Abstract
Planococcus halocryophilus strain Or1, isolated from high Arctic permafrost, grows and divides at −15 °C, the lowest temperature demonstrated to date, and is metabolically active at −25 °C in frozen permafrost microcosms. To understand how P. halocryophilus Or1 remains active under the subzero and osmotically dynamic conditions that characterize its native permafrost habitat, we investigated the genome, cell physiology and transcriptomes of growth at −15 °C and 18% NaCl compared with optimal (25 °C) temperatures. Subzero growth coincides with unusual cell envelope features of encrustations surrounding cells, while the cytoplasmic membrane is significantly remodeled favouring a higher ratio of saturated to branched fatty acids. Analyses of the 3.4 Mbp genome revealed that a suite of cold and osmotic-specific adaptive mechanisms are present as well as an amino acid distribution favouring increased flexibility of proteins. Genomic redundancy within 17% of the genome could enable P. halocryophilus Or1 to exploit isozyme exchange to maintain growth under stress, including multiple copies of osmolyte uptake genes (Opu and Pro genes). Isozyme exchange was observed between the transcriptome data sets, with selective upregulation of multi-copy genes involved in cell division, fatty acid synthesis, solute binding, oxidative stress response and transcriptional regulation. The combination of protein flexibility, resource efficiency, genomic plasticity and synergistic adaptation likely compensate against osmotic and cold stresses. These results suggest that non-spore forming P. halocryophilus Or1 is specifically suited for active growth in its Arctic permafrost habitat (ambient temp. ∼−16 °C), indicating that such cryoenvironments harbor a more active microbial ecosystem than previously thought.
Keywords: cryophile, permafrost, cold-active, cold/osmotic adaptation, subzero environments
Introduction
Cold temperatures provide a finite limit in which life can remain active in the majority of ecosystems found on Earth and are a defining factor of ‘habitability' in the search for life on Mars and elsewhere in our solar system (Fairén et al., 2010; McKay et al., 2012). To date, the coldest reported temperatures for microbial growth and metabolism are −12 °C and −32 °C, respectively (Breezee et al., 2004; Bakermans and Skidmore, 2011). Limiting factors to biological activity at subzero temperatures include the availability of liquid water, a decrease in molecular motion and energetics, as well as the stress of high solute concentrations coinciding with any available liquid water in otherwise frozen habitats. Known cryophilic bacteria are also halophilic, indicating that subzero growth and osmotolerance are closely linked (Bakermans, 2012) and that osmotolerance expands the limit for microbial survival at subzero temperatures (Chin et al., 2010). As such, the microorganisms that inhabit Earth's cryoenvironments are the best candidates for studying the cold temperature limits to life and the mechanisms required for remaining metabolically active within subzero habitats.
Permafrost cryoenvironments experience constant subzero temperatures typically between −10 to −20 °C in Arctic regions. Permafrost harbors diverse microbial communities (Steven et al., 2006; Margesin and Miteva, 2011), however, only recently has their metabolic activity been demonstrated below 0 °C in pure culture (Bakermans et al., 2003; Panikov and Sizova, 2007) and soil microcosms (Rivkina et al., 2002; Panikov et al., 2006; Steven et al., 2007). Subzero brine veins surrounding soil particles (Gilichinsky et al., 1993; Jakowsky et al., 2003) are likely habitats for metabolically active microorganisms within permafrost (Rivkina et al., 2000) as has been shown in ice brine veins (Junge et al., 2004; Bakermans and Skidmore, 2011). Better characterization of subzero metabolic activity of cold-active permafrost microorganisms is critical for defining their contribution to biogeochemical cycles in melting permafrost (Singh et al., 2010; reviewed in Vishnivetskaya et al., 2012).
To date, there are few publications on permafrost bacteria or draft genomes and analyses of their cold adaptive mechanisms, including Psychrobacter arcticus 273-4 (Bergholz et al., 2009; Ayala-del-Río et al., 2010), Exiguobacterium sibiricum (Rodrigues et al., 2008), and Psychrobacter cryohalolentis K5 (Bakermans et al., 2006). Highlights from these psychrophilic/psychrotrophic (referred to here as ‘cryophile') genomes, include cold-adaptive mechanisms that are common to mesophilic bacteria such as chaperone expression, increased membrane fluidity and compatible solute uptake or synthesis. In addition, cryophile-specific cold-adaptive traits, include global resource efficiency, amino acid substitution in cold-active enzymes and increased substrate transport (reviewed in Bakermans et al., 2012b). Ever-expanding sequencing projects on permafrost isolate genomes and metagenomes will yield greater insights into subzero growth.
In our laboratory we screened over 200 isolates from the Canadian high Arctic in search of the most cold and solute tolerant bacteria. Among these isolates, we recently described Planococcus halocryophilus Or1, a non-spore forming aerobic heterotroph isolated from active layer soil from Eureka (Ellesmere Island, Nunavut, Canada) (Mykytczuk et al., 2012) where the Planococcus genus was observed to compose 5.8% of the total bacterial community (by 16S rRNA gene pyrosequencing) (unpublished data). P. halocryophilus Or1 is the first type strain of its genus to be isolated from Arctic permafrost. P. halocryophilus Or1 is tolerant of high salinities (19% NaCl) and grows over a broad temperature range, as high as 37 °C, and before this study, as low as −10 °C (Mykytczuk et al., 2012). The purpose of this study was to investigate the growth capacity of P. halocryophilus Or1 at temperatures down to −15 °C by characterizing its physiology, subzero respiration, genomic cold-adaptive traits, along with transcriptomic analysis under cold (−15 °C) and osmotic (18% NaCl) stress during acclimated growth, to determine the adaptations required for subzero growth and activity at the coldest bacterial growth temperatures described to date.
Materials and Methods
Bacterial growth and viability/activity assays
P. halocryophilus Or1 (type strain DSM 24743, JCM 17719) was isolated as described in (Mykytczuk et al., 2012). Growth characteristics were determined in high salinity (10 to 18% NaCl) media from subzero (−5, −10, −15 °C) to above freezing temperatures (5, 10, 15, 20, 25, 30, 37 °C) in thermostated incubators (±0.5 °C). For this work, we developed a growth medium that did not freeze at −15 °C but froze inconsistently between −16 and −18 °C, comprised of Trypticase Soy Broth (TSB; Difco, BD Diagnostics, Franklin Lakes, NJ, USA) supplemented with 18% NaCl and 7% glycerol, referred to here as TSB-cryo. Growth was measured by optical density at 600 nm or 480 nm for TSB and TSB-cryo, respectively. Viable counts were determined on TSB solid media incubated at room temperature and reported as colony forming units/ml of liquid culture. Growth rate constants were defined by the Arrhenius equation.
Radio-respiration assays were performed in microcosm analyses using 5 g samples of sterile (autoclaved) permafrost from the same active layer core samples obtained in Eureka, Ellesmere Island, from which P. halocryophilus Or1 was isolated. The permafrost was inoculated with 0.5 ml, 100 000 dpm of 1-C14 labeled acetate and 0.5 ml of a 48 h, exponential-phase culture grown at 25 °C. Microcosms were incubated at various temperatures (25, 5, 0, −5, −10, −15, −20, −25 °C) and monitored over a 7-month period for 14CO2 evolved during the microbial-mediated mineralization as described in (Steven et al., 2007). Sterile controls were also completed in triplicate and incubated under the same conditions.
Fatty acid analyses
Cellular fatty acid profiles were obtained from cultures of Or1 harvested from TSB and TSB-cryo media in exponential-growth phase at each given temperature. Culture temperature and total incubation times were as follows: 25 °C -TSB media (48 h), 25 °C-TSB-cryo media, −5 °C (20 days), −10 °C (90 days), −15 °C (130 days). Fatty acids were extracted and fatty acid methyl esters (FAMEs) were prepared and analyzed according to the standard protocol of the MIDI/Hewlett Packard Microbial Identification System (Sasser, 1990) at Keystone Laboratories, Edmonton, Canada. The relative abundance of FAMEs is reported as mol % of the total composition.
Microscopy
Samples for scanning electron microscope were fixed in 2.5% glutaraldehyde followed by dehydration through a graded ethanol series (50%, 70%, 90%, 95%, 100%), critical point dried (Tousimis Samdri-PVT-3B, Rockville, MD, USA), coated with osmium, and imaged on a Leo (Zeiss, Thornwood, NY, USA) 1540XB scanning electron microscope. Samples for transmission electron microscopy were washed in 0.1 ℳ sodium cacodylate buffer (pH 7.3), postfix-stained in 0.1% osmium tetroxide/0.1 ℳ sodium cacodylate buffer, dehydrated through a graded ethanol series and embedded in LR white acrylic resin before ultrathin sectioning (Reichert Ultracut E ultramicrotome, Buffalo, NY, USA) for imaging on a Philips CM 10 transmission electron microscope (FEI, Hilsboro, OR, USA). Samples for ultra-thin section transmission electron microscopy were poststained with 2% aqueous uranyl acetate and lead-citrate.
