Abstract
Intestinal epithelial cells provide an essential line of defense for Caenorhabditis elegans against ingested pathogens. Because nematodes consume microorganisms as their food source, there has presumably been selection pressure to evolve and maintain immune defense mechanisms within the intestinal epithelium. Here we review recent advances that further define the immune signaling network within these cells and suggest mechanisms used by the nematode to monitor for infection. In reviewing studies of pathogenesis that use this simple model system, we hope to illustrate some of the basic principles of epithelial immunity that may also be of relevance in higher order hosts.
Introduction
The coordinated regulation of immune defenses is of paramount importance to combat infection and prevent the deleterious effects of unchecked immune responses on host cells. This is of particular importance in the mammalian intestine where potential pathogens must be differentiated from innocuous bacterial species that are part of the normal flora of humans. One approach to understand the mechanisms of immune detection at epithelial surfaces is to use invertebrate hosts, such as the microscopic nematode Caenorhabditis elegans, to examine evolutionarily conserved aspects of innate immunity and pathogen virulence [1–13]. In its natural habitats, C. elegans animals consume bacteria and fungi as their food source and encounter numerous threats from ingested microorganisms. Thus, there has presumably been a strong selection pressure to evolve and maintain a defense system within the intestinal epithelium of the nematode that is able to mount targeted defense responses toward pathogens, but not against innocuous food sources.
Because of the wealth of genetic and genomic tools available for C. elegans research, we are beginning to understand immune mechanisms in the nematode, which has offered important insights into the origins and fundamental principles of immunity. This topic has been reviewed previously on a number of occasions [1–13], most recently in 2010 [7]. In this review, we focus on studies that use C. elegans to study the immune mechanisms in intestinal epithelial cells and highlight several exciting developments that have emerged in the last two years.
Key differences between the nematode and vertebrate innate immune systems
Several key features of the mammalian innate immune response are not encoded in the C. elegans genome, including homologs of the transcription factor NF-κB or the Toll-like receptor (TLR) adaptor protein MYD88. In addition, the sole TLR homolog in C. elegans does not appear to play a major role in activating the innate immune response by functioning either directly or indirectly as a receptor for pathogen-associated molecular pattern (PAMP) molecules. C. elegans also does not produce homologs of the known vertebrate cytokines. Interestingly, many of these prominent features of the mammalian immune response appear to have been lost from the nematode lineage during evolution since they are present in more primitive metazoans such as the sea anemone Nematostella vectensis, suggesting that the common eumetazoan ancestor of cnidarian (sea anemone and hydra) and bilaterian (worms, arthropods, vertebrates) metazoans had these innate immune signaling components [7]. What then can we learn from the study of C. elegans innate immunity? Although the nematode lacks NF-κB, MYD88 and other components of the TLR signaling pathway, it mounts an immune response that utilizes several evolutionarily conserved signaling pathways, including p38 mitogen-activated protein kinase (MAPK), β-catenin, and FOXO transcription factors, which surprisingly appear to function in parallel to activate at least partially non-overlapping sets of effector genes. A question of primary importance to immunologists is whether these conserved C. elegans immune signaling pathways are also involved in the mammalian innate immune response. From this perspective, it can be argued that nematodes offer an excellent opportunity to identify TLR-independent and NF-κB-independent features of the metazoan innate immune response that may be difficult to identify and study in vertebrate models.
C. elegans anatomy facilitates study of host–pathogen interactions in the intestine
C. elegans does not have an adaptive immune system or mobile immune cells, such as professional phagocytes. Thus, defenses mounted by intestinal epithelial cells are critically important to defend the nematode against ingested pathogens. Importantly, key anatomical features of the C. elegans intestinal epithelium are conserved in mammals (Figure 1). Both cell types have a polarized structure with apical microvilli attached to a terminal web composed of actin and intermediate filaments. Moreover, the nematode is transparent, which allows direct microscopic observation of invading pathogens in an intact host. Finally, the worm intestine in its entirety consists of only 20 non-renewable cells, greatly simplifying the analysis of an entire infectious process, which can be monitored in real time using a variety of microscopic techniques. Two groups have recently taken advantage of these features of C. elegans intestinal cell anatomy to study novel, naturally occurring pathogens of nematodes.
