Abstract
Microbiologists are challenged to explain the origins of enormous numbers of bacterial species worldwide. Contributing to this extreme diversity may be a simpler process of speciation in bacteria than in animals and plants, requiring neither sexual nor geographical isolation between nascent species. Here, we propose and test a novel hypothesis for the extreme diversity of bacterial species—that splitting of one population into multiple ecologically distinct populations (cladogenesis) may be as frequent as adaptive improvements within a single population's lineage (anagenesis). We employed a set of experimental microcosms to address the relative rates of adaptive cladogenesis and anagenesis among the descendants of a Bacillus subtilis clone, in the absence of competing species. Analysis of the evolutionary trajectories of genetic markers indicated that in at least 7 of 10 replicate microcosm communities, the original population founded one or more new, ecologically distinct populations (ecotypes) before a single anagenetic event occurred within the original population. We were able to support this inference by identifying putative ecotypes formed in these communities through differences in genetic marker association, colony morphology and microhabitat association; we then confirmed the ecological distinctness of these putative ecotypes in competition experiments. Adaptive mutations leading to new ecotypes appeared to be about as common as those improving fitness within an existing ecotype. These results suggest near parity of anagenesis and cladogenesis rates in natural populations that are depauperate of bacterial diversity.
Keywords: anagenesis, bacteria, cladogenesis, ecotype, laboratory evolution, speciation
Introduction
Molecular surveys of bacterial diversity have revealed enormous numbers of species, both within a single community and worldwide. Microbial ecologists and evolutionary biologists are challenged to explain the origins and coexistence of at least tens of thousands (Roesch et al., 2007; Elshahed et al., 2008; Huse et al., 2010) and perhaps millions (Curtis et al., 2002; Gans et al., 2005) of bacterial species within a single community. While similar issues also vex zoologists and botanists trying to explain animal and plant diversity, microbiologists have orders of magnitude more species to accommodate within ecological and evolutionary theory (May, 1988).
Many features of bacterial population dynamics contribute to the rapid evolution of bacteria. These include large population sizes (Levin and Bergstrom, 2000), the promiscuity of genetic exchange between taxa (Cohan, 2001; Popa et al., 2011), the small size of recombined segments (Zawadzki and Cohan, 1995) and the simplicity and modularity of their physiology and development (Lawrence, 1999; Doyle et al., 2007). These advantages should act to accelerate not just the rate at which lineages split to form new species (cladogenesis), but also the rate at which one species lineage improves its adaptations without splitting (anagenesis).
One feature of bacterial population dynamics specifically fosters cladogenesis. Owing to the rarity of recombination in bacteria (occurring usually within an order of magnitude of the mutation rate) (Vos and Didelot, 2009), a population may diverge into two ecologically distinct populations that can coexist and diverge indefinitely, without evolving sexual isolation (Cohan, 1994; Vos, 2011; Wiedenbeck and Cohan, 2011). That is, the adaptive divergence of bacterial populations into different ecological niches is not hindered by the rare recombination between them. Thus, while divergence of animals into species-like populations (that are ecologically distinct and irreversibly separate) (de Queiroz, 2005) requires both ecological and sexual divergence, irreversible divergence in bacteria requires only the divergence of ecological features; moreover, geographic isolation is not required (Cohan, 2001; Cohan and Koeppel, 2008; Vos, 2011). Indeed, there is a growing literature of sympatric splitting of bacterial lineages in nature (Sikorski and Nevo, 2005; Coleman et al., 2006; Ward et al., 2006; Hunt et al., 2008; Koeppel et al., 2008; Miller et al., 2009; Oakley et al., 2010) and in laboratory microcosms (Rainey and Travisano, 1998; Treves et al., 1998; Rozen and Lenski, 2000; Maharjan et al., 2006; Blount et al., 2008).
Here, we propose and test a novel hypothesis to explain the extreme diversity of bacterial species—that cladogenetic splitting of lineages into multiple, ecologically distinct populations may be nearly as frequent as anagenetic adaptations within a single population's lineage. This hypothesis runs counter to the traditional view of speciation of animals and plants, influenced by Ernst Mayr (Mayr, 1963). In this view, macroorganismal speciation requires certain rare circumstances, including geographic isolation, which allow newly divergent populations to break free of recurrent, high-frequency genetic exchange. The consequence is that cladogenesis is expected to be much less frequent than quotidian adaptation within a lineage.
Nevertheless, zoologists and botanists have recently challenged the notion that speciation is rare and requires special circumstances. James Mallet and other proponents of ecological speciation have hypothesized that ordinary populations within ordinary animal species may be poised to speciate as easily as a single population might evolve a new adaptation—‘speciation is easy' (Mallet, 2008; Schluter, 2009; Doebeli, 2011). Here we test whether this is true for bacteria.
We have employed an experimental microcosm to address the relative rates of adaptive cladogenesis and anagenesis among the descendants of a clone of Bacillus subtilis, in the absence of competing species. We build on studies where microcosms were inoculated with a single clone, without access to the DNA of other organisms (Rainey and Travisano, 1998; Treves et al., 1998; Rozen and Lenski, 2000; Maharjan et al., 2006; Blount et al., 2008). In these previous experiments, descendants of the starting clone diverged, through mutation alone, into multiple ecologically distinct populations, either through specialization on different chemical (Treves et al., 1998) or spatially limited (Buckling et al., 2000) resources.
We have added to previous approaches a method to compare the rates of adaptive cladogenesis and anagenesis in a microcosm. Our strategy for identifying adaptive cladogenesis and anagenesis events is based on the dynamics of natural selection in populations where recombination is rare, as in B. subtilis (Roberts and Cohan, 1995) and most other bacteria (Vos and Didelot, 2009). Owing to the rarity of recombination, natural selection favoring an adaptive mutation within a population brings to 100% frequency not only the mutation but also nearly the entire genome in which the mutation occurred. Thus, each anagenetic event within a population is marked by the purging of diversity within the population, a process known as periodic selection (Koch, 1974; Levin, 1981).