Genome sequencing and annotation
Cultures of P. halocryophilus Or1 were grown in TSB at 25 °C and DNA was extracted using the Qiagen Blood and Tissue kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol for gram-positive bacteria. A total of 4 μg purified DNA was used for two separate rounds of genome sequencing, performed by both 454-pyrosequencing (GS-Flx; 454 Life Sciences, Branford, CT, USA) and Illumina (Hi-Seq 2000; Illumina, San Diego, CA, USA) paired-end sequencing at the Center for Applied Genomics at The Hospital for Sick Children (Toronto, Canada). The resulting 454 and Illumina sequences were assembled using Newbler (454 Life Sciences, Branford, CT, USA) and Mira (http://sourceforge.net/apps/mediawiki/mira-assembler), respectively. The final hybrid assembly was performed using Mira, Abyss (Simpson et al., 2009), and Consed (Gordon et al., 1998) resulting in a 3.4 Mbp consensus sequence comprised of 35 contigs. Additional assembly methods are summarized in Supplemental Information. Annotation was initiated using automated identification of coding regions within all six open reading frames using Glimmer (Delcher et al., 1999) and RAST (Aziz et al., 2008) followed by manual validation of each coding sequence, functional assignment by comparing against multiple databases including Swiss-Prot (Gasteiger et al., 2003), clusters of orthologous groups, evolutionary genealogy of genes: Non-supervised Orthologous Groups; (Powell et al., 2011), and the NCBI non-redundant protein (Pruitt et al., 2009) databases using a basic local alignment search tool for proteins (BLASTp) (Altschul et al., 1997).
Five parameters used to determine cold adaptation at the protein level, including the arginine/lysine ratio, abundance of acidic residues, aliphacity, proline and hydropathicity (hydrophilicity) determined as the Grand Average of Hydropathicity were applied as previously described (Grzymski et al., 2006; Ayala-del-Río et al., 2010) and outlined in the Supplementary Information. Each of the 2038 proteins was classified as significantly cold or hot adapted for a given parameter using χ2 tests for the sum of cold adapted proteins vs total hot adapted proteins and was used to define the cold adaptation ratio for P. halocryophilus Or1 genes.
Transcriptomic analyses
Transcriptomic analyses were conducted on samples grown at 25 °C (control, TSB), 25 °C and high salt (TSB-cryo) and −15 °C (TSB-cryo). Optical density measurements were used as described above to monitor cultures up to exponential phase of growth for standardized harvesting. As cell numbers were lowest in the −15 °C cultures (∼105 cells/ml), 100 × volume (200 ml) of −15 °C cultures had to be harvested to maintain comparable cell numbers between growth conditions. Total RNA was extracted using TRIzol (Invitrogen, Burlington, ON, Canada) following the manufacturer's protocol with subsequent DNase (Invitrogen) treatment following the manufacturer's protocol.
To remove rRNA, a total of 10 ug RNA was treated with MICROBExpress (Ambion, Life Technologies Inc., Burlington, ON, Canada) to enrich for mRNA. Total purified mRNA (0.5 ug) was converted to complementary DNA (cDNA) using iScript (Biorad, Hercules, CA, USA) following manufacturer's protocols. Degradation of RNA from the cDNA reactions was completed with RNase H (Invitrogen) treatment following manufacturer's protocols and the cDNA was cleaned using the Amicon Ultra (YM-30, Millipore, Billerica, MA, USA) micro-concentrator filters. To improve the yield of purified cDNA, several 0.5–1.0 μg from each of the 25 °C, high salt and −15 °C cDNA samples was subjected to random amplification using the GenomiPHI V2 method (GE Healthcare, Waukesha, WI, USA) following manufacturer's protocols and the resulting products were pooled per sample.
The final amplified cDNA concentrations were between 2.5–4.0 μg and were sequenced on a single lane of Illumina (Hi-Seq 2000) paired-end sequencing at the Centre for Applied Genomics at The Hospital for Sick Children (Toronto, Canada) following their library preparation protocols. The resulting paired-end libraries were analyzed via FastQC to determine the read quality and were filtered for low quality reads and exact duplicate reads using PRINSEQ (Schmieder and Edwards, 2011). The reads were assembled by aligning them against the P. halocryophilus Or1 genome (35 contig set) using Bowtie2 (Langmead and Salzberg, 2012). For each library, read counts were generated for each open reading frames using htseq-count (v0.5.3p6) (S. Anders EMBL Heidelberg: http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). Pairwise comparisons of differential expression for each gene were completed between libraries using the DESeq package (Anders and Huber, 2010) for R 2.15.0 (http://www.R-project.org). Significant fold change was determined at a minimum threshold of ± twofold for each gene (P<0.05). Fold values were log2 transformed and a comparative heat map was created using MeV (v. 4.8.1) (Saeed et al., 2006) with increased expression measured in the direction of cold stress (−5 °C vs 25 °C and −15 °C vs high salt comparisons), and for osmotic stress (high salt vs 25 °C).
Nucleotide sequence accession number
The draft genome sequence of P. halocryophilus Or1 has been deposited in the GenBank database under the accession ANBV00000000. The version described in this paper is the first version, ANBV01000000. It is also available in the RAST database under accession 6666666.17822.
Results and Discussion
Growth and physiological characteristics
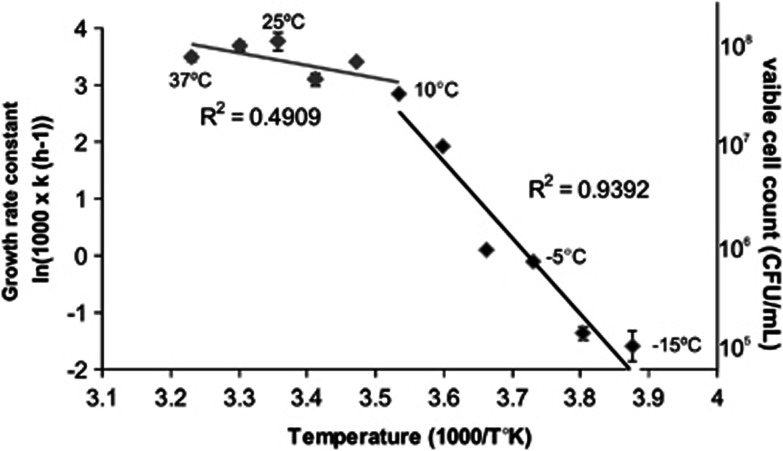
Growth characteristics of P. halocryophilus Or1, as defined by an Arrhenius plot, depict biphasic growth at optimal and suboptimal growth temperatures, where generation times occur from 1 h (25 °C) to every 40 and 50 days at −10 and −15 °C, respectively (Figure 1). The generation times for P. halocryophilus Or1 are consistent with other cryophiles at similar temperatures down to −12 °C, including Psychromonas ingrahamii that has a generation time of 10 days at −12 °C (Breezee et al., 2004) and 39 days for Psychrobacter arcticus at −10 °C (Bakermans et al., 2003). If it were possible to maintain stable liquid media at even colder temperatures, given the generation times indicated above, it is probable that P. halocryophilus Or1, and other extreme cryophiles, would defy the predicted generation times of several years for growth at −20 °C (Jakosky et al. 2003). Viable cell counts reached a maximum of 105 colony forming units/ml at lower temperatures of −10 and −15 °C, while 108 colony forming units/ml were obtained at optimal growth temperatures (Figure 1; Supplementary Figure S1).
Figure 1.
Arrhenius plot of log transformed growth rate constants for P. halocryophilus Or1 over the span of growth temperatures (37, 30, 25, 20, 15, 10, 5, 0, −5, −10, −15 °C). A characteristic biphasic growth trend is shown with the first phase (optimal) denoted in gray and the second phase (suboptimal) in black. Viable cell counts summarized to order of magnitude are shown on the secondary axis and correspond to maximum counts obtained in each growth rate experiment.
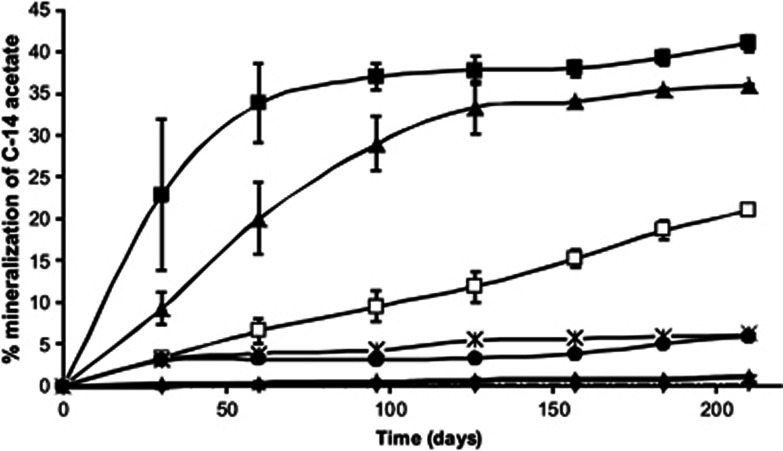
Although it was not possible to show growth below −15 °C, P. halocryophilus Or1 cells were metabolically active in permafrost microcosm experiments, mineralizing 1-C14 labeled acetate into CO2 at cold temperatures resulting in carbon mineralization totaling 40% at 5 and 0 °C, 20% at −5 °C, 4% at −10 and −15 °C and 1% at −20 and −25 °C, after 210 days of incubation (Figure 2). Mineralization in sterile controls at each incubation temperature was significantly lower (0.02–0.3%) over the incubation period. The results indicate that low levels of metabolic activity in P. halocryophilus Or1 occurs in frozen permafrost down to temperatures at least as low as −25 °C. Stable isotope probing could be used to further investigate the contribution of P. halocryophilus Or1 to the microbial oxidation of organic matter at low and subzero temperatures, which is important in refining global flux models where microbial mineralization dominates the flux of CO2 emitted from permafrost and tundra (Zak and Kling, 2006).