Figure 1.
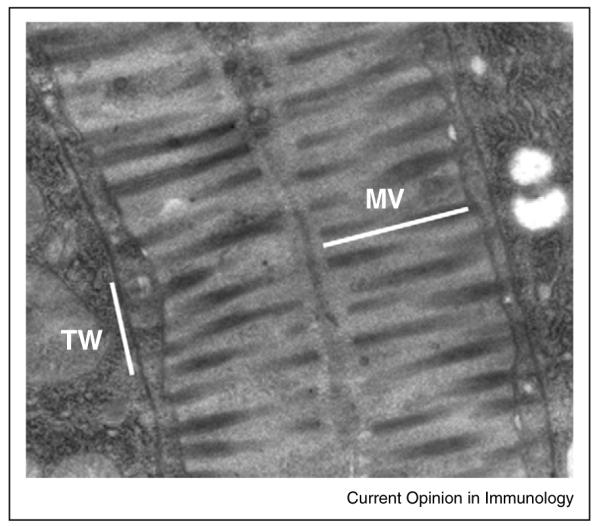
Features of C. elegans intestinal epithelial cells are strongly conserved. C. elegans intestinal epithelial cells bear a striking resemblance to human intestinal cells. Both cells have a brush border composed of microvilli (MV) anchored to a cellular structure called the terminal web (TW).This image was originally published by Troemel et al. [15] and is reproduced here with permission from Emily Troemel.
Troemel et al. identified a microsporidial pathogen in a wild-caught C. elegans strain isolated from a compost pit near Paris, France, which they found comprised a new genus and species [14••,15,16]. This organism, named Nematocida parisii, establishes an intracellular infection within the intestinal epithelium of the nematode and eventually kills the animal. Infected nematodes actively shed spores into the environment, which cause infection in neighboring animals. By direct visualization of infected nematodes in which the actin cytoskeleton and terminal web were engineered to express YFP or CFP, respectively, Estes et al. show that N. parisii creates gaps in the normally contiguous terminal web by promoting the cellular redistribution of actin toward the basolateral side of the cell away from its normal apical location in the terminal web and microvilli [14••]. They postulate that N. parisii spores exit through these gaps into the intestinal lumen.
In another important study of pathogens identified in wild-caught nematodes, Félix et al. characterized the first two viruses able to infect Caenorhabditis species [17••]. From animals with unusual morphological features in their intestines, these researchers identified two single-stranded RNA viruses that are distantly related to nodaviruses and capable of infecting a variety of laboratory nematode strains. Interestingly, host RNAi machinery was implicated in the defense against these viruses. The mechanisms used by C. elegans epithelial cells to defend against intracellular pathogens are incompletely understood, but are now the subject of focused investigations using these new and interesting natural C. elegans pathogens.
The C. elegans p38 MAP kinase PMK-1: one central regulator, multiple immune outputs
Principal among the immune regulators in C. elegans is the NSY-1/SEK-1/PMK-1 MAP kinase pathway, which was identified in a forward genetic screen for mutants with enhanced susceptibility to infection with the Gram-negative bacteria Pseudomonas aeruginosa [18]. This pathway is orthologous to the ASK1 (MAP kinase kinase kinase)/MKK3/6 (MAP kinase kinase)/p38 (MAP kinase) pathway in mammals, and its identification in C. elegans provided an important clue about the evolutionary origins of innate immunity. Activation of this signaling cassette is complex. An ortholog of mammalian SARM called Toll-interleukin-1 receptor (TIR-1) [19–22], and the protein kinases Cδ (PKCδ) [23] and D (PKD) [24] act upstream of NSY-1. A recent study found that a signaling module formed by the G protein alpha subunit (Gqα) and the signal transducer phospholipase Cβ (PLCβ) modulate the activity of the p38 MAP kinase cassette within the intestine [25••]. Interestingly, however, stimulation of the p38 MAP kinase cassette occurs in a manner independent of the single TLR homolog in C. elegans (tol-1). Thus, dissection of the p38 MAP kinase cassette enables analyses of immune mechanisms that are important in the absence of TLR signaling.