However, the diversity-purging effect of periodic selection is limited to a population of ecologically homogeneous organisms. If two populations are utilizing different sets of resources and/or conditions, their ecological distinctness will protect them from the periodic selection events occurring within the other population (Cohan, 2005; Maharjan et al., 2006). We have defined an ecotype as an ecologically homogeneous population that is ecologically distinct from other such populations, thereby representing the domain of competitive superiority of an adaptive variant causing periodic selection (Cohan and Perry, 2007). Thus, the fate of an adaptive mutation within a community descended from a clone will depend on whether the community has diversified through cladogenesis into multiple ecotypes.
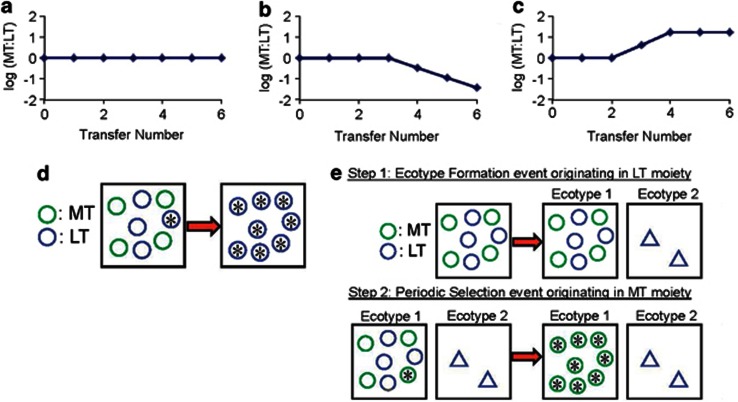
We enabled detection of anagenetic and cladogenetic events by inoculating each of several microcosms with two metabolically marked clones of B. subtilis that were isogenic and otherwise ecologically interchangeable (Atwood et al., 1951; Barrick et al., 2010). The rationale is that if a periodic selection event were to occur within the original ecotype before any new ecotypes were founded (single-ecotype model), the periodic selection event would yield extinction of one of the markers from the community (Figure 1b and d). Alternatively, if a microcosm community were to produce at least one new ecotype before a periodic selection event occurs in the original ecotype, a subsequent periodic selection event within one ecotype would extinguish only the diversity within that ecotype (multi-ecotype model). The cladogenetic origin of multiple ecotypes thus prevents periodic selection from extinguishing a marker from the community (Figures1c and e); instead, the relative frequencies of the markers reach a stable point determined by the relative abundance of the resources of the two populations. We have developed and analyzed statistical tests to identify the model that best fits each community's evolutionary trajectory (Tables 1 and 2; Figure 2).
Figure 1.
The expected trajectory of the MT:LT ratio (r) over time, under three models. (a) The null model, where strains are of equal fitness and in the same ecotype. Here, the initial r ratio is maintained indefinitely. (b) The single-ecotype community model. Here, the ratio stays at its initial value during a ‘wait time' (tw), to be estimated, until the ratio starts changing at a slope (m), to be estimated, leading to elimination of one of the markers. (c) In the multi-ecotype community model, there is a wait time, to be estimated, with no change in ratio, followed by a change in the ratio at a slope, to be estimated, followed by stabilization at a new end ratio (r∞), to be estimated. (d, e) Each box represents an ecotype; each circle or triangle represents an individual organism within an ecotype; the different colors indicate the selectable markers MT and LT; an asterisk indicates a mutant strain that is adaptive within its ecotype's niche. (d) Interpretation of the events observed in (b), where an adaptive mutant within one marker moiety (LT in this case) becomes fixed in the original ecotype, and thereby eliminates the other marker from the single-ecotype community. (e) One likely explanation for the events in (c) is that an ecotype formation event occurs within one marker, and a periodic selection event occurs in the original ecotype, stemming from an adaptive mutation in a genome with the other marker. We note that this method will not detect all cladogenesis events, especially if new ecotypes do not reach appreciable frequencies. In the absence of periodic selection, a new low-frequency ecotype would go unnoticed.
Table 1. Models of the evolutionary trajectory of a community.
| Model |
Parameters |
Expected ratio (r) at time t | |||
|---|---|---|---|---|---|
| Initial MT:LT ratio (r0) | Waiting time (tw) | Slope of change in ratio (m) | Steady-state ratio of two ecotypes (r∞) | ||
| Null | Yes | No | No | No | r=r0 over all times |
| Single-ecotype | Yes | Yes | Yes | No | r=r0 until time tw; log r=log r0+m (t - tw) after tw |
| Multi-ecotype | Yes | Yes | Yes | Yes | r=r0 until time tw; log r=log r0+m (t - tw) after tw and before r reaches r∞; r=r∞ after r reaches r∞ |
The initial-ratio parameter r0 was estimated directly from the initial MT and LT colony counts; the parameters for waiting time (tw), slope (m) and the steady-state ratio (r∞) were estimated to fit the observed trajectories of MT:LT ratio with maximum likelihood, as described in the text. The likelihood of the model yielding an observed ratio at a given time was based on the empirical Gaussian distribution of the ratio (Supplementary Figure 1). Yes and no indicate whether a parameter was included within a given model.
Table 2. Estimates of parameters to explain the trajectories of the MT:LT ratios over time in communities A–J.
| Community |
Relative likelihood |
Parameter estimates |
||||
|---|---|---|---|---|---|---|
| Null model | Single-ecotype model | Multi-ecotype model | Wait time (tw) | Slope of MT:LT ratio change (m) | End MT:LT ratio (log r∞) | |
| A | 0.31* | 1.16 | 3.92 | NA | NA | NA |
| B | <1.00E−142 | 70.80 | 5886.10* | 6 | −0.44 | −2.10 |
| C | 3.41E−112 | 2.10E−8* | 2.10E−8 | 3 | −0.18 | NA |
| D | 2.08E−51 | 4.22E−10 | 2.90E−6* | 5 | −0.34 | −1.00 |
| E | <1.00E−142 | 8.19E−18 | 8.81E−14* | 1 | −0.22 | −2.20 |
| F | 2.03E−73 | 3.13E−4 | 10.53* | 6 | 0.72 | 0.90 |
| G | 8.70E−85 | 1.38 | 133.09* | 9 | −0.86 | −2.10 |
| H | 1.10E−31 | 71.24 | 7312.20* | 7 | 0.38 | 0.70 |
| I | 3.21E−142 | 2.34E−8 | 0.50* | 4 | 0.30 | 1.30 |
| J | 2.07E−80 | 1.70E−6* | 1.95E−6 | 5 | −0.28 | NA |
Each trajectory of log10-transformed ratios was analyzed under each of three evolutionary models. The relative likelihood values are based on the deviation of each observed ratio from that expected under each model, assuming independent deviations of each time point from the model's expectation. Entries with asterisks indicate the most complex model consistent with observed data, based on likelihood ratio tests. The parameters represent the number of transfers for which the initial ratio is maintained (tw, wait time), the slope (m) of the MT:LT ratio after the wait time has been reached and the ratio (r∞) at which the slope levels off (all based on log10-transformed ratios).