Figure 2.
Mineralization assay of C-14 labeled acetate showing respiration by P. halocryophilus Or1 at subzero temperatures. Each point represents the mean cummulative mineralization (% 14CO2 recovered) from triplicate assays with s.e. shown. Curves are shown for 5 °C (▪), 0 °C (▴), −5 °C (□), −10 °C (*), −15 °C (●), −25 °C (♦) and the sterile controls represented by a single curve for the −5 °C control with a dotted line. All other control curves were similar to the one shown and were omitted from this figure for clarity. The line for −20 °C is also not shown as it overlapped with the −25 °C curve and was left out for clarity.
Maintenance of membrane fluidity at cold temperatures shown in previous studies of cryophiles and mesophiles (Mykytczuk et al., 2007), is necessary for continued cell viability. In P. halocryophilus Or1 cold-induced shifts in total fatty acids surprisingly favored an increasing saturated fatty acid abundance (C12:0 to C18:0) in place of branched fatty acids (Table 1). Concomitantly, the branched-to-saturated ratio, a predominant mechanism for controlling membrane fluidity (Russell, 1984), shifted from 7.7 (25 °C), to 7.1 (−5 °C), to 1.7 (−10 °C) and to 0.16 (−15 °C); representing a turnover of 142% of the fatty acid species comprising the cytoplasmic membrane between optimal (25 °C) and −15 °C conditions. These observations are contrary to the trend typically observed for Gram-positive bacteria in response to cold, where increased branching is typically employed in cold adaptation to effectively disorder the membrane, thereby increasing fluidity under cold temperatures (Denich et al., 2003). P. halocryophilus Or1also does not synthesize any polyunsaturated fatty acids found in other cryophiles including P. arcticus (Ponder et al., 2005) and Colwellia psychrerthraea 34H that increase polyunsaturated fatty acids at cold temperatures (Methé et al., 2005). The profile of P. halocryophilus Or1 at high salt exposure at optimum temperature yielded an opposite trend, favouring unsaturated over saturated fatty acids (Table 1). Membrane adaptations in halophiles appear to be driven more by phospholipid head group changes favouring anionic lipids (Galinski and Truper, 1994). Explanations as to how a membrane could function with such a high degree of saturation might be maintaining membrane phase as opposed to absolute viscosity, whereby the changes in chain length, chain saturation, and phospholipid head groups help maintain a stable membrane environment (McElhaney, 1974).
Table 1. Cellular fatty acid profile (mean % abundance) for P. halocryophilus Or1 cells grown at control, high salt and subzero temperatures.
| Fatty acid methyl ester | Growth condition | ||||
|---|---|---|---|---|---|
| |
25 °Cb |
High salt (25 °C) |
−5 °C |
−10 °C |
−15 °C |
| Straight-chain fatty acids: | |||||
| C12:0 | 1.1 | 1.5 | 14.3 | 7.4 | |
| C13:0 | 0.3 | 0.8 | — | 4.7 | |
| C14:0 | 0.5 | — | 0.6 | — | 4.2 |
| C15:0 | — | — | — | — | — |
| C16:0 | 7.0 | 2.6 | 3.4 | 7.8 | 27.2 |
| C17:0 | — | 0.3 | 0.4 | — | 1.6 |
| C18:0 | 2.9 | 0.5 | 4.5 | 13.3 | 38.5 |
| Branched fatty acids: | |||||
| Anteiso C14:0 | — | — | — | — | — |
| Iso-C14:0 | 1.2 | 10.6 | 1.2 | — | — |
| Iso-C15:0 | 1.7 | 7.0 | 1.9 | 2.8 | — |
| Anteiso-C15:0 | 46 | 40.3 | 46.5 | 31.6 | 9.0 |
| Iso-C16:0 | 2.9 | 6.6 | — | 3.9 | — |
| Iso-C17:0 | 2.3 | 3.0 | 0.5 | 2.2 | — |
| Anteiso-C17:0 | 18 | 7.5 | 3.8 | 10.5 | 1.8 |
| Iso-C17:1 ω10c | 0.9 | 1.7 | 2.8 | 2.2 | — |
| Iso-C18:0 | 1.2 | — | — | — | — |
| Unsaturated fatty acids: | |||||
| C16:1ω7c alcohol | 2.1 | 9.5 | 3.4 | 2.7 | 2.6 |
| C16:1 ω11c | 4.9 | 2.6 | 2.3 | 3.2 | — |
| C17:1 ω9c | — | 0.3 | 1.9 | — | — |
| C18:1 ω9c | — | — | 1.6 | — | — |
| Summed feature 4a | 5.2 | 3.9 | 21.0 | 5.7 | 3.0 |
| Total Saturated | 10.4 | 4.8 | 11.0 | 35.4 | 83.6 |
| Total Unsaturated | 7.0 | 12.4 | 9.0 | 5.9 | 2.6 |
| Total Branched | 79.9 | 80.6 | 77.8 | 58.9 | 13.8 |
| Branched to Saturated ratio | 7.7 | 16.8 | 7.1 | 1.7 | 0.2 |
Values reflect the percent of the mean for each fatty acid with s.d. <0.5%, except for anteiso-C15:0 where s.d. <2%. Fatty acid groups that accounted for <0.5% across all growth conditions are not show. –, not detected.
Summed feature four represents iso-C17:1 I and/or anteiso-C17:1 B, which could not be separated by GC with the MIDI system.
Data for control conditions previously published (Mykytczuk et al., 2012).
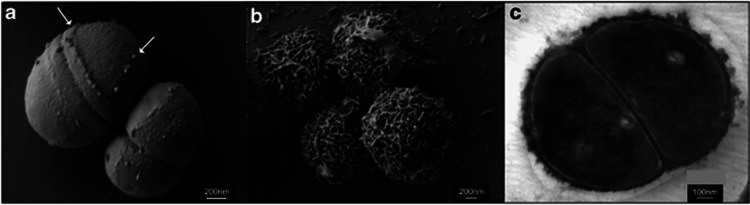
When P. halocryophilus Or1 cells were observed using scanning and transmission electron microscopes, unusual cell envelop features distinguished smooth cells at ambient/optimal growth temperatures (25 °C, Figure 3a) vs densely encrusted subzero (−15 °C, Figure 3b) grown cells. The nodular and sheet-like crust appeared very minimally on 25 °C cells as singular features along division planes. The orientation of this extracellular material along cell wall septation band planes suggests concentration and preservation of either peptidoglycan or secreted exopolysaccharide (EPS) during cell division. Although the material does remain closely associated with the cells during subzero growth and division (Figure 3c), it is unclear if this feature imparts additional protection to the cells to support growth at these temperatures.
Figure 3.
(a) Scanning electron microscope of cells grown at ambient temperatures, white arrows denote nodular feature being preserved along previous cell division planes. (b) Scanning electron microscope of cells grown at −15 °C in 18% NaCl and 7% glycerol, encrusted in dense nodular material. (c) Transmission electron microscopy of cross-section of dividing cells grown at −15 °C, noting the outer cell envelope with the nodular crust material in close association with the cell wall.
Genome analyses
Overview
The P. halocryophilus Or1 draft genome sequence was assembled from a consensus of 454-pyrosequencing and Illumina Hi-Seq paired-end reads resulting in a 3.4 Mbp sequence, likely comprising a single replicating unit with characteristics described in Table 2. Of the 35 final contigs, 24 of them involved rRNA or transposase sequences on their 5′ or 3′ ends, but further manual gap closures were not completed. The annotation resulted in 3548 protein coding sequence encoded by 85.1% of the total sequence length. The genome contains no plasmid related genes but does have 12 different transposases or insertion sequences distributed throughout the genome and belonging to six families: IS231 (two copies), IS150 (four copies), IS1182 (one copy), ISL3 (two copies), IS21 (two copies) and ISNCY (one copy). The genome sequence homology of P. halocryophilus Or1 does not have high similarity to any single available genome. However it is most similar to members of the Bacillaceae, with best hit protein matches (>70% identity) to the marine Bacillus spp. B14905 (502), the mesophilic Planococcus donghaensis MPA1U2 (250), and the cryophilic permafrost isolate Exiguobacterium sibiricum 255-15 (160) (Supplementary Table S1).
Table 2. General features of the P. halocryophilus Or1 genome sequence assembly.
| Characteristic | Value |
|---|---|
| Genome size (35 contigs) | 3 428 475 bp |
| G+C content | 40.5% |
| Number of coding bases | 2 919 305 (85.1%) |
| Protein coding genes (CDS) | 3548 |
| % coding positive strand | 53.3% |
| % coding negative strand | 46.7% |
| RNAs | 86 |
| Hypothetical genes | 907 |
Abbreviation: CDS, coding sequence.
