons and statistical analysis
Pairwise comparisons were done using the STAMP (Statistical Analysis of Metagenomic Profiles) package (Parks and Beiko, 2010). Routinely a two-sided Fisher's exact test was implemented for hypothesis testing, whereas the difference in proportions (DPs) and confidence intervals (CIs) for P=0.95 were calculated using the Newcombe-Wilson method. Multiple test corrections were done using the Storey q-value (Storey and Tibshirani, 2003) or the Benjamini–Hochberg false discovery rate (Benjamini et al., 2001). To compare the distribution of antibiotic and heavy metal resistance proteins between samples, we examined the hypothesis of independence between the origin of the sample and the proportion of resistance proteins using a Pearson's χ2 test as implemented in R (http://www.r-project.org/). The Pearson residuals were used as a measure of excess or depletion in resistance proteins relative to the mean distribution.
Results
Plasmid metagenomes contain among the largest fractions unknown DNA
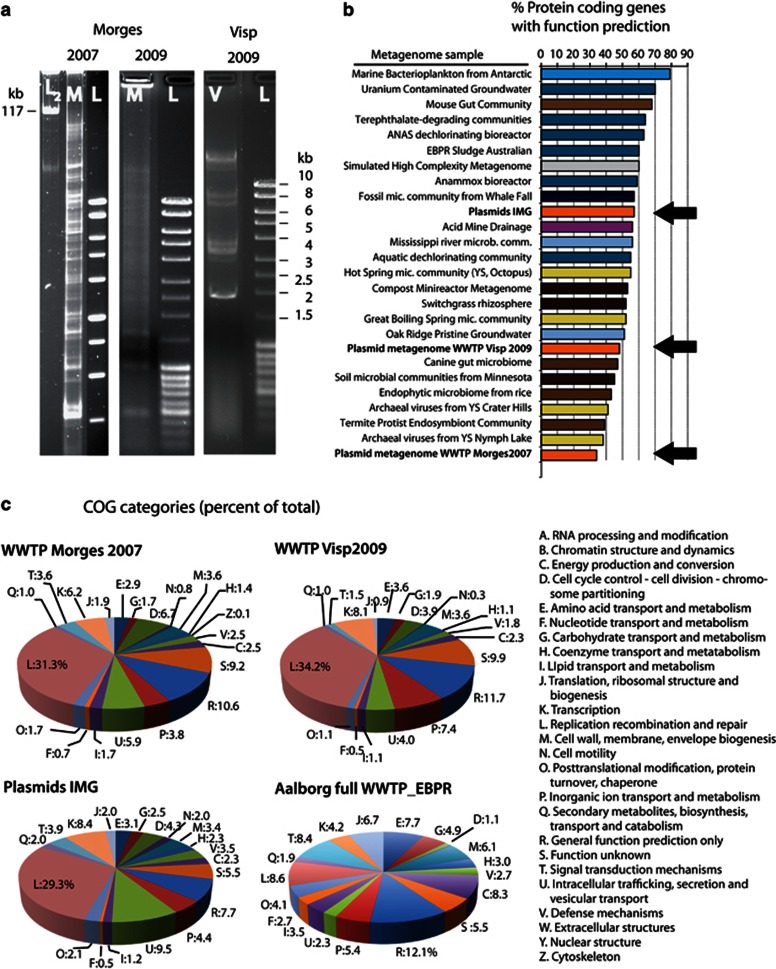
We use physical separation of CCSD from total prokaryotic community DNA and a rigorous metagenomic approach to describe and compare the functional content of the WWTP plasmid metagenomes. The plasmid isolation methods visibly resulted in cleared lysates from washed WWTP sludge samples with few non-lysed cells, as verified microscopically. Nevertheless, there may have been some bias in the types of microorganisms, from which plasmid DNA could be recovered (see further below). Pools of CCSD were prepared from activated sludge samples taken from the Morges municipal WWTP in 2007 and 2009, yielding 71 225 trimmed Sanger reads totaling 56.0 Mbp, and 180 458 GS FLX Titanium chemistry reads totaling 48.7 Mbp of sequence, respectively. The CCSD from the Visp WWTP was isolated in 2009 and yielded 250 115 GS FLX Titanium reads (142.6 Mbp) plus 14 592 Sanger reads (12.1 Mbp). Visual inspection of the Morges 2007 and 2009 CCSD samples by agarose gel electrophoresis and EB staining showed at least 45 discrete bands ranging in size from 2–100 kbp, as well as a large number of diffuse bands corresponding to less abundant DNA molecules (Figure 1a). In contrast, the Visp CCSD sample was visually less variable. The low abundance of sequence matches to 16S rRNA genes in two of three CCSD preparations (Morges 2007 and Visp 2009), indicated little contaminating prokaryotic chromosomal DNA (Supplementary Table S1). This was used to empirically define a functional assignment threshold of 0.05% abundance, below which a predicted gene function could no longer be reliably attributed to the CCSD pool.
Figure 1.