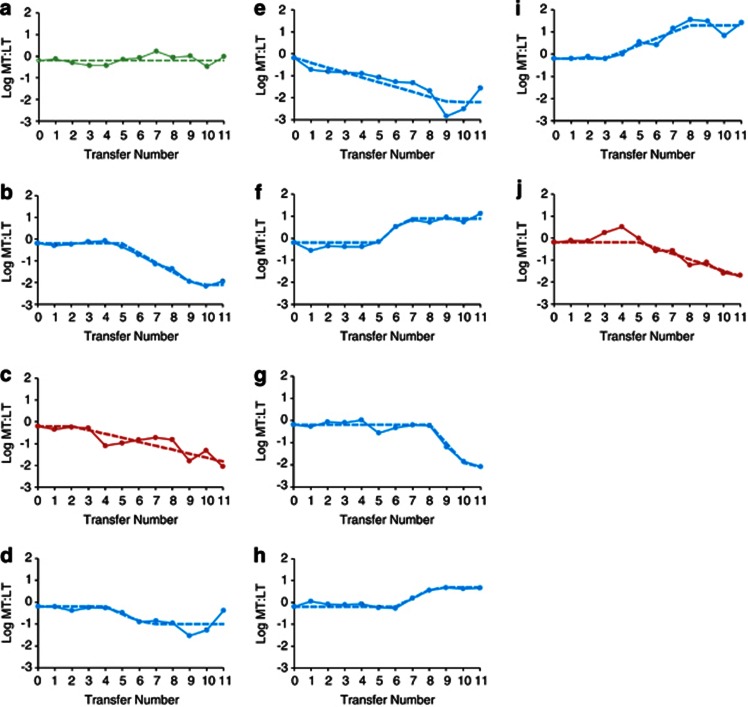
Figure 2.
Fit of the evolutionary trajectories to the null model, the single-ecotype model and the multi-ecotype model. Panels (a–j) Correspond to replicate communities A–J, respectively. The observed trajectory and the expected trajectory for the best-fitting model in each case are shown by solid and dashed lines, respectively. The trajectory of community A was best fit to the null model (green). The trajectories of communities C and J were best fit to the single-ecotype model (red). The trajectories of the remaining communities were best fit to the multi-ecotype model (blue).
We will consider the origins of ecotypes tracked in this experimental system as the origins of bacterial species, as increasing numbers of microbial ecologists and population biologists now view the origins of the most newly divergent, ecologically distinct populations as speciation (Hunt et al., 2008; Sikorski, 2008; Ward et al., 2008; Fraser et al., 2009; Retchless and Lawrence, 2010; Kopac and Cohan, 2011; Vos, 2011). In contrast, the ‘species' formally recognized by taxonomy usually contain an enormous diversity at the genomic, physiological and ecological levels and so are already far too diversified for use in studies of speciation (Staley, 2006; Connor et al., 2010). Moreover, the ecotypes as defined here share the quintessential properties of species (de Queiroz, 2005): the ecotypes are each cohesive, in that their diversity is limited by periodic selection, and different ecotypes are irreversibly separate because they are out of range of one another's periodic selection events and because recombination is too infrequent to prevent their adaptive divergence (Cohan and Perry, 2007; Wiedenbeck and Cohan, 2011).
Our microcosm experiment was designed to address the relative rates of adaptive anagenesis and cladogenesis in B. subtilis, by assaying the likelihood that a community founded with one ecotype would be swept by periodic selection before a second ecotype was formed in the community. We aimed to simulate the potential for resource partitioning within B. subtilis, a heterotrophic, soil-dwelling bacterium, and to this end we provided a chemically and spatially diverse microcosm. We will show that in this environment, anagenetic and cladogenetic adaptations occurred at a similar pace, contrary to the prevailing Mayrian view that the pace of anagenesis should greatly outstrip that of cladogenesis.
Materials and Methods
Modification of strains
Our experimental protocol required strains that were ecologically interchangeable but selectably marked. Starting with strain 1E32 from the Bacillus Genetic Stock Center, which was auxotrophic for lysine, methionine and tryptophan (genotype LMT) and resistant to kanamycin, we generated prototrophic revertant mutants for lysine (MT) and methionine (LT) by selection on plates with minimal medium (including Spizizen salts, glucose and 10 μg ml−1 required amino acids) (Anagnostopoulos and Spizizen, 1961). Each colony was streaked for isolation three times on modified Luria broth plates (Krukonis, 1994), and selectable markers (LT or MT) were reconfirmed on selective minimal media. We confirmed that the revertants were of equal fitness in medium containing all three required amino acids (Supplementary Information; Supplementary Figure 1).
Inoculation of evolution microcosms
Ten microcosms were established from a single LT and a single MT colony in Brain Heart Infusion (BHI; Difco) supplemented with the required amino acids for each genotype and kanamycin, and after each 3.5 days of incubation the microcosms were transferred in serial batch culture (Supplementary Information). A total of 11 transfers were performed, resulting in ∼110 generations for each microcosm. Ratios of MT:LT genotype abundances were determined for each replicate line upon each transfer.