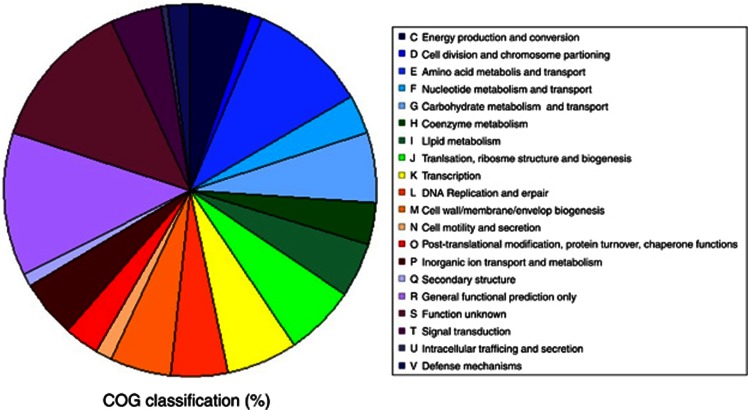
An overview of the P. halocryophilus Or1 genome based on clusters of orthologous groups shows an even distribution between most of the categories related to cellular metabolism (∼5–6% each), except amino acid metabolism and transport, which has the highest abundance (10%) of genes with assigned functions (Figure 4). The largest proportion of genes (26%), were classified as only having a general functional prediction or unknown function. Many of these genes are large open reading frames clustered into groups within the genome (that is, Phal2076—Phal2091). Similar proportions of such genes have been found in other cryophile genomes, including P. ingrahamii (Riley et al., 2008), E. sibiricum 255-15 (Rodrigues et al., 2008) and P. arcticus 273-4 (Ayala-del-Río et al., 2010) and are suspected to include novel stress-related and cold-adaptation proteins.
Figure 4.
Functional distribution of genes within the P. halocryophilus Or1 genome classified by clusters of orthologous groups.
Genomic traits linked to cold and osmotic adaptation
Among the known adaptations to cold and/or osmotic stress, several chaperones, shock proteins, regulation and repair mechanisms were found within the P. halocryophilus Or1 genome and are summarized in Table 3, and in the following categorical descriptions.
Table 3. Summary of known cold and salt stress response proteins found in P. halocryophilus Or1.
| Category and Locus | Description |
|---|---|
| DNA replication and repair | |
| Phal2444 | DNA/RNA helicase of DEAD/DEAH box family |
| Phal1938/2515 | ATP-dependent DNA helicase RecQ |
| Phal2058 | RecA protein |
| Phal3192 | RecD-like DNA helicase YrrC |
| Phal0311 | DNA gyrase subunit A GyrA |
| Membrane/cell envelope alteration | |
| Phal0457/2960 | Fatty acid desaturase |
| Phal2530 | Phytoene synthase |
| Phal0637/2527 | Phytoene desaturase, neurosporene or lycopene producing |
| Phal2528 | Dehydrosqualene desaturase |
| Phal1023 | Tyrosine-protein kinase EpsD |
| Phal1170 | Tyrosine-protein kinase transmembrane modulator EpsC |
| Cold shock proteins | |
| Phal1601 | Cold shock protein CspB |
| Phal3416 | Cold shock protein CspD (RNA chaperone) |
| Phal3050 | Cold shock DEAD-box protein A |
| Phal2936 | Cold shock protein |
| Phal1600 | Cold shock protein ScoF (CspA family) |
| Sigma factors | |
| Phal2274 | RNA polymerase heat shock sigma factor SigI |
| Phal3064 | RNA polymerase sigma factor SigB |
| Phal0492/0870/ | RNA polymerase sigma-70 factor |
| 1943/2010/ | |
| Phal3459 | RNA polymerase sigma factor SigW |
| Osmotic stress/salt adaptation | |
| Phal0682/2544 | Glycine betaine ABC transport system, ATP-binding protein OpuAA |
| Phal0683/2545 | Glycine betaine ABC transport system, permease protein OpuAB |
| Phal0684/2546 | Glycine betaine ABC transport system, glycine betaine-binding protein OpuAC |
| Phal0771/1845/ | Glycine betaine transporter OpuD |
| 2866/2904/ | |
| 2965 | |
| Phal2982/2983 0227 | L-proline glycine betaine ABC transport system permease protein ProV |
| Phal0228 | L-proline glycine betaine ABC transport system permease protein ProW |
| Phal2984/2689 | L-proline glycine betaine binding ABC transporter protein ProX |
| Phal1405 | L-Proline/Glycine betaine transporter ProP |
| Phal1457 | High-affinity choline uptake protein BetT |
| Phal2510/1417 | Osmosensitive K+ channel histidine kinase KdpD |
| Phal1203 | Osmotically inducible protein C, OsmC/Ohr family protein |
| Phal2964 | Proline/sodium symporter PutP |
| Phal1127 | Glycerol uptake facilitator protein |
| Phal0635 | Ferredoxin-dependent glutamate synthase |
| Phal2516/2517 | Glutamate synthase (NADPH) |
| Phal2648/0279/ 1830 | Proton/glutamate symport protein or Sodium/glutamate symport protein |
| Phal0995 | Sodium/alanine symporter |
| Phal1208 | Sodium/glycine symporter GlyP |
| Phal0874 | sodium/hydrogen exchanger family protein |
| Phal3266 | Sodium-dependent phosphate transporter, NhaA |
| Phal0708/1247 | Na+/H+ antiporter NhaC |
| Phal2664/1788 | Sodium-dependent transporter |
| 1815 | |
| Phal1793–1799 | Na(+) H(+) antiporter subunits A through G |
| 2503–2509 | |
| Translation factors | |
| Phal1462 | Heat shock protein 60 family chaperone GroEL |
| Phal1463 | Heat shock protein 60 family cochaperone GroES |
| Phal3226 | Heat shock protein GrpE |
| Phal1502 | Translation elongation factor Tu, Ef-Tu |
| Phal2082 | Translation initiation factor 2, IF2 |
| Phal0525 | Translation initiation factor 3, IF3 |
| Phal2080 | Ribosome-binding factor A, RbfA |
| Phal3303 | Transcription termination protein NusB |
| Phal2085 | Transcription termination protein NusA |
| Miscellaneous | |
| Phal0109 | ATP-dependent Clp protease proteolytic subunit |
| Phal1971 | SOS-response repressor and protease LexA |
| Phal2780 | Low temperature requirement B protein |
| Phal2291 | Low temperature requirement C protein |
| Phal3007/0038/ | Universal stress protein |
| 0629/1240/ | |
| 1246/1301 | |
| Phal2613 | Stress response protein CsbD |
Transcription, translation and regulation
In P. halocryophilus Or1, DNA/RNA replication is probably maintained at cold temperatures by helicases, which belong to the DEAD-box and recombination factors Rec (RecA/D/Q) families (Table 3). These proteins were shown to be active at cold temperatures in a variety of cryophiles and mesophiles at low temperatures and induce cold sensitivity in deletion mutants (Cavicchiolli et al., 2001; Cartier et al., 2010). Transcription and translation factors that could assist P. halocryophilus Or1 at cold temperatures are involved in transcription termination (NusA/B), ribosomal binding (RbfA), translation initiation (IF2, IF3) and elongation (Ef-Tu) along with general chaperone activity (GroEL, GroES, GrpE). Several sigma factors are present (SigB, SigI, SigW) and may act as part of alternative signal transduction for specific genes or regulons under both cold and osmotic stress response as has been observed in other Bacillus spp. and other gram-positive species (Budde et al., 2006; Hecker et al., 2007; Hahne et al., 2010). Multiple copies of the general stress response sigma-70 are also encoded in P. halocryophilus Or1 similar to the multiple copies found in P. ingrahamii (Riley et al., 2008). Several conserved cold shock proteins are also encoded (that is, CspB, and CspD and ScoF (a CspA family protein) (Table 3).
Cell envelope modifications
Two fatty acid desaturases are encoded in the P. halocryophilus Or1 genome (Table 3), although as observed above from physiological data (Table 1), it appears that at −15 °C, P. halocryophilus Or1 does not increase the proportion of unsaturated or branched fatty acids as is commonly observed in other cold-adapted strains. Complete fatty acid synthesis and degradation pathways are present, including several multiple copy enzymes (isozymes) including enoyl-CoA hydratase/isomerase and 15 copies of 3-oxoacyl-(ACP) reductase are encoded, which may allow different specificity or activity under different growth conditions. β-keto-acyl carrier proteins, including KAS-II and two copies of KAS-III were also found and are key to the synthesis and regulation of the straight and branched-chain lipid ratios in the cell membrane, as is found in C. psychrerythreae (Methé et al., 2005). It is also possible that carotenoid production, linked to cold adaptation in numerous polar microbes (Dieser et al., 2010), may contribute to adjustments in membrane fluidity under low temperature conditions (Jagannadham et al., 2000). The P. halocryophilus Or1 genome encodes synthesis primarily for the carotenoid phytoene (colorless) that is likely used in the secondary production of neurosporene (yellow-orange) and lycopene (red-orange) through the activity of phytoene desaturases to impart the bright orange coloration observed in P. halocryophilus Or1 cultures.