The p38 MAP kinase pathway acts cell autonomously in the intestinal epithelium [26] to coordinate defense against a wide variety of ingested pathogens. C. elegans carrying loss-of-function mutations in pmk-1 are hypersusceptible to infection with the Gram-negative pathogens P. aeruginosa [18,27], Salmonella enterica [28], Yersinia pestis [29•] and Serratia marcescens [30••]; the Gram-positive pathogens Enterococcus faecalis [30••] and Staphylococcus aureus [31]; and the fungus Candida albicans [32••]. Moreover, activity of the p38 MAP kinase PMK-1 declines with age and was recently shown to underlie the increased susceptibility to bacterial killing that occurs in older C. elegans [33••]. Troemel et al. used global transcriptional profiling analyses of nematodes growing under normal laboratory conditions to show that PMK-1 regulates the expression of putative antimicrobial effectors, including ShK toxins, C-type lectins and genes carrying a CUB-like domain [27], in the absence of pathogen challenge. This has been termed ‘basal regulation,’ to distinguish it from the induction of immune effectors that occurs during challenge with a pathogen. In addition, a variety of genes that are induced during pathogen attack by diverse pathogens require PMK-1 [18,27,29•,32••]; however, the full spectrum of genes that are activated by pathogens in a PMK-1-dependent manner has not been determined. Interestingly, transcriptional profiling experiments have demonstrated that the immune effectors upregulated by divergent pathogens, including several genes that require PMK-1, are largely non-overlapping [27,29•,32••,34••]. These data suggest that the PMK-1 cascade coordinates the induction of multiple immune effectors that differ depending on the infecting organism.
Work by Shivers et al. has recently shed light on a mechanism downstream of PMK-1 that accounts for part of the immune specificity mediated by this protein [30••]. Using an approach that highlights some of the advantages of working with a model genetic host such as C. elegans, these researchers fused the promoter for a PMK-1-dependent putative antimicrobial peptide with the gene encoding GFP and integrated the array into the C. elegans genome, thereby creating an in vivo sensor for the transcriptional activation of this gene. By conducting a forward genetic screen for C. elegans mutants that were both hypersusceptible to P. aeruginosa infection and exhibited diminished expression of this transcriptional reporter, they uncovered mutations in each of the four genes in the p38 MAP kinase cassette (TIR-1, NSY-1, SEK-1 and PMK-1) and also in ATF-7, a transcription factor orthologous to mammalian ATF2/ATF7, which did not previously have a described immune function in C. elegans. Subsequent characterization of ATF-7 in the C. elegans antibacterial immune response revealed that it functions as a repressor of p38 MAP kinase PMK-1-dependent genes in C. elegans when worms are feeding on E. coli. However, ATF-7 switches to become a transcriptional activator of immune response genes when it is directly phosphorylated by PMK-1 during P. aeruginosa infection. Interestingly, it seems that ATF-7 is not a positive regulator of resistance to E. faecalis, despite the fact that PMK-1 is required for normal defense against this Gram-positive pathogen. These data suggest that there are PMK-1-dependent signaling regulators that are downstream of PMK-1 and independent of ATF-7 that are differentially activated during E. faecalis infection. How one pathway coordinates such disparate outputs remains an open and very interesting question.
Evolutionarily ancient mechanisms of pathogen detection
A remarkable feature of C. elegans innate immunity that has emerged from a number of transcriptional profiling experiments is that the nematode is able to mount pathogen-specific immune responses. The genes induced by infection with P. aeruginosa [27], S. aureus [34••], Microbacterium nematophilum [35], and C. albicans [32••] are remarkably distinct. Moreover, we recently found that the nematode selectively represses the transcription of putative antibacterial immune effectors during infection with the pathogenic fungus C. albicans [32••], an observation that was supported by a separate study of two nematode fungal pathogens [36•]. Marsh et al. also reported that a single antimicrobial peptide required for normal defense against a fungal pathogen of the nematode acts as a susceptibility factor for a bacterial pathogen [37•]. Taken together, these data imply the existence of mechanisms that enable the nematode to distinguish between invading microbes to coordinate pathogen-specific defense responses.
