Introduction
Oncostatin M (OSM), belonging to the IL-6 family of cytokines (Heinrich et al. 2003), was first reported and purified from U937 monocytic cells (Zarling et al. 1986; Ensoli et al. 1999; Hasegawa et al. 1999). In normal physiological condition, OSM is associated with multiple biological processes and cellular responses including growth, differentiation, and inflammation However, anti-proliferative activity of OSM against breast cancer cell line generated the interest of biomedical community on this molecule (Douglas et al. 1997, 1998). OSM was also found associated with pathological conditions such as proliferation of ovarian cancer cells (Taga and Kishimoto 1997), prostate cancer 22Rv1 cells (Hoffman et al. 1996), up-regulation of the ER chaperone Grp78/BiP in the liver cells, atherosclerotic lesions, ischemic heart disease and rheumatoid arthritis (Linsley et al. 1990; Dunham et al. 1999). The dual role of OSM in either inducing or inhibiting the proliferation of various types of cells called upon the scientific community to investigate role of OSM in various physiological and experimental contexts in detail. However, diverse molecular level information pertaining to OSM signaling is not available in a centralized resource. Therefore, we have systematically gathered and curated molecular information from literature and created a public resource for OSM induced signaling events. We integrated OSM signaling pathway into NetPath (Kandasamy et al. 2010), which is a public resource of human signaling pathways.
OSM is known to mediate its biological effects by binding to two distinct heterodimers of gp130 with either leukemia inhibiting factor receptor (LIFR) or OSM receptor-beta (OSMR-beta) (Thoma et al. 1994). Former heterodimer between gp130 and LIFR is called type I receptor complex and the latter between gp130 and OSMR-beta is called type II receptor complex. Type I receptor can bind to either OSM or leukemia inhibiting factor, whereas type II receptor has more affinity towards OSM (O’Hara et al. 2003). The binding of OSM to either gp130/OSMR-beta or gp130/LIFR induces the activation of Janus Kinase family members through tyrosine phosphorylation (Tanaka and Miyajima 2003). The activated JAK family members in turn induce the activation of Signal Transduction and Activator of Transcription (STAT) proteins (Schaefer et al. 2000). Alternatively, the activated receptors can also activate mitogen-activated protein kinase (MAPK) pathway (Van Wagoner et al. 2000) and PI3K/AKT pathways (Arita et al. 2008). It was also reported that OSM bring about ligand-induced receptor degradation of gp130, OSMR-beta, and LIFR before enhancing the synthesis of the receptor subunits (Blanchard et al. 2001).
Materials and methods
Literature search was carried out using ‘oncostatin’ as a keyword. The interpreted information regarding OSM signaling cascade was recorded in ‘PathBuilder,’ a software utility developed by Kandasamy and colleagues to annotate signaling pathways (Kandasamy et al. 2009). In order to generate an OSM specific signaling pathway, we have considered only those reactions that are specifically initiated by OSM stimulation. Molecular reactions initiated by OSM in combination with other cytokines were thus, not considered in the signaling cascade depicted here. We considered NetPath annotation criteria, as previously described in (Goel et al. 2011; Nanjappa et al. 2011; Raju et al. 2011a) to generate the OSM signaling cascade and for categorizing the involved reactions into protein-protein interactions, post-translational modifications (PTMs), protein activation-inhibition reactions, protein translocation events and gene regulation events. In case of PTMs, we have mapped site and residue information of the PTMs given in the literature to specific and longest isoform of the corresponding protein as given in the RefSeq database. In addition to the NetPath criteria, we applied more stringent NetSlim criteria to choose more confident and core pathway reactions, among larger set of OSM pathway reactions (Raju et al. 2011b). Using this subset of pathway reactions, we depicted a core signaling cascade map of OSM, using ‘PathVisio’ (http://www.PathVisio.org).
After an initial round of internal review, we uploaded OSM signaling pathway information into NetPath. Further, we requested the ‘Pathway Authority,’ who has considerable expertise on OSM signaling, for expert opinion. The pathway authority, in this case SKL, who is also a co-author in this manuscript, has reviewed the listed reactions pertaining to the OSM signaling cascade. We have incorporated suggestions and opinion of the pathway authority.