Oxidative stress
Antioxidant defense against reactive oxygen species (ROS) is also a requirement for growth at low temperatures due to an increase in the solubility of gases and an accumulation of reactive oxygen species with colder temperatures (Chattopadhyay, 2002). P. halocryophilus Or1 encodes several copies of catalases (three copies), two different superoxide dismutases cofactored to manganese and copper-zinc, as well as additional H2O2 scavengers and antioxidants, including thiol- and glutathione peroxidases and thioredoxin reductase. Other bacteria that also increase antioxidant capacity through reactive oxygen species scavengers include C. psychrerythraea and Desulfotalea psychrophila, each encoding several copies of catalases and superoxide dismutases (D'Amico et al., 2006), although not as many as encoded in the P. halocryophilus Or1 genome. P. halocryophilus Or1 is also able to enhance protection of the reactive oxygen species-sensitive amino acid, methionine, by encoding five copies of MsrA, a methionine sulfoxide reductase involved in reverting any oxidative damage to methionine (Weissbach et al., 2002).
Osmotic stress
Either in its natural permafrost habitat or during freezing conditions that would accumulate solutes in the extracellular environment, P. halocryophilus Or1 would have to synthesize or accumulate compatible solutes. The genome suggests it is well equipped for solute uptake, and encodes multiple copies of osmolyte uptake genes for glycine betaine, including two copies of ProVWX, two to three copies of OpuA operon genes and five copies of the OpuD glycine betaine transporter, although only two OpuD copies appeared complete in a single open reading frames (Table 3). Additional proline (ProP, PutP), glycerol, glycine (GlyP) and choline betaine uptake/transport proteins were also found although there appear to be no genes for betaine synthesis (that is, betaine aldehyde dehydrogenase (BetB) is absent). Such high copy numbers of osmolyte transporters in P. halocryophilus Or1, which are not found in other cryophilic bacterial genomes (Supplementary Table S3), likely reflect the higher degree of halotolerance exhibited by P. halocryophilus Or1 and would impart a synergistic adaptation to osmotic and cold stress by balancing the osmotic pressure exerted over the cell envelope, and freezing depression in the cytoplasm by creating a solute rich intracellular environment (Wood et al., 2001).
Genome-wide protein adaptations
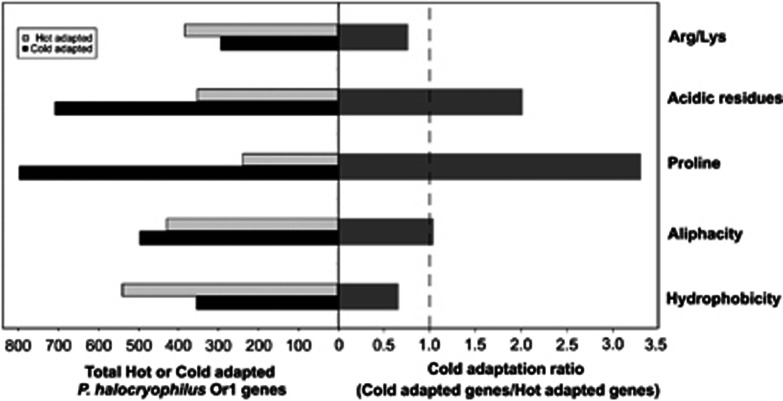
Assessing genome-wide cold adaptation has effectively been used to describe global adaptive features of cryophilic proteins and can be defined by the cold adaptation ratio that distinguish proteins with greater flexibility or thermal stability at colder temperatures based on the amino acid content (Ayala del Rio, 2010). The cold adaptation ratio for 2038 of the P. halocryophilus Or1 coding sequence in the five cold-adaptive amino acid features (Arg to Lys content, acidic residues, proline residues, aliphatic index and hydrophobicity) were 0.76, 2.01, 3.31, 1.04 and 0.66, respectively (Figure 5; Supplementarty Supplementary Table S2). Thus three of the five indicators showed P. halocryophilus Or1 has significantly more cold-adapted proteins containing less proline, fewer acidic residues, and to a lesser degree, fewer aliphatic residues. The lower abundance of these amino acid residues would favor increased flexibility of the proteins by avoiding the increased stability imparted by N-Cα bonds and salt bridges formed by proline and acidic (glutamate, aspartate) amino acids, respectively (Fields, 2001). These features are consistent with the cold adaptive ratios described for P. arcticus (Ayala-del-Río et al., 2010), P. cryohalolentis K5, C. psychrerythraea 34H and P. ingrahamii 37 (Metpally and Reddy, 2009). What distinguishes P. halocryophilus Or1 is that only three of the five indicators show significant cold adaptation, which is likely related to its broad growth temperature range where ‘hot-adapted' proteins would also be required to maintain protein stability and sustain growth at its upper limit of 37 °C.
Figure 5.
Genome-wide cold adaptation defined by five parameters that are used to determine cold adaptation at the protein level. The cold adaptation ratio (total cold adapted proteins/total hot adapted proteins) is shown to be significant above a threshold of 1.0 (right). The total number of proteins found to be significantly hot or cold adapted among 2038 of the P. halocryophilus Or1 genome coding sequence are shown per category (left).
The thermal flexibility apparent from amino acid distribution is also supported by the genomic redundancy whereby P. halocryophilus Or1 has many isozymes, with a total of 302 genes with one or more (a maximum of 15) isozymes encoded within the genome, accounting for ∼17% of the genome. The role of isozymes, specifically temperature-dependent isozyme exchange, may be important in broad growth temperature range cryophiles (or eurypsychrophiles) that may rely on cold or hot-adapted isozymes to catalyze similar reactions at different growth temperatures, as described in (Maki et al., 2006).
Transcriptomic analyses
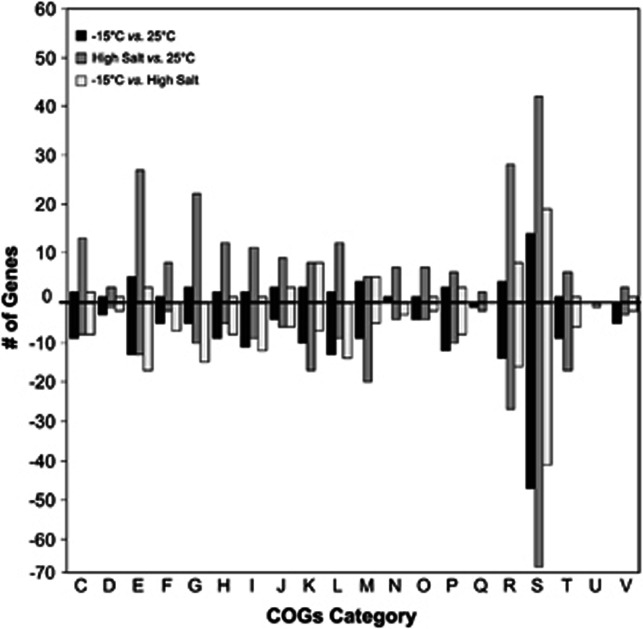
To further investigate the features of the P. halocryophilus Or1 genome that are actively involved in cold and/or osmotic adaptation, we conducted transcriptomic analyses comparing optimal conditions (25 °C), subzero (−15 °C and 18% NaCl) and high salt (25 °C and 18% NaCl) (Supplementary Table S4). From all the current literature, this is the coldest subzero temperature (−15 °C ) to date that has been used in transcriptomic analyses, while the previous transcriptomic studies of cryophiles were conducted at temperatures down to −6 °C. The results identified a total of 677 unique genes that were significantly (P<0.05) differentially expressed across all three experiments. The pairwise comparisons between transcriptome profiles (−15 vs 25 °C, high salt vs. 25 °C, −15 °C vs. high salt) are summarized according to clusters of orthologous group functional categories (Figure 6) illustrating the greatest number of changes was observed in the largest functional categories comprising genes with unknown or basic functional prediction only (Figure 6, bars R and S). In general, decreased expression was observed across most functional categories at −15 °C indicating a broad suppression of cellular functions. However, increases in what appear to be specific subsets of functional genes occurred under cold growth, with notable increases for transcription and cell wall modifications (Figure 6, bars K and M),, amino acid metabolism, and inorganic ion transport (Figure 6, bars E and P). The opposite trend in global expression is observed at high salt, inducing significant increases in particular for amino acid, carbohydrate and energy metabolism, along with DNA replication and repair mechanisms (Figure 6, bars E, G, C and L, respectively).
Figure 6.
Numbers of differentially expressed genes arranged by clusters of orthologous groups at −15 °C vs. control (25 °C), high salt vs. control (25 °C), and −15 °C vs. high salt. The data for upregulated and downregulated genes are shown above and below the line, respectively. clusters of orthologous group categories are the same as described in Figure 3.