Tests of periodic selection and ecotype formation
Our analyses of evolutionary trajectories focused on the log of the MT:LT ratio, as selection yields a linear change in the log of the genotype ratio (r) (Marée et al., 2000). We used a maximum likelihood approach to test three models of evolution based on the MT:LT trajectory for each of the 10 communities (A–J) (Figure 1; Table 1). In the null model no evolution occurs, yielding the expectation that the initial MT:LT ratio (r0=0.638; Figure 1a) would be sustained throughout the experiment. In the single-ecotype model (Figure 1b), a periodic selection event purges the diversity within the original ecotype before a second ecotype is formed. In this case, a community retains the initial MT:LT ratio (r0) for a waiting time (tw), to be estimated, and then with a periodic selection event, the log-transformed ratio changes linearly at a slope (m) (Marée et al., 2000), to be estimated. Finally, in the multi-ecotype community model (Figure 1c), a second ecotype forms before a periodic selection event sweeps the microcosm's diversity. Here, the community is expected to stay at the initial ratio (r0) for a waiting time (tw), to be estimated, and then the log ratio moves linearly at a slope (m) to be estimated, until it reaches an end ratio (r∞), to be estimated. This end ratio reflects the near-steady-state relative densities of the two ecotypes, presumably determined by the relative abundances of the ecotypes' resources and/or possibly antagonistic effects (Figure 1). We analyzed the changes in the MT:LT ratio over time to identify the model best explaining the dynamics of ecotype formation and periodic selection in each community (Supplementary Information).
Mat formation and colony morphology
We utilized changes in colony morphology as one of the several methods to detect the origin of new, ecologically distinct populations in a microcosm (Braun, 1965; Bukholm et al., 1997; Rainey and Travisano, 1998; Treves et al., 1998; Rozen and Lenski, 2000). We hypothesized that mutants with colony morphologies distinct from the ancestor's might represent either novel populations or more-adaptive variants of the ancestral population. In other cases, we inferred ecological distinctness from microhabitat differences (Rainey and Travisano, 1998). Individual isolates from the A, B and H communities were assayed for mat formation and colony morphology (Supplementary Information).
Confirmation of ecological distinctness
We hypothesized that strains with different combinations of marker genotype, colony morphology and microhabitat association were members of different ecotypes. We tested for ecological distinctness of putative ecotypes by placing member strains into competition.
We inoculated microcosm tubes with one strain from each of two putative ecotypes, at a high and a low starting ratio of MT:LT (rhi0 and rlo0). The tubes were incubated and transferred with MT:LT ratios determined at inoculation and at each transfer. These short-term competition experiments involved 3–4 transfers, during which time we assumed that changes in the MT:LT ratio were due to differences between isolates at the beginning of the competition experiment, and that ratio changes were not due to mutations occurring during the experiment.
For each pair of strains, we tested the simultaneous fit of the two trajectories (that is, starting at the high and low MT:LT ratios) to each of three models: a null model, with no fitness differences between strains; the same-ecotype competition model, where the two strains are members of the same ecotype but differ in fitness; and the different-ecotypes competition model (Supplementary Information; Supplementary Figure 2).
Results
Evolutionary trajectories of the 10 communities
The 10 communities showed a striking diversity of evolutionary trajectories (Figure 2), and it was apparent that a simple set of models would not fully explain all the evolutionary changes. Nevertheless, we attempted to rule out the simple model of anagenesis without cladogenesis, such that a periodic selection event in the original ecotype would purge the MT-LT diversity before a second ecotype could be founded.
This single-ecotype model was supported in only two of the 10 communities, based on the MT:LT trajectories (Table 2; Figures 2c and j). In communities C and J, the anagenesis-only model gave a significant increase in likelihood of the observed trajectory over the null model, but the multi-ecotype model failed to yield significant improvement over the single-ecotype model. In a third community (A), there was no evidence that either periodic selection or ecotype formation was needed to fit the trajectories (Figure 2a). Evidence for the multi-ecotype model was found in the remaining seven of the 10 communities' trajectories (Table 2; Figure 2). For each of these communities, the multi-ecotype model estimated the marker genotypes' ending ratios to range between 4.5:1 and 158:1 (non-log-transformed ratios, with ratios <1 converted here to their reciprocals).
However, a more complex single-ecotype model involving clonal interference may also generate a trajectory consistent with the patterns observed (Gerrish and Lenski, 1998). Specifically, recurring adaptive anagenesis in opposite markers could result in a curve that fit our expectations for the multi-ecotype model. To account for this possibility, we tested more explicitly for the existence of multiple ecotypes within two communities fitting the multi-ecotype model, B and H. We also investigated whether the A community, having shown no apparent adaptive evolution, might have cryptically evolved additional ecotypes.
Test of ecotype diversity within community A
Despite the flat trajectory of the MT:LT ratio in community A (Figure 2a), variation in colony morphology suggested the possible existence of multiple ecotypes. The LT moiety contained two colony variants, the ancestral type (morphotype 1, deemed putative ecotype A1a) and a variant with much larger colonies (morphotype 2, putative ecotype A2) (Figure 3; Table 3; Supplementary Table 2); all MT isolates (putative ecotype A1b) were of the ancestral morphology. We next sought evidence that these putative ecotypes were ecologically distinct from one another and from the original ecotype (ecotype α).
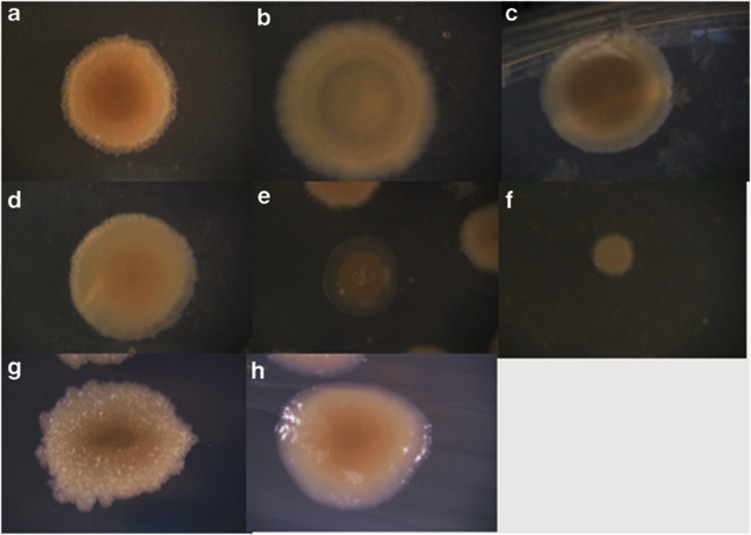
Figure 3.