Analysis and comparison of the WWTP plasmid metagenomes. (a), Agarose gel separated recovered CCSD pools from WWTP Morges (M) in 2007 and 2009, and WWTP Visp (V) in 2009, compared with regular DNA size standards (L) and the TOL plasmid (L2). (b), Percent protein coding genes with function prediction of total nonredundant contigs plus singletons annotated uniformly using IMG/M (Markowitz et al., 2008), for a variety of prokaryotic community metagenomes, viral and plasmid metagenomes (indicated with a black arrow). For metagenome accession numbers and for a complete ranking of 291 metagenomes see Supplementary Information, Supplementary Table S3. (c), COG category predictions (percentages of total) for protein coding genes uniformly annotated using IMG/M in the Morges 2007 and Visp 2009 plasmid metagenomes compared with collective plasmids in IMG/M and the full WWTP metagenome from Aalborg (DK) (Albertsen et al., 2012).
Surprisingly, the largest part of both CCSD metagenomes did not match any known DNA in the collective nonredundant nucleotide and genome databases (for example, only 18% of all predicted CCSD proteins matched existing GenBank entries at 80% amino acid similarity). Only 34% of the predicted coding sequences in the Morges 2007 and 48% in the Visp nonredundant plasmid metagenomes (that is, all assembled contigs plus remaining singletons) could be functionally annotated using IMG/M (Markowitz et al., 2008) (Figure 1b), with even slightly lower proportions of significant hits to known COGs or protein families (Pfam and TIGRfam, Supplementary Table S2). In addition, about one-fifth of the functionally classifiable coding sequences fell into the poorly characterized COG categories R of ‘general function prediction only' and S of ‘function unknown' (Figure 1c). The Morges 2007 nonredundant plasmid metagenome ranked among the lowest in the proportion of functionally assignable DNA in comparison to a range of available metagenomic data sets (Supplementary Table S3) representing microbial communities of a wide variety of environments, lower than various virus metagenomes, thermophilic Archaea (38–41% classifiable) or termite protist endosymbionts (39%, Figure 1b). In comparison, 57% of the coding sequences predicted from an artificial assembly of 1148 plasmids available in IMG/M (Supplementary Table S4) are functionally characterizable (Figure 1b). This indicates that WWTP plasmid metagenomes occupy a significant part of unknown sequence space, comparable with virus metagenomes (Breitbart et al., 2002). Therefore, plasmids may not only provide their prokaryotic hosts with known auxiliary functions of ecological and adaptive importance, but may also act as a source of new functional DNA that may at some point be co-opted to improve cellular fitness.
Plasmids capture most types of known bacterial gene functions
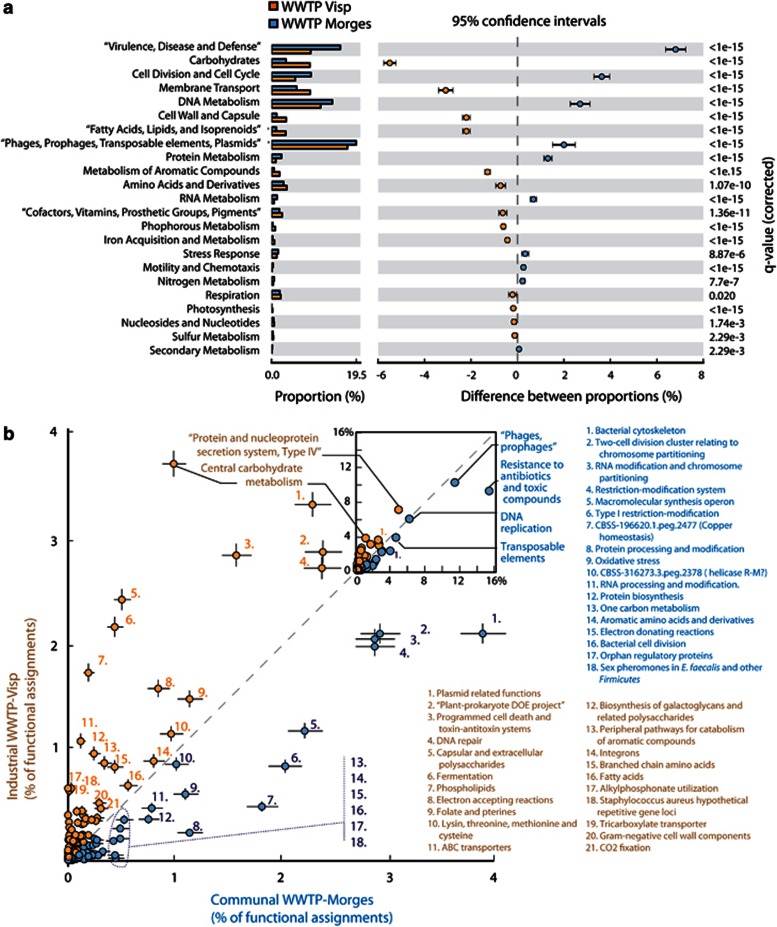
The 34–48% functionally characterizable part of the CCSD metagenomes comprises a wide-range of cellular functions, supporting the hypothesis that most types of functions can be captured on plasmid DNA and become mobilized (Figure 2). In comparison to a complete (chromosomal and plasmid) WWTP metagenome (Albertsen et al., 2012) the genes encoding DNA replication, recombination and repair (COG-L) were highly overrepresented in the plasmid metagenomes (ca. fivefold difference, Figure 1c). This is mostly due to the high density of transposable elements in the plasmid metagenomes. Functions categorized in COG-D (Cell cycle control, cell division and partitioning) and -U (Intracellular trafficking, secretion and vesicular transport), which broadly associate with replicon maintenance and transfer, were also proportionally more abundant in the plasmid metagenomes. In terms of functional subsystems classification by SEED (Meyer et al., 2008), the Morges 2007 CCSD compared with the full WWTP metagenome is highly significantly (corrected P-value <1·10−15) enriched in genes associated with resistance to antibiotics and toxic compounds, stress response and type II and IV secretion systems (SEED categories Virulence, disease and defense, and Membrane transport, Supplementary Figure. S1). Interestingly, 19.5% of predicted proteins of the plasmid metagenome showed discernable signal peptides, suggesting extracellular transport. A further 8.9% of genes were predicted to encode transmembrane proteins (Supplementary Figure S1), which is consistent with recent observations that secreted proteins are an adaptive and cooperative prokaryotic trait selected on mobile DNA elements (Nogueira et al., 2009; Rankin et al., 2011).