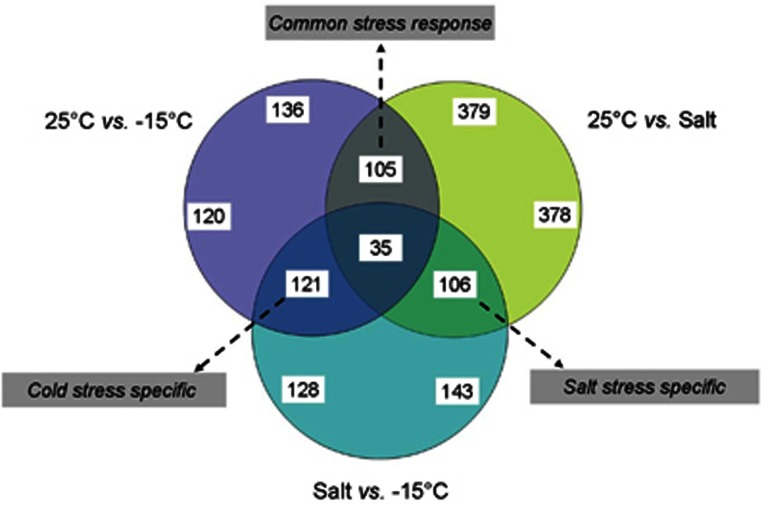
The pairwise comparisons of the transcriptome data were also used to find the genes that might be specific to cold or osmotic stress versus those that appear to be equally expressed and could be synergistic in their response to both stressors (Figure 7). The overlap between transcriptome profiles indicated that only 35 genes changed expression across all three comparisons, however,∼100–120 genes appear specific to cold and/or osmotic response. Specific descriptions of these cold or salt adapted genes is summarized as follows and illustrated categorically in Figure 8. A global summary of key adaptive features within P. halocryophilus Or1 gleaned from both the genomic data are summarized in Figure 9a while transcriptomic data are illustrated in 9b (salt-specific) and 9c (cold specific).
Figure 7.
Venn diagram illustrating the comparison between genes showing significant changes in expression among the three transcriptomic datasets for P. halocryophilus Or1 grown at control (25 °C), high salt (25 °C and 18% NaCl), and low temperature (−15 °C+18% NaCl) conditions. The number of genes found to be unique within and between treatments is shown denoting groups of genes that appear to be linked to either cold, salt, or common stress response.
Figure 8.
Heat map of selected differentially expressed genes grouped by clusters of orthologous group category. Comparisons between transcriptomic data sets are shown in columns: (a) high salt vs −15 °C, (b) high salt vs 25 °C, (c) −15 °C vs 25 °C. Values are expressed as log2 transformed fold changes. All chosen genes were differentially expressed in at least one data set with P<0.05.
Figure 9.
Summary of genome encoded adaptive traits (a), and the transcriptomic results for adaptive mechanism that were increased in expression and appear to be induced by high salt (b), or −15 °C (c). Traits that were similarly induced over basal levels under both stresses are outlined in black.
Cold specific response
Energy metabolism
At −15 °C, the majority of enzymes involved in energy metabolism were repressed, with the exception of the final components of the electron transport chain—cytochrome C oxidase (polypeptide III) and ATP synthase (F0-A chain)—that were both increased in expression. Several genes that shuttle substrates towards the tricarboxylic acid cycle cycle, including succinic semialdehyde dehydrogenase, alcohol dehydrogenase and several oxidoreductases were also increased in expression (Figure 8, group C). Although some of these appear to be induced under high salt conditions as well (see below), the higher levels at −15 °C suggest that an increased abundance of these proteins may be induced to maintain cellular energy metabolism and ATP levels. This is consistent with the greater energy requirement needed per unit biomass that is synthesized during cold temperature growth (Bakermans and Nealson, 2004).
Transcriptional regulation and translation
Growth at −15 °C in P. halocryophilus Or1 induced higher expression of nine transcriptional regulators that were not induced under high salt conditions. These could be functioning as alternate sigma factors regulating the expression of cold specific genes or suppression of energy demanding cellular processes (Figure 8, group K). These include increased expression of general stress response regulator σ70, and alternate factor σ28 (FliA) that could function to positively regulate flagellar synthesis as observed in B. subtilis (Helmann et al., 1988). Increased expression of several transcriptional regulators—TetR, ThiJ, TipA and a MerR family regulator—were induced at −15 °C. Although these regulators are observed primarily in antibiotic-sensing, they can also act in response to other environmental stress, such as the TetR family of regulators that includes a choline-sensing repressor of the bet regulon involved in osmotic stress (Lamark et al., 1991).
Cell envelope precursors, modifications and synthesis
Active division at −15 °C would require that P. halocryophilus Or1 have cold-active cell wall, membrane, and envelope-component synthesis. From the transcriptomic data it is evident that cell envelope synthesis and cell division at −15 °C is mediated by one of three genomic copies of the peptidoglycan synthetase, along with proteins involved in precursor synthesis: glucosamine-fructose 6-phosphate aminotransferase, teichoic acid biosynthesis protein, GgaB and UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA), the enzyme responsible for production of UDP-N-acetylmuramic acid (Figure 8, group M). This increase in cell wall biosynthesis transcripts in P. halocryophilus Or1 is similar to the observations of E. sibiricum increasing cell wall synthesis at −2.5 °C (Rodrigues et al., 2008) while different cell envelope modifications were induced in the gram-negative P. arcticus (Bergholz et al., 2009). With similar results observed in both P. halocryophilus Or1 and E. sibiricum, these observed cell wall modifications may be characteristic of gram-positive cryophiles although differences in general cultivation conditions may also have a role in cell wall response (Bakermans, 2012). Membrane lipid metabolism also continues at −15 °C through the increased expression of cardiolipin synthetase and one of the 15 genomic copies of 3-oxoacyl-(ACP) reductase involved in fatty acid biosynthesis (Figure 8, group I). This copy of 3-oxoacyl-(ACP) reductase has a significant cold adaptation signature with less acidic residues and lower aliphacity than 6 of the 15 counterpart isozymes that have distinctly warm adapted signatures. Again, with these multi-copy genes there appears to be cold-induced isozyme exchange occurring in P. halocryophilus Or1.
Protein modification and turnover
In addition to having a significant proportion of cold-adapted proteins encoded within its genome, P. halocryophilus Or1 also actively employs protein turnover at −15 °C likely to increase the conservation of biosynthetic precursors during slower growth rates. In particular, evidence for arginine degradation through increased expression of RocB, lysine deamination through L-lysine dehydrogenase, along with upregulation of several amidases, and a trypsin-like serine protease suggest that individual amino acids along with proteins may be broken down for use as nitrogen sources, or in new protein synthesis, respectively (Figure 8, group E, J and O); all supporting that P. halocryophilus O r1 uses resource efficiency during growth at −15 °C.
Salt specific response
Osmotic balance
The most obvious stress response to osmotic upshift is the induction of osmoregulatory functions. However, once adapted to high salt conditions P. halocryophilus Or1 had only a fraction of the genomically encoded halotolerance mechanisms upregulated over basal (25 °C) levels. For example, all of the genes from the multiple genomic clusters of Opu or Pro operon genes remained at basal levels. The acclimated vs shock-like response to osmotic and/or stress would induce different levels of osmolyte transporters, and they may not be required once osmotic balance is obtained within P. halocryophilus Or1. Such is the case with one copy of the ProX gene, indicating suppressed levels from both the high salt and −15 °C conditions (Figure 8, group M). The time-resolved picture in response to sudden osmotic stress vs salt acclimation was shown to follow induction followed by rapid suppression of osmoregulatory genes in numerous salt tolerant bacteria (Marin et al., 2004; Hahne et al., 2010); thus, an analysis would have to be performed shortly after osmotic upshift, in order to get a more comprehensive picture of the P. halocryophilus Or1 osmotic response.
Energy and carbohydrate metabolism
As general metabolic functions become suppressed under stressful growth conditions, accumulation or production of cellular stores of carbohydrates are important for cell survival under nutrient limited conditions. Several genes from a putative operon for glycogen synthesis (Phal2841-Phal2844) are induced at high salt conditions and likely respond to P. halocryophilus Or1 need for carbohydrate reserves under stress. Accumulation of glycogen has been found in salt-acclimated cyanobacteria (Page-Sharp et al., 1998) and would be useful stores of glucose that could be recycled as stressful conditions persist. Gluconate uptake is also functioning at higher levels through the upregulation of gluconate symporter and gluconate permease, which indicates P. halocryophilus Or1 is scavenging additional carbon sources under high salt conditions (Figure 8, group G).
Lipid synthesis and cell wall modifications
Although peptidoglycan synthesized-related genes were upregulated in response to −15 °C, a significant subset of lipid metabolism-related genes were induced specifically under salt stress (Figure 8, group I). Fatty acid breakdown is primarily important to remove lipids that may be impeding membrane fluidity homeostasis via induction of the enzymes 3-ketoacyl-CoA thiolase, and long-chain-fatty-acid-CoA ligase, which appear to be involved under high salt conditions. Synthesis of new fatty acids is induced under both cold and salt stress, whereby cells incubated under high salt conditions had upregulated levels of enoyl-(ACP) reductase and one of the 15 genomic copies of 3-oxoacyl-(ACP) reductase (a unique isozyme from the one induced at −15 °C). Synthesis of new phospholipid head groups was also upregulated via increased levels of phosphatidylserine decarboxylase and CDP-diacylglycerol-serine O-phosphatidyltransferase. It appears that the large turnover in membrane lipids in P. halocryophilus Or1 observed in vivo (described above; Table 1) may be the result of a specific tailored membrane in response to both stresses, and results in a unique lipid composition that has not previously been described in other cryophilic or halophilic bacterial species.