The morphotypes arising in communities A, B and H. Morphotype 1 was the ancestral morphotype. All other morphotypes were unique to a particular marker moiety (LT or MT) and a single community (Table 3; Supplementary Table 2). (a) Morphotype 1 (ancestral morphotype present in all three communities). (b) Morphotype 2, from community A. (c, d). Morphotypes 3 and 4, respectively, from community B. (d–h). Morphotypes 5 through 8, respectively, from community H. All morphotypes were streaked for isolation and were found to breed true; in the case of morphotype 5, the picture was taken before isolation.
Table 3. The distinguishing characteristics of the putative ecotypes identified in communities A, B and H.
| Putative ecotype | Community | Marker moiety | Morphotype | Mat formation |
|---|---|---|---|---|
| α | A, B and H | LT and MT | 1 | None |
| A1a | A | LT | 1 | None |
| A1b | A | MT | 1 | None |
| A2 | A | LT | 2 | Weak |
| B1 | B | LT | 1 | None |
| B3 | B | MT | 3 | None |
| B4 | B | LT | 4 | Strong |
| H5a | H | LT | 5 | None |
| H5b | H | LT | 5 | Weak |
| H6 | H | LT | 6 | None |
| H7 | H | MT | 7 | None |
| H8 | H | MT | 8 | None |
‘Weak' mat formation indicates the presence of a delicate surface mat, easily disrupted by agitation of the microcosm tube; ‘strong' indicates a thick and highly durable mat, resistant even to extended vortexing of the microcosm tube.
We set up pairwise competition experiments among the three evolved putative ecotypes (A1a, A1b and A2) from community A. In each experiment, two strains representing different putative ecotypes were placed in competition, starting from two initial ratios. For each pair of putative ecotypes, the high-initial-ratio and low-initial-ratio experiments approached a similar MT:LT ratio and remained there (Supplementary Table 3; Figure 4), indicating that they were distinct ecotypes. In all cases, the different-ecotypes model fit the experimental competition results significantly better than the same-ecotype model.
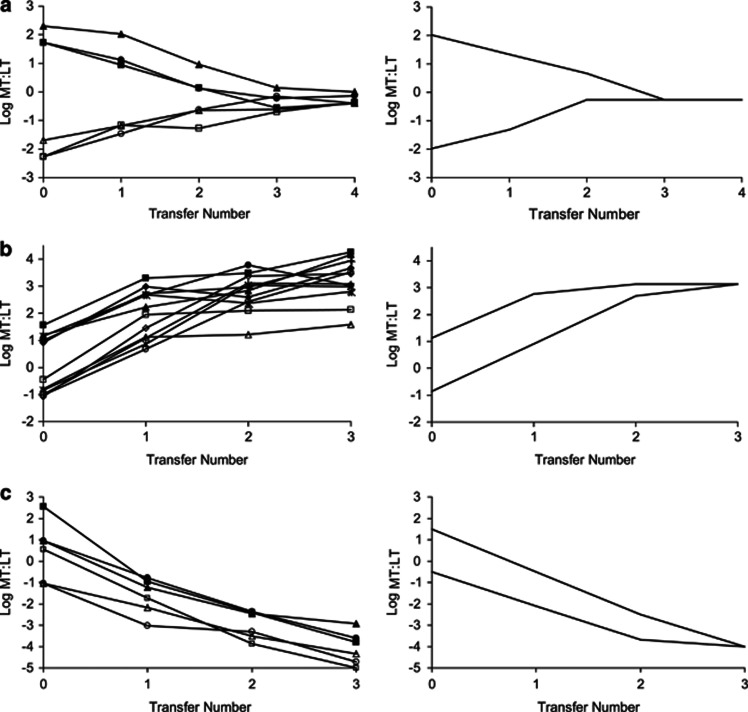
Figure 4.
Results of representative competition experiments among putative ecotypes. Each pair of strains is indicated by a unique symbol; the symbol is filled for experiments started at a high MT:LT ratio, and the symbol is open for experiments started at a low ratio. All competition results are summarized in Supplementary Tables 3–5 and are shown in Supplementary Figures 3–5. In each panel, the left figure shows the results for each pair of strains, and the right figure shows the best-fitting model, based on the average results over all strain pairs. The three pairs of putative ecotypes displayed, all from community A, are as follows: putative ecotypes A1a and A1b (a); putative ecotypes A1a and A2 (b); putative ecotypes A2 and A1b (c).
In each competition experiment between the three evolved putative ecotypes and the ancestral ecotype α, the different-ecotypes model gave a significantly better fit than the same-ecotype model (Supplementary Table 3; Supplementary Figure 3). These results were unanimous across strain pairs for a given pair of putative ecotypes, except for one of the six strain pairs in the A1b versus α comparison. Each of the evolved ecotypes increased at the expense of the ancestor, but they did not extinguish the ancestor as would have been expected for strains of the same ecotype. Nevertheless, evolved ecotypes A1a and A2 left the ancestor at extremely low frequencies.
Beyond the competition experiments, we found that the A2 ecotype differed from the others when grown in isolation. This ecotype formed a light mat on the surface, unlike the ancestor and the other evolved ecotypes of this community, suggesting niche partitioning among ecotypes by microhabitat specialization.
Test of ecotype diversity within community B
The trajectory of the MT:LT ratio in community B fit the expectation of the multi-ecotype model (Table 2; Figure 2b). After showing little change for four transfers, the ratio shifted in favor of the LTs, reaching an MT:LT ratio around 0.01. In community B we observed two new colony morphotypes (morphotypes 3 and 4), distinct from both the ancestral morphotype 1 and from community A's morphotype 2 (Figure 3; Supplementary Table 2).