Results and discussion
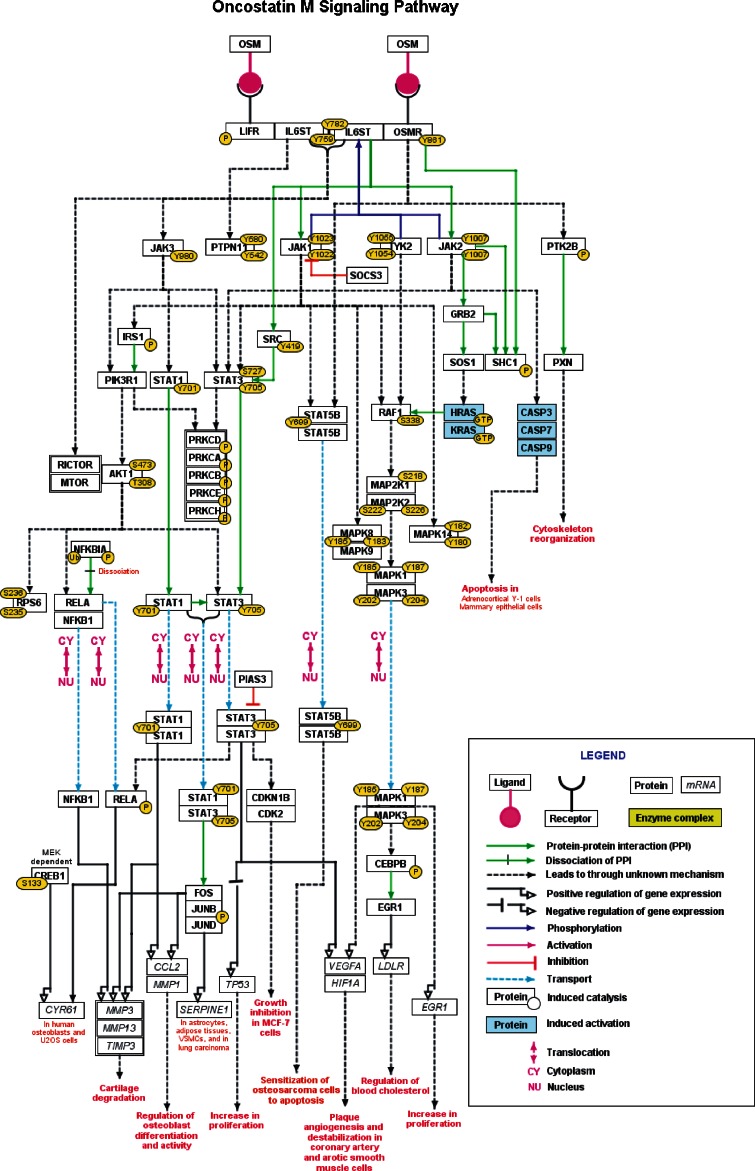
The information pertaining to OSM signaling was found in over 1,100 scientific research articles. A manual analysis through the available and relevant scientific literature led to the identification of 78 molecules involved in the OSM signaling cascade reactions. Among these, 78 molecules were part of 35 protein-protein interaction and 54 PTM events. OSM signaling pathway was also found to regulate the expression of 322 unique genes in humans. Stimulated by OSM signaling pathway, 8 proteins were found to be translocated between different sub-cellular compartments. We provided these pathway reactions and their associated descriptions in NetPath (http://www.netpath.org/pathways?path_id=NetPath_114). The molecule page in NetPath was made dynamic and informative by linking represented molecules to external repositories including HPRD (Goel et al. 2012; Prasad et al. 2009), OMIM (Hamosh et al. 2005), Entrez Gene (Maglott et al. 2005) and Swiss-Prot (Boeckmann et al. 2003). A graphical depiction of OSM pathway reactions chosen by NetSlim criteria is provided in Fig. 1. The OSM reaction map can also be accessed at NetSlim through the link provided (http://www.netpath.org/netslim/NetSlim_114).