DNA recombination
Two copies of transposases (IS4 family) and a site-specific recombinase/resolvase induced at high salt could be involved in site-specific re-ordering of the genome that may impart a selective advantage for bacteria such as P. halocryophilus Or1 in their natural habitat (Aziz et al., 2010). The importance of transposition in the closely related permafrost bacterium E. sibiricum was inferred (Vishnivetskaya and Kathariou, 2005), through comparisons to mesophilic strains showing a higher abundance of IS elements (nine times as many) in the cold adapted E. sibiricum (Bakermans, 2012). The mesophilic P. donghaensis MPA1U2 genome encodes only four transposases compared with the 12 found in the P. halocryophilus Or1 genome, which lends support to the potential role of transposases in evolutionary and adaptation processes for bacteria in cold/hypersaline environments.
Common stress response
General stress response was apparent within the P. halocryophilus Or1 transcriptome data sets with concomitant changes in the expression of 105 genes observed in response to both sustained salt and −15 °C growth conditions. All of the genes undergoing increased expression (a total of 27) in response to both stresses are illustrated in Figures 9b or c (highlighted) and indicate a synergistic pattern in the cellular mechanisms allowing P. halocryophilus Or1 to grow under these conditions.
Energy and carbohydrate metabolism
Components of carbohydrate metabolism that were similarly induced under both cold and salt stress indicated the induction of metabolite transporters including a sugar ABC transporter, substrate-binding protein and a permease of the drug/metabolite transporter superfamily (Figure 8, group G). Also, in line with the ATP synthase components that appeared to respond to the cold temperatures, both stresses appear to induce the ATP synthase F1-alpha and beta chain subunits, lending further support that under stress conditions, ATP production is essential for maintaining cellular stores of energy.
Motility and cell wall modifications
Motility appeared to be important for both salt and cold stress responses and could assist P. halocryophilus Or1 cells to localize along the solute gradients or solute exclusion freezing fronts in brine growth media, or natural solute-rich habitats. The data suggest that flagellar synthesis is favored at −15 °C while synthesis of pili is induced under high salt conditions (Figure 8, group N). Microscopic observations confirmed that cells remained motile under all the growth conditions (data not shown). In the closely related E. sibirium that produces both motility apparatuses, expression analyses showed that pilus and flagellar synthesis was repressed at −2.5 °C compared with growth at 39 °C (Rodriguez et al., 2008). The motility observed in P. halocryophilus Or1 cells may be a direct result of long-term static incubation in the liquid brine media that likely required the cells remain mobile both in high salt and cold conditions, no doubt imparting high energetic costs for the cells.
Although we observed EPS increasing in P. halocryophilus Or1 cultures down to −5 °C, EPS was not found to be a major constituent of the cell envelope at −15 °C (Mykytczuk et al., in preparation). However, genes involved in EPS synthesis including, tyrosine kinase (EpsD) and glucose-1-phosphate thymidylyltransferase, were found to increase in expression at −15 °C and to a lesser extent under high salt conditions (Figure 8, group M). EPS has been implicated in cold adaptation in cryophilic and mesophilic bacteria in response to cold stress (Krembs and Deming, 2008).
Maintained stress response
Although many of the genomically encoded stress response genes described in Table 3 were found to be maintained at basal expression levels in the acclimated cultures tested, certain universal stress response functions remained induced. Not surprisingly, increased chaperonin activity was maintained under stress through higher expression of heat shock protein 33 and GroEL (Figure 8, group O). As continued survival of the cells requires sensing any changes in environmental conditions, signal transduction mechanisms remained upregulated under both stresses including sensor histidine kinases and several distinct genes encoding diguanylate cyclase/phosphodiesterases (with GGDEF and EAL domains) involved in production of second messenger di-GMP (Figure 8, group T). Multiple copies of these signaling domains involved in two-component signal transduction are found in other bacteria and are involved in functions including secretion, biofilm formation, cell differentiation and targeted transcriptional regulation (Galperin et al., 2001). Several hypothetical proteins were also increased in expression and are among the large gene clusters observed throughout the genome (Supplementary Figure S2, groups R and S). These types of putative gene clusters have also been identified to change in expression under subzero growth in E. sibiricum (Rodrigues et al., 2008) and P. arcticus (Ayala-del-Río et al., 2010) and together represent novel cryophilic gene targets worthy of further investigation.
Synergistic adaptation in P. halocryophilus Or1 and conclusions
This first genomic/transcriptomic analysis of P. halocryophilus Or1 not only separates the cellular response to extreme subzero and hypersaline stress, but also supports the general hypothesis that cryophiles from terrestrial environments must be equally adapted to cold temperatures and dynamic solute concentrations that would characterize the thin water film habitats present in permafrost as well as maintain the ability to grow at warmer temperatures because of the greater temperature fluctuations typically seen in permafrost environments (Rivkina et al., 2000). Together the data indicate that P. halocryophilus Or1 effectively uses genomic plasticity and specific response strategies in parallel, rather than antagonistically, to employ resource efficiency and selective induction and regulation of metabolic functions. The combination of increased flexibility and stability of proteins and uniquely adapted cell envelope compensates against the ordering/disordering effects of a cold and solute-rich environment, while effective osmolyte uptake balances the osmotic pressure exerted over the cell envelope creating a solute-rich cytoplasm capable of remaining liquid at −15 °C. All of these suggest that P. halocryophilus is highly suited for survival and growth in its Arctic permafrost habitat (ambient temp. ∼−16 °C), indicating that these cryoenvironments harbor active microbial ecosystems. Given that permafrost represents ∼26% of Earth's soil ecosystem and undergoing rapid melting with global warming, microbial activity could release the large carbon reservoir as CO2 and CH4, even at subzero temperatures.
Extremophiles such as P. halocryophilus Or1 will expand our insights into molecular traits required for subzero growth and provide a model for understanding how biomolecules function in viable and dividing bacterial cells at the coldest growth temperatures described to date. Cold-active and osmotically stable enzymes or metabolites from P. halocryophilus Or1 carry high interest for future investigations. Lastly, as the search for evidence of life continues in our solar system, organisms such as P. halocryophilus Or1 provide an analog of how life can function in subzero brines; being one of the possible liquid water habitats thought to exist on Mars (McEwan et al., 2011) and other solar system targets including Enceladus (Postberg et al., 2011).
Acknowledgments
The authors would like to thank Ross Overbeek from the RAST team in helping develop a frameshift correction tool to improve the gene calls in the original 454 assembly, and to Roland Wilhelm for laboratory assistance. Funding for this research was provided by the Natural Sciences and Engineering Research Council (NSERC), Canada Research Chair program (CRC), Canadian Foundation for Innovation (CFI), Polar Continental Shelf Program (PCSP), the Canadian Space Agency (CSA) Canadian Analogue Research Network (CARN) Program, and NSERC CREATE postdoctoral research/rotation support grants to NCSM and CRO.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholtz PW, et al. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium reveals mechanisms for adaptation to low temperature growth. Appl Environ Microbiol. 2010;76:2304–2312. doi: 10.1128/AEM.02101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakermans C, Ayala-del-Rio HL, Ponder MA, Vishnivetskaya T, Gilichinsky D, Thomashow MF, et al. Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int J Syst Evol Microbiol. 2006;56:1285–1291. doi: 10.1099/ijs.0.64043-0. [DOI] [PubMed] [Google Scholar]
- Bakermans C, Bergholz PW, Rodrigues DF, Vishnivetskaya TA, Ayala-del-Río HL, Tiedje JM.2012bGenomic and expression analyses of cold-adapted microorganismsIn: Polar Microbiology: Life in a Deep Freeze Miller RV, Whyte LG, (eds).ASM Press: Washington, D.C.126–155. [Google Scholar]
- Bakermans C, Nealson KH. Relationship of critical temperature to macromolecular synthesis and growth yield in Psychrobacter cryopegella. J Bacteriol. 2004;186:2340–2345. doi: 10.1128/JB.186.8.2340-2345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakermans C, Skidmore M. Microbial metabolism in ice and brine at −5 °C. Environ Microbiol. 2011;13:2269–2278. doi: 10.1111/j.1462-2920.2011.02485.x. [DOI] [PubMed] [Google Scholar]
- Bakermans C, Tsapin A, Souza-Egipsy V, Gilichinsky D, Nealson K. Reproduction and metabolism at −10 °C of bacteria isolated from Siberian permafrost. Environ Microbiol. 2003;5:321–326. doi: 10.1046/j.1462-2920.2003.00419.x. [DOI] [PubMed] [Google Scholar]
- Bakermans C.2012Psychrophiles: life in the coldIn: Anitori R, (eds),Extremophiles: Microbiology and Biotechnology Caister Academic Press: Beaverton, Oregon [Google Scholar]
- Bergholz PW, Bakermans C, Tiedje JM. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol. 2009;191:2340–2352. doi: 10.1128/JB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breezee J, Cady N, Staley JT. Subfreezing growth of the sea ice bacterium Psychromonas ingrahamii. Microb Ecol. 2004;47:300–304. doi: 10.1007/s00248-003-1040-9. [DOI] [PubMed] [Google Scholar]
- Budde I, Steil L, Scharf C, Volker U, Bremer E. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology. 2006;152:831–853. doi: 10.1099/mic.0.28530-0. [DOI] [PubMed] [Google Scholar]
- Cartier G, Lorieux F, Allemand F, Dreyfus M, Bizebard T. Cold adaptation in DEAD-Box proteins. Biochemistry. 2010;49:2636–2646. doi: 10.1021/bi902082d. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Saunders N, Thomas T.2001Cold shock response in microorganisms In: Extremophiles: Basic Concepts. Gerday C, Glansdorff N, (eds). UNESCO Encylopedia of Life Support Systems. Eolss publishers, Oxford www.eolss.net .