We set up competition experiments between putative ecotype B3 (marked by MT and morphotype 3) and putative ecotype B4 (marked by LT and morphotype 4) and between putative ecotypes B3 and B1 (the latter marked by LT and morphotype 1). These experiments each showed a significant fit to the different-ecotypes model, with ecotype B3 reaching a near-steady frequency of 0.07 in competition with B4 and 0.55 in competition with B1 (Supplementary Table 4; Supplementary Figure 4). Competition between the evolved B3 ecotype and the ancestral ecotype also supported the different-ecotypes model, but with the ancestral ecotype brought to an extremely low frequency (Supplementary Table 4; Supplementary Figure 4).
When grown in isolation, strains from the B4 putative ecotype formed a thick mat at the surface of the medium (Figure 5), thicker than observed with the mat-forming A2 ecotype of community A. None of the other putative ecotypes of community B was observed to form mats, but grew exclusively in the broth phase, indicating a microhabitat-based niche partitioning among putative ecotypes of this community.
Figure 5.
Differences in mat-forming ability of putative ecotypes B4 and B1 within community B. Putative ecotype B4 (LT, morphotype 4) formed a dense mat on the surface of the medium, while putative ecotype B1 grew in the broth phase, suggesting niche partitioning on the basis of microhabitat specialization.
Test of ecotype diversity within community H
Community H's trajectory of MT:LT ratio was also consistent with a multi-ecotype model, and showed the MT and LT genotypes reaching a steady MT:LT ratio near 4.5 (Table 2; Figure 2h).
The evolved strains of Community H displayed more variation in colony morphology than either the A or B communities (Figure 3; Table 3; Supplementary Table 2). Four new morphotypes arose in this community (morphotypes 5–8), none of which was found in communities A or B or in the ancestral genotype.
Strains sampled from morphotypes 6–8 failed to form a mat when grown in isolation. Of the four isolates of morphotype 5 tested, two formed a mat and two did not. This suggests the possibility that morphotype 5 may be subdivided into two ecotypes, which we hypothesize as H5a (not forming a mat) and H5b (forming a mat).
We performed competition experiments between putative ecotypes that could be distinguished by their auxotrophic markers. In these experiments, all putative ecotypes tested were found to represent different ecotypes (Supplementary Table 5; Supplementary Figure 5).
Discussion
Our microcosm model has introduced a system for estimating the relative rates of cladogenesis and anagenesis during the early stages of adaptation to a novel habitat. By inoculating each microcosm community with two selectably marked but ecologically interchangeable strains of equal fitness, we were able to distinguish evolutionary trajectories that were consistent with a single-ecotype, anagenesis-only model versus a multi-ecotype, cladogenetic model (Figure 1). In only two of the 10 communities were the markers' trajectories consistent with the single-ecotype model. One community's trajectory suggested no adaptive evolution. Remarkably, seven of the 10 community trajectories showed a pattern consistent with the multi-ecotype model, with new ecotypes formed before a single periodic selection event was completed within the ancestral ecotype. Because a periodic selection event in the ancestral ecotype is required to view evidence of formation of a new ecotype (Figure 1), these results are consistent with an approximate parity of rates for anagenesis and cladogenesis.
However, we note that an alternative, more complex single-ecotype model could possibly explain the trajectories of the seven communities that apparently fit the multi-ecotype model. For example, with ‘clonal interference' within a single ecotype (Gerrish and Lenski, 1998), the appearance of one adaptive mutation within one marker, followed by an adaptive mutation of nearly equal fitness in the other marker, would cause fluctuations in the MT:LT ratio; however, neither marker would become fixed. It was therefore necessary to confirm that the trajectory patterns we observed were caused by formation of novel ecotypes.
We directly investigated the ecological diversity within two of the communities that had approached a nearly steady state (B and H), to test whether the single-ecotype model could explain their steady states. Similarly, we tested whether the trajectory consistent with no adaptive evolution (in community A) may have belied cryptic ecological diversity. We hypothesized that each unique combination of marker genotype, colony morphotype and microhabitat association within a community was a distinct ecotype.
Following previous approaches (Rainey and Travisano, 1998; Blount et al., 2008), we tested for ecological distinctness of pairs of putative ecotypes by placing them in competition. In every case, pairs of putative ecotypes were confirmed as ecologically distinct by reaching a single, steady frequency when placed in competition at different starting ratios. Also, each of the evolved putative ecotypes was confirmed to be a different ecotype from the ancestor, by not being able to extinguish the ancestor through competition. However, the evolved ecotypes frequently brought the ancestral ecotype to an extremely low frequency, which suggests that the ancestral ecotype may also have been present in microcosms at the end of the original evolution experiment, but at very low frequency.
We found evidence of recurrent divergence in one ecological dimension. In each community, one ecotype evolved the ability to utilize the air–liquid interface by producing a floating mat, while the other ecotypes were restricted to the broth phase. Recurrent evolution of a mat-forming ecotype occurred also in non-stirred microcosms of Pseudomonas evolution (Rainey et al., 2000). We predict that in our system the non-mat ecotypes have diverged in other, yet unrecognized, ecological dimensions, perhaps by utilizing different chemical resources or by utilizing the more hypoxic, conic bottom of the tube (Mahendran et al., 2006).
Our experiments have shown the importance of using a variety of methods for identifying putative ecotypes, and then confirming their ecological distinctness through competition experiments. For example, the A1a and A1b ecotypes would have been invisible to us were it not for their associations with different selectable markers, as they showed the same colony morphology and microhabitat association. Also, the H5a ecotype would have been unnoticed were it not for its mat-forming ability, as H5a and H5b shared the same marker genotype and colony morphology. Our experiments support the hypothesis that colony morphology evolution is more likely to indicate a new ecotype than an anagenetic improvement in the ancestor ecotype, as each evolved colony morphotype tested was shown to be a different ecotype.
While some previous laboratory evolution experiments have demonstrated a potential for rapid adaptive evolution within a lineage (Cooper and Lenski, 2000; Riley et al., 2001), and others have shown a potential for rapid diversification of ecotypes (Rainey and Travisano, 1998; Treves et al., 1998; Maharjan et al., 2006), ours is the first to compare the pace of anagenesis versus cladogenesis, and we have found evidence for a near parity of these two processes of adaptive evolution in a laboratory microcosm.