Fig. 1.
Schematic representation of Oncostatin M signaling pathway- NetSlim (http://www.netpath.org/netslim/NetSlim_114) map generated to graphically represent the different reactions induced by OSM in various cell types
Role of OSM in inhibiting and inducing proliferation
OSM was observed to promote and inhibit cell proliferation in different cell types. It was found that OSM employs different downstream elements to bring about these effects. Studies pertaining to OSM induced inhibitory effects in MCF-7 cell lines revealed a STAT3 activated module of signaling. This activation of STAT3 by OSM was in turn found to influence various genes associated with migration and cell differentiation (Zhang et al. 2003). In human proximal tubule cells, OSM was found to have anti-fibrotic effects, through the attenuation of transforming growth factor -β1 initiated events and inhibition of CTGF expression (Sarkozi et al. 2011). In case of SKOV3 ovarian cancer cell line, proliferating effect of OSM have been traced to the phosphorylation and activation of STAT3, MAPK1/2 and p38 signaling cascade (Li et al. 2011). However, proliferative effects of OSM on OSA cell lines were shown to be associated with activation of STAT3, Src and JAK2 cascade and to increased expression of MMP2 and VEGF genes, which in turn associates OSM with tumor invasion and angiogenesis (Fossey et al. 2011).
Role of OSM in other pathological conditions
OSM has been associated with multiple sclerosis of microglia and astrocytes through upregulation of associated molecules including intercellular adhesion molecule-1 in human cerebral epithelial cells (Ruprecht et al. 2001). Regulation of transcription of LDLR gene by OSM has been associated with the regulation of blood cholesterol level. OSM was also reported to utilize JAK/STAT and MAPK signaling cascades in HepG2 cells and cause a decreased expression of PCSK9 gene, which influences LDLR expression in liver (Cao et al. 2011). Albasanz-Puig and colleagues demonstrated the dual roles of OSM, brought about through interplay of STAT1 and STAT3, in both promoting as well as inhibiting atherosclerosis. OSM had also been found to inhibit VEGF expression through STAT1, whereas STAT3 module induced by OSM was found to increase the expression of VEGF expression (Albasanz-Puig et al. 2012). The ability of OSM to influence different genes including CYR1, PGLF and IL6, in various cells and cell lines, through either JAK/STAT, MAPK or CREB signaling cascades, associated OSM to various pathological conditions in rheumatoid arthritis (Migita et al. 2011; Tu et al. 2012; Kok et al. 2011).
Role of OSM in repair and remodeling
Studies on adult human cardiac myocytes, fibroblasts and bone marrow-derived mesenchymal stem cells showed the recruitment of STAT3 or MAPK1/2 by OSM, which initiated remodeling in pathological conditions such as arthritis and osteoporosis, as well as aiding in repair of fractures (Weiss et al. 2005; Nicolaidou et al. 2012).
Availability of data
OSM signaling pathway data can be freely downloaded from NetPath (http://www.netpath.org/pathways?path_id=NetPath_114) and NetSlim (http://www.netpath.org/netslim/NetSlim_114), for unrestricted non-commercial use under the adaptive Creative Commons License (http://creativecommons.org/licenses/by-nc/2.5). The data available in both the resources will be periodically updated. We have also submitted OSM pathway to Wikipathways (http://www.wikipathways.org/index.php/Pathway:WP2374).
Conclusions
OSM is emerging as an important molecule with potential clinical and therapeutic values in multiple human diseases. However, a variety of biological effects induced by OSM signaling pathway remain poorly defined so far. Therefore, we tried to gather publicly available experimentally derived information on OSM induced events and depicted these molecular events in the form of a signaling pathway. We believe that OSM signaling pathway information, as freely available will soon be integrated into various global resources and gene set enrichment analysis tools, which will allow discovery of biological processes and pathological conditions where OSM signaling is involved. OSM signaling pathway developed in this study will also provide a platform for biomedical community to design further discovery oriented high-throughput studies for this signaling pathway. OSM pathway map generated by us would aid and accelerate future studies of OSM related processes.