- Chattopadhyay MK. Low temperature and oxidative stress. Curr Sci. 2002;83:109. [Google Scholar]
- Chin JP, Megaw J, Magill CL, Nowotarski K, Williams JP, Bhaganna P, et al. Solutes determine the temperature windows for microbial survival and growth. PNAS. 2010;107:7835–7840. doi: 10.1073/pnas.1000557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denich TJ, Beaudette LA, Lee H, Trevors JT. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J Microbiol Meth. 2003;52:149–182. doi: 10.1016/s0167-7012(02)00155-0. [DOI] [PubMed] [Google Scholar]
- Dieser M, Greenwood M, Foreman CM. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct Antarct Alp Res. 2010;42:396–405. [Google Scholar]
- D'Amico S, Collins T, Marx J-C, Feller G, Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Rep. 2006;7:385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairén AG, Davila AF, Lim D, Bramall N, Bonaccorsi R, Zavaleta J, et al. Astrobiology through the ages of mars: the study of terrestrial analogues to understand the habitability of mars. Astrobiology. 2010;10:821–843. doi: 10.1089/ast.2009.0440. [DOI] [PubMed] [Google Scholar]
- Fields PA. Review: protein function at thermal extremes: balancing stability and flexibility. Comp Biochem Physiol A. 2001;129:417–431. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- Galinski EA, Truper HG. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiol Rev. 1994;15:95–108. [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilichinsky DA, Soina VS, Petrova MA. Cryoprotective properties of water in the Earth cryolithosphere and its role in exobiology. Orig Life Evol Biosph. 1993;23:65–75. doi: 10.1007/BF01581991. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grzymski JJ, Carter B, DeLong E, Feldman R, Ghadiri A, Murray A. Comparative analysis of genome fragments from 6 uncultured Antarctic marine, planktonic bacteria reveals widespread amino acid modification towards cold adaptation. Appl Env Micro. 2006;72:1532–1541. doi: 10.1128/AEM.72.2.1532-1541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne H, Mäder U, Otto A, Bonn F, Steil L, Bremer E, et al. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J Bacteriol. 2010;92:870–882. doi: 10.1128/JB.01106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Pané-Farré J, Völker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Márquez LM, Chamberlin MJ. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988;170:1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannadham MV, Chattopadhyay MK, Subbalakshmi C, Vairamani M, Narayanan K, Rao CM, et al. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch Microbiol. 2000;173:418–424. doi: 10.1007/s002030000163. [DOI] [PubMed] [Google Scholar]
- Jakosky BM, Nealson KH, Bakermans C, Ley RE, Mellon MT. Subfreezing activity of microorganisms and the potential habitability of Mars' polar regions. Astrobiology. 2003;3:343–350. doi: 10.1089/153110703769016433. [DOI] [PubMed] [Google Scholar]
- Junge K, Eicken H, Deming JW. Bacterial activity at −2 to −20 °C in Arctic wintertime sea ice. Appl Environ Microbiol. 2004;70:550–557. doi: 10.1128/AEM.70.1.550-557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krembs C, Deming JW.2008The role of exopolymers in microbial adaptation to sea iceIn Mergesin R, Schinner F, Marx J-C, Gerday C, (eds),Psychrophiles: From Biodiversity To Biotechnology Springer-Verlag: Berlin; 247–264. [Google Scholar]
- Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S, Yoneta M, Takada Y. Two isocitrate dehydrogenases from a psychrophilic bacterium, Colwellia psychrerythraea. Extremophiles. 2006;10:237–249. doi: 10.1007/s00792-005-0493-9. [DOI] [PubMed] [Google Scholar]
- Margesin R, Miteva V. Diversity and ecology of psychrophilic microorganisms. Res Microbiol. 2011;162:346–361. doi: 10.1016/j.resmic.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Marin K, Kanesaki Y, Los DA, Murata N, Suzuki I, Hagemann M. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 68031. Plant Physiol. 2004;136:3290–3300. doi: 10.1104/pp.104.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhaney RN. The effect of alterations in the physical state of the membrane lipids on the ability of Acholeplasma laidlawii B to grow at various temperatures. J Mol Biol. 1974;84:145–157. doi: 10.1016/0022-2836(74)90218-6. [DOI] [PubMed] [Google Scholar]
- McEwen AS, Ojha L, Dundas CM, Mattson SM, Byrne S, Wray JJ, et al. Seasonal flows on warm Martian slopes. Science. 2011;333:740–743. doi: 10.1126/science.1204816. [DOI] [PubMed] [Google Scholar]
- McKay C, Mykytczuk NCS, Whyte LG.2012Chapter 14: Life in Ice on Other Worlds In Miller R, Whyte LG, (eds),Polar Microbiology: Life in a Deepfreeze ASM Press: Washington; 290–304. [Google Scholar]
- Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. PNAS. 2005;102:10913–10908. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metpally RP, Reddy BV. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genomics. 2009;10:11. doi: 10.1186/1471-2164-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytczuk NCS, Trevors JT, Leduc LG, Ferroni GD. Fluorescence polarization in studies of bacterial cytoplasmic membrane fluidity under environmental stress. Prog Biophys Mol Biol. 2007;95:60–82. doi: 10.1016/j.pbiomolbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Mykytczuk NCS, Wilhelm RC, Whyte LG. Planococcus halocryophilus sp. nov., an extreme sub-zero species from high Arctic permafrost. Int J Syst Evol Microbiol. 2012;62:1937–1944. doi: 10.1099/ijs.0.035782-0. [DOI] [PubMed] [Google Scholar]
- Page-Sharp M, Behm CA, Smith GD. Cyanophycin and glycogen synthesis in a cyanobacterial Scytonema species in response to salt stress. FEMS Microbiol Lett. 1998;160:11–15. [Google Scholar]
- Panikov NS, Flanagan PW, Oechel WC, Mastepanov MA, Christensen TR. Microbial activity in soils frozen to below −39 °C. Soil Biol Biochem. 2006;38:785–794. [Google Scholar]
- Panikov NS, Sizova MV. Growth kinetics of microorganisms isolated from Alaskan soil and permafrost in solid media frozen down to −35 °C. FEMS Microbiol Ecol. 2007;59:500–512. doi: 10.1111/j.1574-6941.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Ponder MA, Gilmour SJ, Bergholz PW, Mindock CA, Hollingsworth R, Thomashow MF, et al. Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol Ecol. 2005;53:103–115. doi: 10.1016/j.femsec.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Postberg F, Schmidt J, Hillier J, Kempf S, Srama R. A saltwater reservoir as the source of a compositionally stratified plume on Enceladus. Nature. 2011;474:620–622. doi: 10.1038/nature10175. [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2011;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy, and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Staley JT, Danchin A, Wang TZ, Brettin TS, Hauser LJ, et al. Genomics of an extreme psychrophile. BMC Genomics. 2008;9:210. doi: 10.1186/1471-2164-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkina EM, Friedmann EI, McKay CP, Gilichinsky DA. Metabolic activity of permafrost bacteria below the freezing point. Appl Environ Microbiol. 2000;66:3230–3233. doi: 10.1128/aem.66.8.3230-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D, Ivanova N, He Z, Huebner M, Zhou J, Tiedje J. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics. 2008;9:547. doi: 10.1186/1471-2164-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell NJ. Mechanisms of thermal adaptation in bacteria, blueprints for survival. Trends Biochem Sci. 1984;3:108–112. [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101 Newark DE, MIDI Inc. Schmieder R, Edwards R, (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics. 1990;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Steven B, Leveille R, Pollard WH, Whyte LG. Microbial ecology and biodiversity in permafrost. Extremophiles. 2006;10:259–267. doi: 10.1007/s00792-006-0506-3. [DOI] [PubMed] [Google Scholar]
- Steven B, Niederberger TD, Bottos EM, Dyen MR, Whyte LG. Development of a sensitive radiorespiration method for detecting microbial activity at subzero temperatures. J Microbiol Methods. 2007;71:275–280. doi: 10.1016/j.mimet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya T, Kathariou S. Putative transposases conserved in Exiguobacterium isolates from ancient Siberian permafrost and from contemporary surface habitats. Appl Environ Microbiol. 2005;71:6954–6962. doi: 10.1128/AEM.71.11.6954-6962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya TA, Liebner S, Wilhelm R, Wagner D.2012Microbial carbon cycling in permafrostIn: Whyte L, Miller R, (eds),ASM Polar Microbiology Textbook ASM Press: Washington, DC; 183–200. [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews B, et al. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- Wood JM, Bremer E, Csonka LN, Kraemer R, Poolman B, van der Heide T, et al. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol A Mol Integr Physiol. 2001;137:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- Zak DR, Kling GW. Microbial community composition and function across an Arctic tundra landscape. Ecology. 2006;87:1659–1670. doi: 10.1890/0012-9658(2006)87[1659:mccafa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.