Figure 2.
Community level functional bias of protein coding genes in the communal (Morges 2007) versus industrial WWTP (Visp) plasmid metagenomes. (a), Percent SEED categorizable protein coding genes and pairwise proportional differences calculated using STAMP (Statistical Analysis of Metagenomic Profiles). (b), Functional overrepresentation of SEED subcategories in Morges versus Visp plasmid metagenomes, as percent of total functional assignments. Whiskers denote calculated 95% confidence intervals.
We investigated the possible phylogenetic origin of the known genes encoded in the plasmid metagenome, using the protein coding regions in the most conservative and five times covered ‘MIRA CONTIGS' data set derived from both WWTP Morges CCSD pools and a BLASTP-MEGAN approach (Huson et al., 2007), and compared this with a prokaryotic community analysis on the Morges 2007 sludge by in-situ hybridization (FISH) using rRNA-directed fluorescent probes (Supplementary Table S5). Among the 2828 predicted proteins, 1739 were assignable to the domain Bacteria and 3 to Archaea. A further 8 could not be unequivocally assigned and the remaining 1077 did not match anything at the selected thresholds (Supplementary Figure S2). Assignments to Betaproteobacteria comprised 32%, with 34% of cells in sludge hybridizing to the Betaproteobacteria selective probe (Supplementary Table S5). In contrast, Gammaproteobacterial genes (15.2%) were overrepresented in the plasmid metagenome compared with FISH-results (3.0%), whereas Alphaproteobacterial (5.1% in metagenome versus 36.0% by FISH) and Actinobacterial (2.6% versus 12.0%) sequences were underrepresented (Supplementary Table S5). Around one-fifth of the sequences could not be assigned more precisely than at the domain (16.6%, Bacteria) or phylum (21.6%, Proteobacteria) level (Supplementary Figure S2). Taken from single reads, the inferred phylogenetic distributions were similar for both Morges 2007, Morges 2009 and the Visp 2007 plasmid metagenomes, and close to an artificial pool of known NCBI plasmids. In contrast, plasmid metagenome phylogenies were rather distinct from most other complete microbial metagenomes including a full WWTP metagenome, except for the Whale fall metagenome (Supplementary Figure S3). This suggests that plasmids maintain a clear phylogenetic signature despite mobilizing a large variety of genes from prokaryote host chromosomes.
Replicon diversity
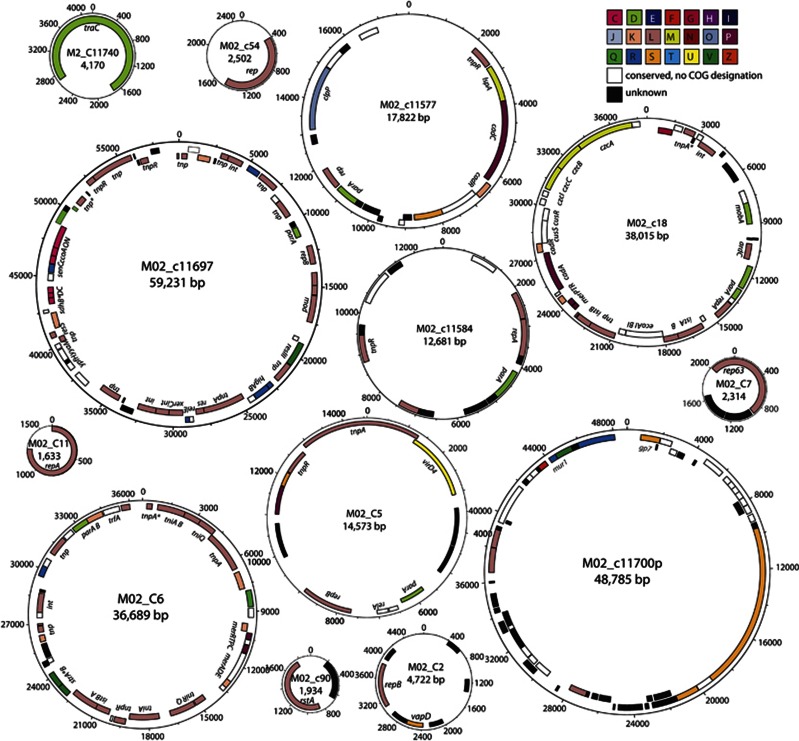
The combination of Sanger and 454 sequencing technologies permitted assembly of a subset of contigs from the Morges 2007 sample with at least fivefold coverage (‘MIRA CONTIGS', 649 contigs covering 2 205 428 bp in total). Of those, a total of 40 replicons could be completely closed and were verified by PCR for correct assembly (Supplementary Table S6). The smallest replicon (M02_C11) was 1633 bp and encoded only a single repA plasmid replication protein (Figure 3). A number of other extremely small plasmids were assembled with different types of replication proteins (Figure 3). The largest replicon that could be assembled was 59 231 bp (M02_c11697) and appeared to be a highly complex plasmid with plasmid partitioning and maintenance control functions (for example, higAB), and multiple different transposons (Figure 3). The general high density of transposable elements on the various replicons, the wide-range of genes for conserved hypothetical functions and of genes for unknown predicted proteins illustrate and emphasize the statements made above on the basis of nonredundant reads only. In addition, among the assembled plasmid replicons several carried genes for heavy metal resistances (for example, mercury, cadmium and zinc; M02_c11577, M02_c18, M02_C6, Figure 3) or putative antibiotic resistance genes (for example, strAB for streptomycin resistance, M02_C6). In contrast, very few conspicuous tra or mob functions were detectable (for example, mobA on M02_c18, virD4 on M02_c5), although in general a high proportion (up to 7%) of 'protein and nucleoprotein secretion system, type IV' was detected in the complete data sets, suggesting mobilization and transfer capacity of many replicons (Figure 2). Interestingly, also three putative circular phage replicons were assembled (12.3 kb, 22.4 kb and 48.8 kb), the largest of which was manually curated and is depicted in Figure 3 (M02_c11700p). The high sequence coverage of the phage genomes suggests that they are highly abundant in the WWTP system.