Transcriptional profiling data have revealed that C. elegans induces the transcription of putative defense effectors following exposure to heat-killed, avirulent C. albicans and S. aureus [32••,34••]. These data suggest that surveillance for invading microorganisms in the nematode may be mediated, at least in part, through ‘pattern recognition,’ an ancient immune surveillance mechanism able to detect conserved microbial molecules [so-called microbial-associated or pathogen-associated molecular patterns (MAMPs/PAMPs)] [38]. However, no direct evidence that C. elegans can detect MAMPs/PAMPs has been published. Interestingly, pattern recognition may not play as important a role in the detection of P. aeruginosa infection. Heat-killed P. aeruginosa fail to elicit an immune response [34••] and the induction of a particular anti-pseudomonal defense gene was dependent on the pathogenic potential of the infecting bacterial strain [39••]. In addition, unpublished experiments from the Troemel and Ausubel laboratories suggest that C. elegans may monitor the host effects of bacterial toxins to trigger an immune response. However, the detailed mechanisms by which C. elegans monitors and responds to P. aeruginosa infection are still not known. They may involve context-dependent signals generated during the infection, which in other systems have been called ‘DAMPs’ [40–42] or ‘patterns-of-pathogenesis’ [43]. In mammals, molecules released from damaged tissues including DNA, uric acid, and ATP have been shown to activate innate immunity [40]. Although some details are beginning to emerge [44–46], the receptors and downstream regulators involved in the detection and response to DAMPs have not yet been identified. Interestingly, plants have evolved somewhat analogous mechanisms of immune defense that rely heavily on indirect recognition of pathogen invasion. In addition to surveying for MAMPs/PAMPs, plants utilize a family of conserved intracellular receptor structurally related to NOD-like receptors (NLRs) in mammals [47] to monitor the activities of pathogen-encoded molecules, so-called ‘effectors,’ and activate an immune response when host-derived target molecules are modified [48]. Evidence for such ‘effector-triggered immunity’ is also beginning to emerge in other metazoans, but not yet in C. elegans [49]. The genetic, genomic, and morphological features of C. elegans summarized above make the nematode a powerful model in which to further dissect conserved mechanisms of metazoan pathogen detection.
The unfolded protein response during bacterial infection
Several investigators have demonstrated a specific role for endoplasmic reticulum (ER) unfolded protein response (UPR) pathways in the intestine during infection of the nematode [50••,51••,52,53••]. Richardson et al. found that p38 MAP kinase PMK-1-mediated defenses require compensatory activation of the UPR by the X-box binding protein (XBP-1) to handle the accumulation of unfolded proteins in the ER, which occurs as the host mounts an immune response that is comprised primarily of secreted genes [51••]. In a separate study, these investigators also demonstrated a dynamic requirement for the UPR in the maintenance of cellular homeostasis [54]. The UPR in C. elegans is also required for normal defense against pore-forming toxins produced by pathogenic bacteria [52]. Interestingly, p38 MAP kinase PMK-1 and c-Jun N-Terminal Kinase (JNK)-like MAP kinase form a signaling network that regulates both the UPR and other cellular defenses following exposure to pore-forming toxins [53••]. In addition, Sun et al. found that inputs from the sensory nervous system in the nematode suppress innate immune responses, which they conclude occurs via the downregulation of the UPR in non-neuronal tissues [50••]. It also seems that these sensory neurons receive signals from pathogenic, but not heat-killed bacteria, which raises the intriguing hypothesis that they detect some pattern of pathogenesis to fine-tune the host immune response during infection. An interesting body of literature suggests that the nervous system also regulates a pathogen avoidance program that confers a survival advantage for nematodes during infection [55–57]. The precise roles that neuronal signaling pathways play in the regulation of immune signaling pathways, on the one hand, and pathogen avoidance on the other, have not been fully resolved [57–59]. A detailed discussion of the neural control of immunity and behavioral avoidance of pathogens is outside the scope of this review.