Acknowledgments
We thank the Department of Biotechnology, Government of India for research support to the Institute of Bioinformatics, Bangalore. Gourav Dey and Nazia Syed are recipients of Junior Research Fellowship from the University Grants Commission (UGC), Government of India. Aneesha Radhakrishnan is a recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR), Government of India. We thank G. S Sameer Kumar and Renu Goel for assistance in curation.
Conflict of interest
The author(s) declared no conflicts of interests.
Abbreviations
- AKT
v-akt murine thymoma viral oncogene homolog 1
- JAK
Janus kinase
- LIFR
Leukemia inhibiting factor
- MAPK
Mitogen-activated protein kinase
- OSM
Oncostatin M
- OSMR-beta
OSM receptor-beta subunit
- PI3K
Phosphoinositide 3-kinase
- PTMs
Post-translational modifications
- STAT
Signal transduction and activator of transcription
Footnotes
Gourav Dey and Aneesha Radhakrishnan contributed equally.
Contributor Information
Gourav Dey, Email: gourav@ibioinformatics.org.
Aneesha Radhakrishnan, Email: aneesha@ibioinformatics.org.
Nazia Syed, Email: nazia@ibioinformatics.org.
Joji Kurian Thomas, Email: joji@ibioinformatics.org.
Arpitha Nadig, Email: arpithanadig02@gmail.com.
Kotteazeth Srikumar, Email: srikumarprof@gmail.com.
Premendu Prakash Mathur, Email: ppmathur@yahoo.com.
Akhilesh Pandey, Email: pandey@jhmi.edu.
Sze-Kwan Lin, Email: sklin7400@ntu.edu.tw.
Rajesh Raju, Email: rajesh@ibioinformatics.org.
T. S. Keshava Prasad, Phone: +91-80-28416140, FAX: + 91-80-28416132, Email: keshav@ibioinformatics.org.
References
- Albasanz-Puig A, Murray J, Namekata M, Wijelath ES. Opposing roles of STAT-1 and STAT-3 in regulating vascular endothelial growth factor expression in vascular smooth muscle cells. Biochem Biophys Res Commun. 2012;428(1):179–184. doi: 10.1016/j.bbrc.2012.10.037. [DOI] [PubMed] [Google Scholar]
- Arita K, South AP, Hans-Filho G, Sakuma TH, Lai-Cheong J, Clements S, Odashiro M, Odashiro DN, Hans-Neto G, Hans NR, et al. Oncostatin M receptor-beta mutations underlie familial primary localized cutaneous amyloidosis. Am J Hum Genet. 2008;82:73–80. doi: 10.1016/j.ajhg.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard F, Wang Y, Kinzie E, Duplomb L, Godard A, Baumann H. Oncostatin M regulates the synthesis and turnover of gp130, leukemia inhibitory factor receptor alpha, and oncostatin M receptor beta by distinct mechanisms. J Biol Chem. 2001;276:47038–47045. doi: 10.1074/jbc.M107971200. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A, Wu M, Li H, Liu J. Janus kinase activation by cytokine oncostatin M decreases PCSK9 expression in liver cells. J Lipid Res. 2011;52:518–530. doi: 10.1194/jlr.M010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AM, Goss GA, Sutherland RL, Hilton DJ, Berndt MC, Nicola NA, Begley CG. Expression and function of members of the cytokine receptor superfamily on breast cancer cells. Oncogene. 1997;14:661–669. doi: 10.1038/sj.onc.1200882. [DOI] [PubMed] [Google Scholar]
- Douglas AM, Grant SL, Goss GA, Clouston DR, Sutherland RL, Begley CG. Oncostatin M induces the differentiation of breast cancer cells. Int J Cancer. 1998;75:64–73. doi: 10.1002/(SICI)1097-0215(19980105)75:1<64::AID-IJC11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Ensoli F, Fiorelli V, DeCristofaro M, Santini Muratori D, Novi A, Vannelli B, Thiele CJ, Luzi G, Aiuti F. Inflammatory cytokines and HIV-1-associated neurodegeneration: oncostatin-M produced by mononuclear cells from HIV-1-infected individuals induces apoptosis of primary neurons. J Immunol. 1999;162:6268–6277. [PubMed] [Google Scholar]