Figure 3.
Subset of manually curated assembled plasmids and one phage replicon from Morges 2007 using MIRA on contigs covered more than five times. Replicon closures verified through paired-end reads and by PCR. Locations of protein-encoding open reading frames predicted using the GenDB expert annotation system are indicated (Meyer et al., 2003). Gene names were assigned to open reading frames in case of a functional prediction above 80% identities. Colors depict predicted COG categorization, as indicated. Replicon numbers accessible in GenBank through numbers as specified in Supplementary Table S6.
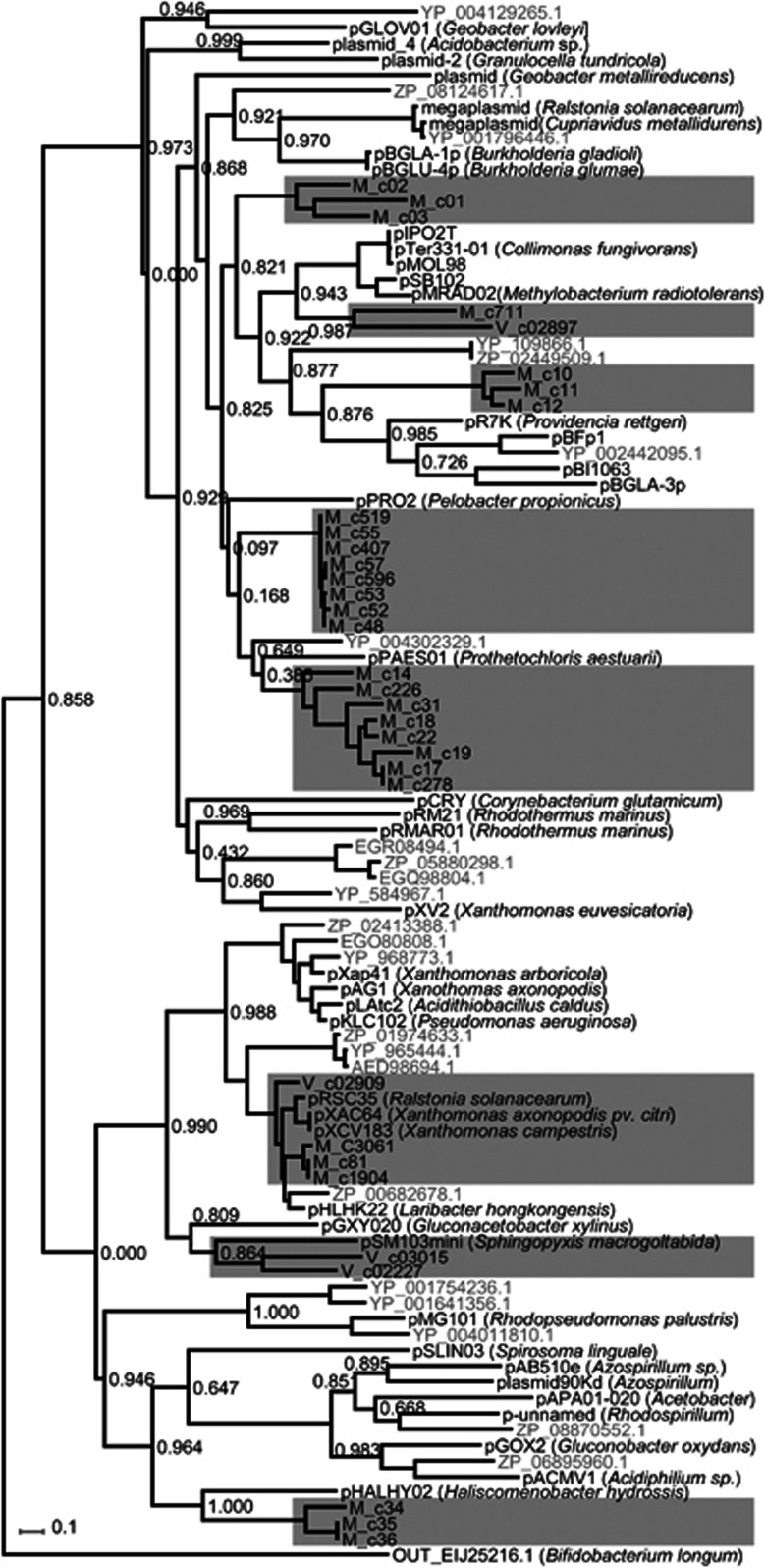
On the basis of replication protein diversity analysis, we detected 13 of 16 known protein families, of which RepA_C (Pfam 04796), Rep_3 (Pfam 01051) and RPA (Pfam10134) were the most abundant (Table 1). The RepA_C family contains among others IncP9 plasmids from Proteobacteria and replicons such as pTP10 from Actinobacteria (Supplementary Information File 1). Replicons with Rep_3 type replication initiator protein often originate in Firmicutes or Beta- and Gammaproteobacteria. RPA-types have few known plasmid members but are mostly found in Alphaproteobacteria (Table 1, Supplementary Information File 1). Interestingly, the plasmid replicon families differed considerably between WWTP Morges and Visp, with RepA_C being the most abundant in Morges (45.5%) and RPA in Visp (69.5%). This suggests strong local niche adaptation by existing plasmids in the activated sludge communities. Within the RepA_C family alone the plasmid replicons from Morges formed a number of new, deeply branching and distinct groups for which the closest characterized neighbor was plasmid pPAESO1 from Prostecochloris aestuarii DSM 271 (Figure 4). Plasmid RepA_C replicons from Morges and Visp were also very distinct, except in one case (v_c02909, M_c3061, M_c81, M_c1904), which formed a distinct cluster with plasmid pXAC64 from Xanthomonas axonopodis (Figure 4).
Table 1. Representation of different replication initiation protein families in the WWTP CCSD metagenomes.
| Pfam | Name | Known repliconsa | Known hostsa |
Abundance % (total)b |
|
|---|---|---|---|---|---|
| Morges | Visp | ||||
| PF04796 | RepA_C | pCAR3, pIPO2T (IncP9), pXAC33, pAP12875, pNL1 | Proteobacteria, Actinobacteria | 45.4 (769) | 2.0 (205) |