Parallel pathways and the evolution of innate immune defenses
The p38 MAP kinase pathway coordinates the basal and infection-induced regulation of immune effectors that are required for defense against most C. elegans pathogens, but it does not act alone (Figure 2). The transcription factor ZIP-2 controls an immune signaling pathway that acts independently of PMK-1 and is induced by only by virulent strains of P. aeruginosa [39••]. C. elegans FSHR-1 is a G-protein coupled receptor and homolog of the mammalian follicle-stimulating hormone receptor controls an immune signaling pathway in C. elegans that is also distinct from the PMK-1 pathway [60]. Likewise, the β-catenin homolog BAR-1 and the homeobox transcription factor EGL-5 are important for defense against S. aureus [34••,61]. A third pathway regulated by the FOXO transcription factor DAF-16 acts downstream of the insulin/insulin-like growth factor receptor DAF-2 to regulate longevity, immunity, and stress resistance [27,62–64]. A recent report presents data that the conserved transcription factor SKN-1, an ortholog of mammalian Nrf proteins, coordinates a transcriptional program that protects the host from its own reactive oxygen species, which are produced to fight bacterial infection of the nematode [65,66]. Using slightly different assay conditions, Shivers et al., however, did not find that SKN-1 was required for defense against P. aeruginosa [30••]. Each of these pathways acts in parallel to coordinate the elaboration of largely non-overlapping immune effectors. We hypothesize that defense pathways in C. elegans evolved in response to environmental threats, further dissection of which may yield clues about evolutionarily conserved immune mechanisms in higher order hosts.
Figure 2.
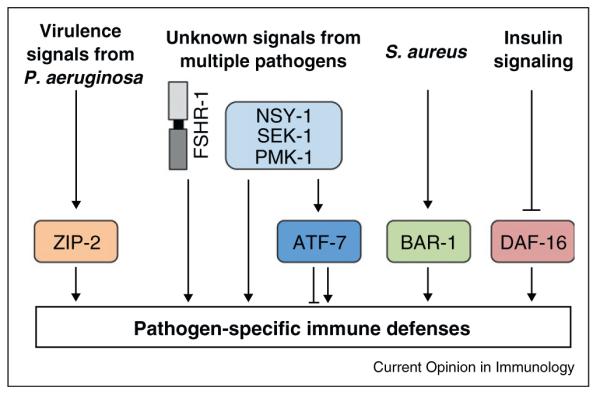
Multiple immune signaling pathways function in C. elegans epithelium. C. elegans coordinate pathogen-specific immune responses toward ingested pathogens through several signaling pathways that act in parallel. The transcriptional outputs from these signaling mediators are unique, but can be overlapping.
Novel anti-infective compounds identified using a C. elegans pathogenesis assay
One potentially interesting application of C. elegans pathogenesis assays involves their use in large-scale screens to identify novel antimicrobials [12]. Such assays are facilitated by the fact that 15–20 adult C. elegans animals fit comfortably in the wells of standard 384-well assay plates and can be lethally infected with a variety of microbial pathogens [1–13]. In the first relatively high throughput study of this kind, Moy et al. tested 37,214 compounds in a high-throughput assay and identified 119 small molecules that prolonged the lifespan of nematodes infected with the Gram-positive human bacterial pathogen E. faecalis, including a number that had no structural relationship to any known antimicrobials [67]. Interestingly, several of these small molecules cured nematodes at doses lower than required to inhibit bacterial growth. In contrast, traditional antibiotics, such as ciprofloxacin, ampicillin and vancomycin cured E. faecalis-infected nematodes only at doses several fold higher than the in vitro minimum inhibitory concentration (MIC) for the bacteria. These data raise the intriguing possibility that a subset of the small molecules identified in the C. elegans-based screen act either by inhibiting virulence factor production in the bacteria or by directly stimulating the host innate immune response. Work is currently underway to characterize the mechanism of action of these interesting compounds.
Conclusions
Dissection of the mechanisms of host defense and pathogen detection in C. elegans holds promise to elucidate the origins and fundamental principles of innate immunity, and may lead to important developments that broaden our understanding of such processes in higher order hosts.