The high rate of cladogenesis seen here does not indicate simply an evolutionary potential to broaden the ecological niche through utilization of new resources. Any mutation that just added a resource without any fitness tradeoff would constitute an anagenetic improvement of the existing ecotype, resulting in a more generalist population (Ochman and Davalos, 2006; Wiedenbeck and Cohan, 2011). Rather, the cladogenesis events we observed required that changes in resource utilization incurred a tradeoff in competitive ability within the parental niche (MacLean and Bell, 2003).
Do our results suggest a general parity of cladogenesis and anagenesis rates outside of experimental microcosms? In some ways our experiments may have underestimated the potential for cladogenesis. We attempted to simulate the opportunities for cladogenesis in nature by providing a medium that is chemically diverse (Brain Heart Infusion, derived from the species Bos taurus) and physically diverse (by not stirring the liquid cultures). However, natural soil microhabitats surely offer a greater diversity of ecological niches, by providing potential resources originating from many species and a diversity of physicochemical gradients (Torsvik and Ovreas, 2002). Also, by not making donor DNA available for uptake by our organisms, the present experiment likely eliminated an important opportunity for the evolution of adaptations.
Because no competing species were present in our microcosms, the experiments most directly predict diversification in environments devoid of potential competitors. For example, the experiments may approximate the potential for diversification in the pathogen Clostridium difficile after colonizing a human gut whose diversity has been reduced by antibiotic therapy (Britton and Young, 2012). The microcosm may also simulate diversification in other severely disturbed habitats, for example soil following fumigation (Ibekwe et al., 2001) or a volcanic eruption. Likewise, the experiments approximate a scenario of bacterial contamination in the processed-food industry, where one or two sporeformers invade an otherwise-sterile food environment (Postollec et al., 2012). On a much grander scale, the experiments may simulate diversification on Mars by Terran bacteria if ‘planetary protection' protocols should fail (Schuerger et al., 2012). In each of these cases, the microcosm experiments plausibly predict rapid diversification from a single clone, through parity of cladogenesis and anagenesis rates.
Our microcosms are less likely to simulate diversification from a clone in a complex natural community, where cladogenesis is likely constrained by a great diversity of potentially competing species (Caporaso et al., 2012). In a complex community a nascent ecotype that utilizes a new resource may have to compete successfully with many species that have long been specialized on that resource (Fukami et al., 2007). Nevertheless, competition between distant relatives has been little studied (Dens et al., 1999), and the role of natural community diversity in quashing the origins of ecotypes merits additional experimental investigation.
Whatever difficulties complex communities present for the origin of ecotypes, new ecotypes appear to originate in complex communities in nature at an extremely fast rate (Doolittle and Zhaxybayeva, 2009). Comparisons of genome content consistently suggest ecological differences among closest relatives, in both complex (Scanlan et al., 2009; Touchon et al., 2009; Shapiro et al., 2012) and depauperate communities (Denef et al., 2010). The potential for rapid speciation even in complex communities intensifies the question of whether cladogenesis reaches a parity of rate with anagenesis in natural communities.
We propose a phylogenetic test to estimate the relative rates of cladogenesis and anagenesis in nature. This is a test of the ‘species-less' model of bacterial diversification (Cohan, 2011). In this model, the diversity within an ecotype is not limited by forces of cohesion, such as periodic selection or drift, but by the short time an ecotype exists from the time of its founding by a single clone until its extinction. A paucity of periodic selection predicts that a mother and daughter pair of most-newly-divergent ecotypes in nature should appear as a paraphyletic and a monophyletic group, respectively (Cohan, 2011); such a result would suggest a parity of cladogenesis and anagenesis rates in nature.
Observations of rapid bacterial speciation, both in our microcosms and in nature, appear to support Mallet's (Mallet, 2008) hypothesis that ‘speciation is easy.' This is not surprising, since bacteria may speciate through ecological divergence alone and in sympatry (Wiedenbeck and Cohan, 2011). Moreover, bacterial speciation can be facilitated in nature by horizontal genetic transfer, and huge population sizes can make possible extremely rare adaptive genetic transfers and mutations (Cohan and Koeppel, 2008). It seems that speciation in bacteria may indeed be quite easy, but it is becoming apparent that even some large, highly mobile animals may share the bacteria's readiness to speciate (Mallet, 2008). While zoology and botany have made many contributions to the development of microbial evolutionary and ecological theory (Ward, 2006), perhaps speciation is a place where paradigms can flow both ways.