| PF01051 | Rep_3 | pRC18, pCL300, pSCFS1, R46 (IncN) | Gamma-, Betaproteobacteria, Firmicutes | 15.7 (266) | 16.2 (1167) |
| PF10134 | RPA | pSD20, pMTH1, pHLHK8 | Mostly Alphaproteobacteria | 7.8 (133) | 69.5 (7145) |
| PF01446 | Rep_1 | pGI1 (RCR group III), pAE78, pLNU5, pLTK2 | Mostly Firmicutes | 6.8 (116) | 6.3 (648) |
| PF08707 | PriCT_2 | pUH24 | Diverse | 6.4 (109) | 2.1 (216) |
| PF07042 | TrfA | pB4, pJP4 (IncP1b), pUO1, R751, pIJB1 | Mostly Proteobacteria | 6.4 (109) | 0.8 (80) |
| PF08708 | PriCT_1 | pLIM (ColE2), pRE25, pEI1, pMG1 | Firmicutes, Proteobacteria, Actinobacteria | 3.7 (62) | 0.9 (97) |
| PF03090 | Replicase | pNC500, pAP2, pA501, pLO2 | Firmicutes, Proteobacteria, Actinobacteria | 2.7 (46) | 0.5 (55) |
| PF09250 | Prim-Pol | pHEN7, pDL10, pCG2 (pBL1 fam) | Firmicutes, Proteobacteria, Actinobacteria | 2.7 (45) | 0.3 (29) |
| PF05144 | Phage_CRI | pNAD1, pVT1, pMM1 | Mostly Gamma- and Betaproteobacteria | 0.8 (14) | 0.0 (2) |
| PF02486 | Rep_trans | pBMB2062, pRE25, pSTK1, pMLU1, pC223 | Firmicutes, Proteobacteria, Actinobacteria | 0.6 (11) | 0.2 (18) |
| PF05732 | RepL | pSMQ1, pKH12, pSN2 | Mostly Firmicutes | 0.6 (11) | 0.3 (29) |
| PF03428 | RP-C | pHCG3, Ti | Mostly Alphaproteobacteria | 0.2 (4) | 0.9 (94) |
| 100 (1695) | 100 (10258) | ||||
Abbreviation: Pfam, protein family.
Non-exhaustive list of example plasmid replicons occurring in the ACLAME database. For a full list, see Supplementary Information-File 1.
Pfam counts (in parentheses) are the estimated number of gene copies from integrated contig coverage data.
Figure 4.
PhyML (Guindon and Gascuel, 2003) tree based on a PRANK alignment (Loytynoja and Goldman, 2005) of full-length RepA_C family protein sequences from WWTP Morges (names beginning with M) and Visp (names beginning with V), highlighted within gray shaded areas, with their most similar orthologs (GenBank entries with existing plasmid names in black; others in gray). Topology search using best of nearest neighbor interchanges and subtree pruning and regraftings, the LG model of amino acid substitution and discrete gamma model with four categories. Numbers at the nodes indicate the minimum of SH-like and χ2-based branch supports (Anisimova and Gascuel, 2006) (closer to one indicates stronger support). Outgroup is the RepA_C|EIJ25216.1| from Bifidobacterium longum subsp. longum 2-2B. A complete list of all assigned full names is available in Supplementary Table S11.