- Fossey SL, Bear MD, Kisseberth WC, Pennell M, London CA. Oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines. BMC Cancer. 2011;11:125. doi: 10.1186/1471-2407-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R, Harsha HC, Pandey A, Prasad TSK. Human protein reference database and human proteinpedia as resources for phosphoproteome analysis. Mol Biosyst. 2012;8:453–463. doi: 10.1039/c1mb05340j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R, Raju R, Maharudraiah J, Kumar GSS, Ghosh K, Kumar A, Lashmi PT, et al. A signaling network of thyroid-stimulating hormone. J Proteome Bioinforma. 2011;4:238–241. doi: 10.4172/jpb.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Sato S, Ihn H, Takehara K. Enhanced production of interleukin-6 (IL-6), oncostatin M and soluble IL-6 receptor by cultured peripheral blood mononuclear cells from patients with systemic sclerosis. Rheumatology (Oxford) 1999;38:612–617. doi: 10.1093/rheumatology/38.7.612. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RC, Moy FJ, Price V, Richardson J, Kaubisch D, Frieden EA, Krakover JD, Castner BJ, King J, March CJ, Powers R. Resonance assignments for oncostatin M, a 24-kDa alpha-helical protein. J Biomol NMR. 1996;7:273–282. doi: 10.1007/BF00200429. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Keerthikumar S, Raju R, Prasad TSK, Ramachandra YL, Mohan S, Pandey A. PathBuilder–open source software for annotating and developing pathway resources. Bioinformatics. 2009;25:2860–2862. doi: 10.1093/bioinformatics/btp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok SH, Hou KL, Hong CY, Wang JS, Liang PC, Chang CC, Hsiao M, Yang H, Lai EH, Lin SK. Simvastatin inhibits cytokine-stimulated Cyr61 expression in osteoblastic cells: a therapeutic benefit for arthritis. Arthritis Rheum. 2011;63:1010–1020. doi: 10.1002/art.27433. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhu J, Sun F, Liu L, Liu X, Yue Y. Oncostatin M promotes proliferation of ovarian cancer cells through signal transducer and activator of transcription 3. Int J Mol Med. 2011;28:101–108. doi: 10.3892/ijmm.2011.647. [DOI] [PubMed] [Google Scholar]
- Linsley PS, Kallestad J, Ochs V, Neubauer M. Cleavage of a hydrophilic C-terminal domain increases growth-inhibitory activity of oncostatin M. Mol Cell Biol. 1990;10:1882–1890. doi: 10.1128/mcb.10.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita K, Komori A, Torigoshi T, Maeda Y, Izumi Y, Jiuchi Y, Miyashita T, Nakamura M, Motokawa S, Ishibashi H. CP690,550 inhibits oncostatin M-induced JAK/STAT signaling pathway in rheumatoid synoviocytes. Arthritis Res Ther. 2011;13:R72. doi: 10.1186/ar3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjappa V, Raju R, Muthusamy B, Sharma J, Thomas K, Nidhina PAH, Harsha HC, Pandey A, Anilkumar G, Prasad TSK. A comprehensive curated reaction map of leptin signaling pathway. J Proteome Bioinforma. 2011;4:184–189. [Google Scholar]
- Nicolaidou V, Wong MM, Redpath AN, Ersek A, Baban DF, Williams LM, Cope AP, Horwood NJ. Monocytes induce STAT3 activation in human mesenchymal stem cells to promote osteoblast formation. PLoS One. 2012;7:e39871. doi: 10.1371/journal.pone.0039871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara KA, Kedda MA, Thompson PJ, Knight DA. Oncostatin M: an interleukin-6-like cytokine relevant to airway remodelling and the pathogenesis of asthma. Clin exp allergy: j Br Soc Allergy Clin Immunol. 2003;33:1026–1032. doi: 10.1046/j.1365-2222.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Prasad TSK, Kandasamy K, Pandey A. Human protein reference database and human proteinpedia as discovery tools for systems biology. Methods Mol Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- Raju R, Balakrishnan L, Nanjappa V, Bhattacharjee M, Getnet D, Muthusamy B, Kurian Thomas J et al. (2011a) A comprehensive manually curated reaction map of RANKL/RANK-signaling pathway. Database (Oxford) 2011: bar021 [DOI] [PMC free article] [PubMed]