Acknowledgments
This research was supported by NSF Grant EF-0328698 and NASA Grant EXB02-0000-0095 to FMC, Howard Hughes Medical Institute fellowships to LB, NG and JOW, and research funds from Wesleyan University.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc Natl Acad Sci USA. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Kauth MR, Strelioff CC, Lenski RE. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol. 2010;27:1338–1347. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun W. Bacterial Genetics. Saunders: Philadelphia; 1965. [Google Scholar]
- Britton RA, Young VB. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 2012;20:313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Kassen R, Bell G, Rainey PB. Disturbance and diversity in experimental microcosms. Nature. 2000;408:961–964. doi: 10.1038/35050080. [DOI] [PubMed] [Google Scholar]
- Bukholm G, Tannaes T, Nedenskov P, Esbensen Y, Grav HJ, Hovig T, et al. Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand J Gastroenterol. 1997;32:445–454. doi: 10.3109/00365529709025079. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012;6:1089–1093. doi: 10.1038/ismej.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am Naturalist. 1994;143:965–986. [Google Scholar]
- Cohan FM. Bacterial species and speciation. Syst Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- Cohan FM.2005Periodic selection and ecological diversity in bacteriaIn: Nurminsky D, (eds)Selective Sweep Landes Bioscience: Georgetown, TX; 78–93. [Google Scholar]
- Cohan FM.2011Are species cohesive?--a view from bacteriologyIn: Walk S, Feng P, (eds)Bacterial Population Genetics: A Tribute to Thomas S. Whittam American Society for Microbiology Press: Washington, DC; 43–65. [Google Scholar]
- Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr Biol. 2008;18:R1024–R1034. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Cohan FM, Perry EB. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- Connor N, Sikorski J, Rooney AP, Kopac S, Koeppel AF, Burger A, et al. The ecology of speciation in Bacillus. Appl Environ Microbiol. 2010;76:1349–1358. doi: 10.1128/AEM.01988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz K. Ernst Mayr and the modern concept of species. Proc Natl Acad Sci USA. 2005;102 (Suppl 1:6600–6607. doi: 10.1073/pnas.0502030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci USA. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dens EJ, Vereecken KM, Van Impe JF. A prototype model structure for mixed microbial populations in homogeneous food products. J Theor Biol. 1999;201:159–170. doi: 10.1006/jtbi.1999.1021. [DOI] [PubMed] [Google Scholar]
- Doebeli M. Adaptive Diversification. Princeton Univ. Press: Princeton; 2011. [Google Scholar]
- Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, et al. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol. 2008;74:5422–5428. doi: 10.1128/AEM.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- Fukami T, Beaumont HJ, Zhang XX, Rainey PB. Immigration history controls diversification in experimental adaptive radiation. Nature. 2007;446:436–439. doi: 10.1038/nature05629. [DOI] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103:127–144. [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibekwe AM, Papiernik SK, Gan J, Yates SR, Yang CH, Crowley DE. Impact of fumigants on soil microbial communities. Appl Environ Microbiol. 2001;67:3245–3257. doi: 10.1128/AEM.67.7.3245-3257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics. 1974;77:127–142. doi: 10.1093/genetics/77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner WA, Ward DM, et al. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci. 2008;105:2504–2509. doi: 10.1073/pnas.0712205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopac S, Cohan FM.2011A theory-based pragmatism for discovering and classifying newly divergent bacterial speciesIn: Tibayrenc M, (eds)Genetics and Evolution of Infectious Diseases Elsevier: London; 21–41. [Google Scholar]
- Krukonis GP. Diversity of naturally occurring bacteriophage of Bacillus subtilis and their interactions with their hosts In Ecology and evolutionary biology Vol. Ph.D. University of Arizona: Tucson; 1994. [Google Scholar]
- Lawrence J. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- Levin BR. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics. 1981;99:1–23. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Bergstrom CT. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc Natl Acad Sci USA. 2000;97:6981–6985. doi: 10.1073/pnas.97.13.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, Bell G. Divergent evolution during an experimental adaptive radiation. Proc R Soc Lond B Biol Sci. 2003;270:1645–1650. doi: 10.1098/rspb.2003.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan R, Seeto S, Notley-McRobb L, Ferenci T. Clonal adaptive radiation in a constant environment. Science. 2006;313:514–517. doi: 10.1126/science.1129865. [DOI] [PubMed] [Google Scholar]
- Mahendran B, Choi NC, Choi JW, Kim DJ. Effect of dissolved oxygen regime on growth dynamics of Pseudomonas spp during benzene degradation. Appl Microbiol Biotechnol. 2006;71:350–354. doi: 10.1007/s00253-005-0152-1. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marée AF, Keulen W, Boucher CA, De Boer RJ. Estimating relative fitness in viral competition experiments. J Virol. 2000;74:11067–11072. doi: 10.1128/jvi.74.23.11067-11072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RM. How many species are there on earth. Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Belknap Press of Harvard Univ Press: Cambridge; 1963. [Google Scholar]
- Miller SR, Williams C, Strong AL, Carvey D. Ecological specialization in a spatially structured population of the thermophilic cyanobacterium Mastigocladus laminosus. Appl Environ Microbiol. 2009;75:729–734. doi: 10.1128/AEM.01901-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BB, Carbonero F, van der Gast CJ, Hawkins RJ, Purdy KJ. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J. 2010;4:488–497. doi: 10.1038/ismej.2009.146. [DOI] [PubMed] [Google Scholar]
- Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postollec F, Mathot AG, Bernard M, Divanac'h ML, Pavan S, Sohier D. Tracking spore-forming bacteria in food: from natural biodiversity to selection by processes. Int J Food Microbiol. 2012;158:1–8. doi: 10.1016/j.ijfoodmicro.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Buckling A, Kassen R, Travisano M. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol Evol. 2000;15:243–247. doi: 10.1016/s0169-5347(00)01871-1. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci USA. 2010;107:11453–11458. doi: 10.1073/pnas.1001291107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MS, Cooper VS, Lenski RE, Forney LJ, Marsh TL. Rapid phenotypic change and diversification of a soil bacterium during 1000 generations of experimental evolution. Microbiology. 2001;147 (Pt 4:995–1006. doi: 10.1099/00221287-147-4-995. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Cohan FM. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution. 1995;49:1081–1094. doi: 10.1111/j.1558-5646.1995.tb04435.x. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- Schuerger AC, Golden DC, Ming DW. Biotoxicity of Mars soils: 1. Dry deposition of analog soils on microbial colonies and survival under Martian conditions. Planet Space Sci. 2012;72:91–101. [Google Scholar]
- Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski J. Populations under microevolutionary scrutiny: what will we gain. Arch Microbiol. 2008;189:1–5. doi: 10.1007/s00203-007-0294-x. [DOI] [PubMed] [Google Scholar]
- Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at ‘Evolution Canyons' I and II, Israel. Proc Natl Acad Sci USA. 2005;102:15924–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans R Soc Lond B Biol Sci. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Vos M. A species concept for bacteria based on adaptive divergence. Trends Microbiol. 2011;19:1–7. doi: 10.1016/j.tim.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Ward DM. Microbial diversity in natural environments: focusing on fundamental questions. Antonie Van Leeuwenhoek. 2006;90:309–324. doi: 10.1007/s10482-006-9090-x. [DOI] [PubMed] [Google Scholar]
- Ward DM, Bateson MM, Ferris MJ, Kühl M, Wieland A, Koeppel A, et al. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Phil Trans Roy Soc Ser B. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kuhl M, Grossman A. Genomics, environmental genomics and the issue of microbial species. Heredity. 2008;100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal gene transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- Zawadzki P, Cohan FM. The size and continuity of DNA segments integrated in Bacillus transformation. Genetics. 1995;141:1231–1243. doi: 10.1093/genetics/141.4.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.