Gene capture and selection at the community level
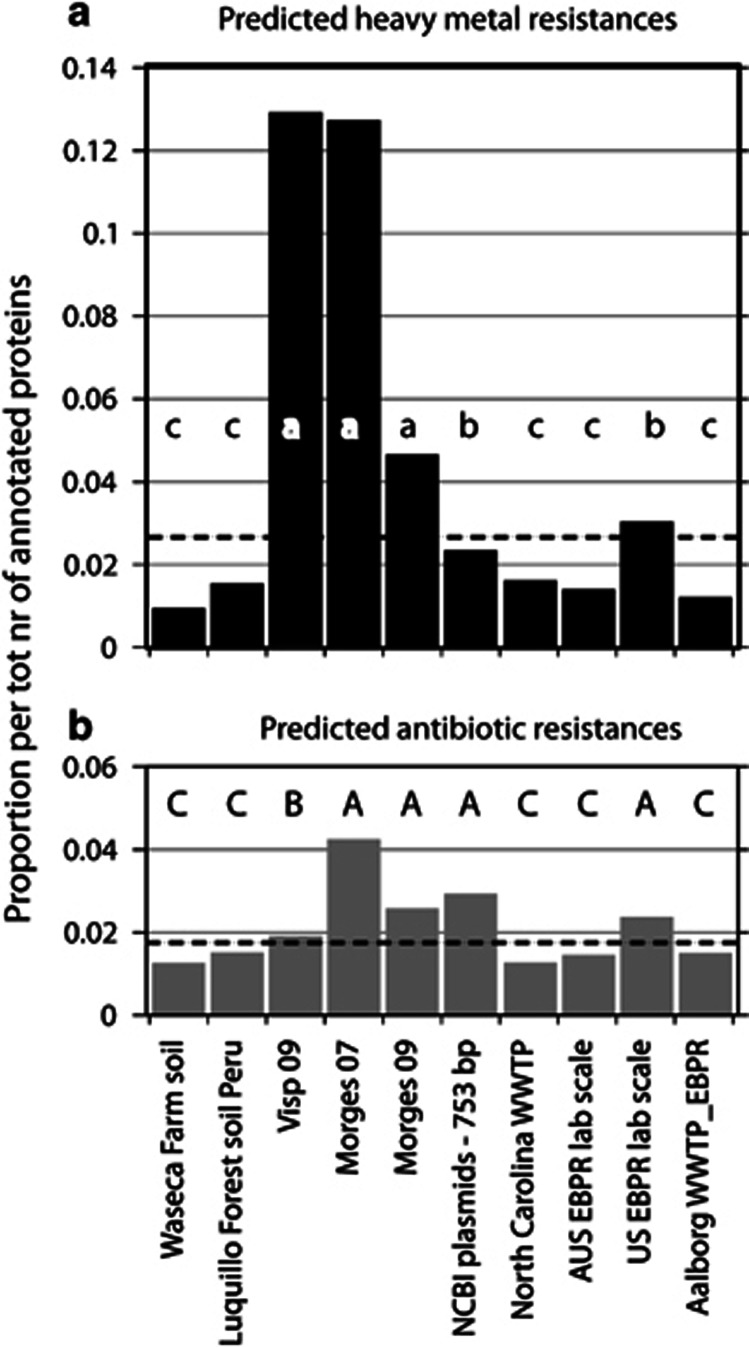
To address the hypothesis that plasmid genes are selected at the level of the microbial community as a whole, we compared the functionally annotated gene content in the Morges 2007 versus Visp 2009 CCSD metagenomes (the Morges 2009 data set being excluded because of slightly higher potential chromosomal contamination). Whereas both metagenomes were almost equally dominated by plasmid DNA replication and maintenance functions plus transposable elements, the Visp 2009 metagenome was significantly enriched in functions involved in carbohydrate metabolism than Morges 2007 (Figure 2). In contrast, the Morges plasmid metagenome was significantly enriched in virulence, defense and stress response functions, although this functional category comprised 10% or more of all assignments in both Morges and Visp metagenomes (Figure 2b). Consequently, both Morges and Visp plasmid metagenomes encode a wide-range of antibiotic (for example, beta-lactamases, spectinomycin/streptomycin 9-O-adenylyltransferase) and heavy metal resistances (for example, mercury, copper, cadmium, zinc and chromium, Supplementary Figure S4). The proportion of arsenic resistance genes was clearly higher in the Visp plasmid metagenome, whereas the combined Morges plasmid metagenomes contained slightly more antibiotic resistance genes (Supplementary Figure S4, Supplementary Tables S7-S9). Compared with both WWTP plasmid metagenomes, the NCBI artificial plasmid metagenome is biased towards specific functions associated with virulence and membrane transport, underscoring that our current knowledge of plasmid functions from individual prokaryotic isolates is not representative for large scale complex microbial communities (Supplementary Figure S5). Both Morges and Visp plasmid metagenomes are highly enriched in heavy metal and antibiotic resistance proteins compared with full WWTP or soil metagenomes (χ2 test, P<2·10−16, Figure 5).