- Raju R, Nanjappa V, Balakrishnan L, Radhakrishnan A, Thomas JK, Sharma J, Tian M, Palapetta SM, Subbannayya T et al. (2011b) NetSlim:high-confidence curated signaling maps. Database (Oxford) 2011:bar032 [DOI] [PMC free article] [PubMed]
- Ruprecht K, Kuhlmann T, Seif F, Hummel V, Kruse N, Bruck W, Rieckmann P. Effects of oncostatin M on human cerebral endothelial cells and expression in inflammatory brain lesions. J Neuropathol Exp Neurol. 2001;60:1087–1098. doi: 10.1093/jnen/60.11.1087. [DOI] [PubMed] [Google Scholar]
- Sarkozi R, Hauser C, Noppert SJ, Kronbichler A, Pirklbauer M, Haller VM, Grillari J, Grillari-Voglauer R, Mayer G, Schramek H. Oncostatin M is a novel inhibitor of TGF-beta1-induced matricellular protein expression. Am J Physiol Renal Physiol. 2011;301:F1014–F1025. doi: 10.1152/ajprenal.00123.2011. [DOI] [PubMed] [Google Scholar]
- Schaefer LK, Wang S, Schaefer TS. Oncostatin M activates stat DNA binding and transcriptional activity in primary human fetal astrocytes: low- and high-passage cells have distinct patterns of stat activation. Cytokine. 2000;12:1647–1655. doi: 10.1006/cyto.2000.0774. [DOI] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Miyajima A. Oncostatin M, a multifunctional cytokine. Rev Physiol Biochem Pharmacol. 2003;149:39–52. doi: 10.1007/s10254-003-0013-1. [DOI] [PubMed] [Google Scholar]
- Thoma B, Bird TA, Friend DJ, Gearing DP, Dower SK. Oncostatin M and leukemia inhibitory factor trigger overlapping and different signals through partially shared receptor complexes. J Biol Chem. 1994;269:6215–6222. [PubMed] [Google Scholar]
- Tu HJ, Lin TH, Chiu YC, Tang CH, Yang RS, Fu WM (2012) Enhancement of placenta growth factor expression by oncostatin M in human rheumatoid arthritis synovial fibroblasts. J Cell Physiol [DOI] [PubMed]
- Van Wagoner NJ, Choi C, Repovic P, Benveniste EN. Oncostatin M regulation of interleukin-6 expression in astrocytes: biphasic regulation involving the mitogen-activated protein kinases ERK1/2 and p38. J Neurochem. 2000;75:563–575. doi: 10.1046/j.1471-4159.2000.0750563.x. [DOI] [PubMed] [Google Scholar]
- Weiss TW, Kvakan H, Kaun C, Zorn G, Speidl WS, Pfaffenberger S, Maurer G, Huber K, Wojta J. The gp130 ligand oncostatin M regulates tissue inhibitor of metalloproteinases-1 through ERK1/2 and p38 in human adult cardiac myocytes and in human adult cardiac fibroblasts: a possible role for the gp130/gp130 ligand system in the modulation of extracellular matrix degradation in the human heart. J Mol Cell Cardiol. 2005;39:545–551. doi: 10.1016/j.yjmcc.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Zarling JM, Shoyab M, Marquardt H, Hanson MB, Lioubin MN, Todaro GJ. Oncostatin M: a growth regulator produced by differentiated histiocytic lymphoma cells. Proc Natl Acad Sci U S A. 1986;83:9739–9743. doi: 10.1073/pnas.83.24.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Li C, Halfter H, Liu J. Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene. 2003;22:894–905. doi: 10.1038/sj.onc.1206158. [DOI] [PubMed] [Google Scholar]