Figure 5.
Comparison of proportional abundances of genes for heavy metal (a) and antibiotic resistance (b) in the Morges and Visp CCSD metagenomes. Proportions are calculated as the number of predicted proteins in each category compared with the total number of annotated proteins in the respective metagenome using MG-RAST subsystems classification (Meyer et al., 2008). The distribution of resistance proteins differs significantly between samples (χ2 test P<2·10−16). Letters indicate strong deficit (c, C), large excess (a, A) or little difference (b, B) compared with the χ2 expectation. The dotted lines indicate the computed average proportion corrected for the sample size.
Discussion
We show for the first time a comprehensive analysis of the metagenomic pool of extrachromosomal DNA in complex microbial communities from WWTPs. Although CCSD contains both plasmid and circular phage DNAs, it can be considered as a reasonable description of the community's 'mobilome'. In contrast to current ultrahigh throughput metagenomic sequencing on total DNA, mobilome metagenomics requires tedious separation and purification of CCSD, and is therefore still more constrained by technical and not sequencing limitations. In addition, only the combined approach of Sanger and 454 sequencing permitted replicon assembly from metagenomic data, which was essential for the claims made in this study but, which also limited the high-throughput of a larger set of samples from more and different WWTPs.
Our results permit one to draw a number of pertinent conclusions. First, plasmids isolated from microbial communities without culturing bias carry a majority (51–68%) of uncharacterized coding sequences, supporting previous hypotheses and observations that plasmids are a driving force for innovation of genetic information that may at some point provide selective advantage to host cells. Second, plasmids effectively encode a wide-range of known functions, and plasmid metagenomes contribute detectably and significantly to the mobilization of antibiotic and heavy metal resistance genes, compared with full metagenomes (Figure 5). As our results originate from culture-independent approaches, we feel it gives a very strong, unique and important demonstration of the general contribution of plasmids to antibiotic resistance gene distribution in complex microbial communities. Third, the plasmid metagenomes from two WWTPs with similar operating conditions but different influent loads are very similar in terms of the proportion of plasmid maintenance functions, but significantly different in terms of auxiliary gene functions. In addition, the plasmid replicons from both systems belong to new, deep-branching and very different families (Figure 4, Table 1).
Given the industrial influent with high organic carbon loads coming into WWTP Visp, it was striking to see a high proportion and coverage of catabolic functions (carbohydrate metabolism) encoded on the Visp plasmid metagenome. This suggests that plasmids with genes for catabolic functions were selected under those conditions and contributed to adaptation of the prokaryotic sludge community as a whole. On the other hand, the broader range of compounds in household wastewater in the WWTP Morges selected for a wide variety of other gene functions, including a high proportion of genes encoding antibiotic and toxic compound resistance. The enrichment was significant in both mobilomes of 2007 and 2009, although the latter contained slightly more contaminating chromosomal material, as a result of which the proportion of genes for antibiotic and toxic compound resistance was slightly lower (Supplementary Table S1, Figure 5). The differences between the functional content of the plasmid metagenomes from the two WWTP are so dramatic, and the sequence depth of the data sets is so high that we feel confident that the general conclusions we draw from the results are statistically sound, even though this should be confirmed by further independent mobilome samples from other WWTP with similar types of influent. Our results thus strongly point to a similar mechanism of adaptation in both WWTP (that is, gene mobilization via plasmids), but very different at the local conditions and systems boundaries (that is, different plasmid replicons present in the system and different selective conditions in terms of carbon inflow). A recent study on a plasmid metagenome from the rumen also provided evidence for specific plasmid functional selection of presumed gut niche advantageous functions (Kav et al., 2012).
So far, metagenomic studies have been very successful in characterizing the genetic and taxonomic diversity of a large number of microbial ecosystems, and have enormously increased our knowledge of the functions of the majority of microorganisms that have not been isolated in pure culture. Such metagenomic approaches generally attempt to sequence as much as possible of the total DNA isolated from the community in question, or focus on sequencing of specifically amplified phylogenetic markers such as the gene for 16S rRNA. Viral metagenomes have also been subject to intensive study, because of the possibility to separate viruses from prokaryotic cells and eukaryotic microorganisms (Breitbart et al., 2002; Bench et al., 2007; McDaniel et al., 2008; Rosario et al., 2009). In contrast, much less is known about the diversity and evolution of plasmid replicons in microbial communities, mostly because of the more cumbersome purification needed to separate plasmid DNA from chromosomal DNA molecules. To address this problem a number of groups have used techniques of ‘exogenous plasmid isolation' to capture plasmids from complex systems in specific known hosts (Top et al., 1994; Miyazaki et al., 2006; Schlüter et al., 2007), with or without specific selective growth conditions (for example, growth in the presence of antibiotics to recover plasmids with genes for antibiotic resistances) (Schlüter et al., 2008, Szczepanowski et al., 2008). More recently, also transposon associated capture (TRACA) was used to capture and sequence roughly two dozen plasmid replicons from the total metagenomic DNA via propagation in Escherichia coli (Jones and Marchesi, 2007; Jones et al., 2010). Finally, purification using plasmid-safe exonuclease treatment was used to specifically enrich plasmid DNA from complex communities and characterize the plasmid metagenome (Kav et al., 2012). In contrast, we used here modifications of classical plasmid alkaline lysis procedures in combination with CsCl-EB density centrifugation to purify CCSD molecules directly from WWTP microbial communities. Although our method may not have been completely effective in cell lysis of all species, and does not permit to isolate linear plasmids, we obtain an unprecedented view of plasmid diversity and abundant double-stranded circular phage DNAs in complex microbial ecosystems. Analysis of different types of replicons detected through replication protein families suggested that we cover plasmids regularly occurring in both Gram-negatives and Gram-positives (Table 1). This is in good agreement with the FISH analysis of phylogenetic groups (Supplementary Table S5). In contrast to previous CCSD metagenome efforts, we are also the first to actually assemble a range of plasmid replicons and a few circular phages directly from the metagenomic sequences, some of which are the largest assembled from metagenomics so far (Figure 3, Supplementary Table S6). Although we detected the presence of replicons related to plasmids that were exogenously recovered from WWTP communities on the basis of antibiotic resistances, such as pB4 or pB10 from the TrfA-type (Schlüter et al., 2003, 2008), they occurred at frequencies of 6% and lower (Table 1). This suggests that the proportionally more abundant plasmids from the RepA_C, Rep_3 and RPA groups have a more important role for the WWTP community. In conclusion, our study shows the technical possibilities to conduct specific plasmid metagenome analysis without culturing bias, and to study system level differences in the mobilome of microbial communities. Our data highlight the importance of plasmids for local adaptation of complex microbial communities, and forms a foundation for new efforts to further understand their evolution.
Acknowledgments
We thank Lutz Krause for his initial help in the project. This work was supported by a grant from the Swiss Infectigen program and by the Community Sequencing Program of the U.S. Department of Energy Joint Genome Institute, supported by the Office of Science of the U.S. Department of Energy under contract No. DE-AC02-05CH11231. We gratefully acknowledge Nina Sanapareddy and Anthony Fodor (UNC Charlotte) for sharing with us the North Carolina WWTP metagenome sequences and for granting access to their data set on MG-RAST. We further thank Erika Yashiro for bioinformatic support. The computations were performed at the Vital-IT Center (http://www.vital-it.ch) for high-performance computing of the SIB Swiss Institute of Bioinformatics.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hansen LB, Saunders AM, Nielsen PH, Nielsen KL. A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J. 2012;6:1094–1106. doi: 10.1038/ismej.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, et al. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S.1999Genome sequence assembly using trace signals and additional sequence information Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 99 German Conference on Bioinformatics: Hannover, Germany; 45–56.Available at http://sourceforge.net/apps/mediawiki/mira-assembler/index.php?title=Main_Page#Papers . [Google Scholar]
- Chou H-H, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Taylor MW, Wagner M. Wastewater treatment: a model system for microbial ecology. Trends Biotechnol. 2006;24:483–489. doi: 10.1016/j.tibtech.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Fondi M, Bacci G, Brilli M, Papaleo M, Mengoni A, Vaneechoutte M, et al. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol Biol. 2010;10:59. doi: 10.1186/1471-2148-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- Jones BV, Sun F, Marchesi JR. Comparative metagenomic analysis of plasmid encoded functions in the human gut microbiome. BMC Genomics. 2010;11:46. doi: 10.1186/1471-2164-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kav AB, Sasson G, Jami E, Doron-Faigenboim A, Benhar I, Mizrahi I. Insights into the bovine rumen plasmidome. Proc Natl Acad Sci USA. 2012;109:5452–5457. doi: 10.1073/pnas.1116410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–1074. doi: 10.1093/molbev/msq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L, Breitbart M, Mobberley J, Long A, Haynes M, Rohwer F, et al. Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS One. 2008;3:e3263. doi: 10.1371/journal.pone.0003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, et al. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki R, Sato Y, Ito M, Ohtsubo Y, Nagata Y, Tsuda M. Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation. Appl Environ Microbiol. 2006;72:6923–6933. doi: 10.1128/AEM.01531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Research. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EP. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol. 2009;19:1683–1691. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Rankin DJ, Rocha EP, Brown SP. What traits are carried on mobile genetic elements, and why. Heredity (Edinb) 2011;106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol. 2009;75:1688–1696. doi: 10.1128/AEM.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Pühler A, et al. The 64 508 bp IncP-1b antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1b group. Microbiology. 2003;149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Krause L, Szczepanowski R, Goesmann A, Pühler A. Genetic diversity and composition of a plasmid metagenome from a wastewater treatment plant. J Biotechnol. 2008;136:65–76. doi: 10.1016/j.jbiotec.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- Siefert JL. Defining the mobilome. Methods Mol Biol. 2009;532:13–27. doi: 10.1007/978-1-60327-853-9_2. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowski R, Bekel T, Goesmann A, Krause L, Kromeke H, Kaiser O, et al. Insight into the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to antimicrobial drugs analysed by the 454-pyrosequencing technology. J Biotechnol. 2008;136:54–64. doi: 10.1016/j.jbiotec.2008.03.020. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- Top E, De Smet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microbiol. 1994;60:831–839. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R, Hsiao WW, Kudo H, Myhre M, Dosanjh M, Petrescu A, et al. Functional characterization of a catabolic plasmid from polychlorinated- biphenyl-degrading Rhodococcus sp. strain RHA1. J Bacteriol. 2004;186:7783–7795. doi: 10.1128/JB.186.22.7783-7795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, et al. Cultivating the uncultured. Proc Natl Acad Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.