Acknowledgements
We are grateful to Emily Troemel for providing us with the image used in Figure 1 and to our many colleagues that allowed us to cite their unpublished results. We thank Dennis Kim, Emily Troemel, Christine Kocks, and Lynda Stuart for their comments on the manuscript. The authors’ research has been supported by the Irvington Institute Fellowship Program of the Cancer Research Institute (to RPW) and by the following grants from the National Institutes of Health: K08 award AI081747 (to RPW), P01 award AI044220 (to FMA), R01 award AI064332 (to FMA), R01 award AI085581 (to FMA), R01 award AI076372 (to FMA), P01 award AI083214 (to FMA), and P30 award DK040561 (to FMA).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Shivers RP, Youngman MJ, Kim DH. Transcriptional responses to pathogens in Caenorhabditis elegans. Curr Opin Microbiol. 2008;11:251–256. doi: 10.1016/j.mib.2008.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulenburg H, Kurz CL, Ewbank JJ. Evolution of the innate immune system: the worm perspective. Immunol Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- 3.Partridge FA, Gravato-Nobre MJ, Hodgkin J. Signal transduction pathways that function in both development and innate immunity. Dev Dyn. 2010;239:1330–1336. doi: 10.1002/dvdy.22232. [DOI] [PubMed] [Google Scholar]
- 4.Millet ACM, Ewbank JJ. Immunity in Caenorhabditis elegans. Curr Opin Immunol. 2004;16:4–9. doi: 10.1016/j.coi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Gravato-Nobre MJ, Hodgkin J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell Microbiol. 2005;7:741–751. doi: 10.1111/j.1462-5822.2005.00523.x. [DOI] [PubMed] [Google Scholar]
- 6.Ewbank JJ. WormBook. The C. elegans Research Community; 2006. Signaling in the immune response. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irazoqui JE, Ausubel FM. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Caenorhabditis elegans as a model to study tissues involved in host immunity and microbial pathogenesis. Clin Exp Immunol. 2010;160:48–57. doi: 10.1111/j.1365-2249.2010.04122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D. Studying host–pathogen interactions and innate immunity in Caenorhabditis elegans. Dis Model Mech. 2008;1:205–208. doi: 10.1242/dmm.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 11.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2011;708:105–121. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 12.Pukkila-Worley R, Holson E, Wagner F, Mylonakis E. Antifungal drug discovery through the study of invertebrate model hosts. Curr Med Chem. 2009;16:1588–1595. doi: 10.2174/092986709788186237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 14 ••.Estes KA, Szumowski SC, Troemel ER. Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog. 2011;7:e1002227. doi: 10.1371/journal.ppat.1002227. This study demonstrates that Nematocida parisii, an intracellular eukaryotic pathogen of the nematode, causes rearrangement of the actin cytoskeleton within intestinal epithelial cells to allow spores to exit.
- 15.Troemel ER, Félix M-A, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:e309–e2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troemel ER. New models of microsporidiosis: infections in zebrafish, C. elegans, and honey bee. PLoS Pathog. 2011;7:e1001243. doi: 10.1371/journal.ppat.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 ••.Félix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. The authors identify two novel viral pathogens of Caenorhabditis species, which they isolated from wild-caught nematodes that had unusual intestinal epithelial cell morphologies.
- 18.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 19.Chuang C-F, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou J-F, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 21.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler K, Kurz CL, Cypowyj S, Couillault C, Pophillat M, Pujol N, Ewbank JJ. Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe. 2009;5:341–352. doi: 10.1016/j.chom.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Ren M, Feng H, Fu Y, Land M, Rubin CS. Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity. 2009;30:521–532. doi: 10.1016/j.immuni.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Kawli T, Wu C, Tan M-W. Systemic and cell intrinsic roles of Gqalpha signaling in the regulation of innate immunity, oxidative stress, and longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:13788–13793. doi: 10.1073/pnas.0914715107. The authors describe a role for a signaling module formed by the G protein alpha subunit (Gqα) and phospholipase Cβ (PLCβ) in the regulation of longevity, immunity and stress resistance.
- 26.Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe. 2009;6:321–330. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aballay A, Drenkard E, Hilbun LR, Ausubel FM. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol. 2003;13:47–52. doi: 10.1016/s0960-9822(02)01396-9. [DOI] [PubMed] [Google Scholar]
- 29 •.Bolz DD, Tenor JL, Aballay A. A conserved PMK-1/p38 MAPK is required in C. elegans tissue-specific immune response to Y. pestis infection. J Biol Chem. 2010;285:10832–10840. doi: 10.1074/jbc.M109.091629. The authors use a combination of genetic and biochemical techniques to show that the p38 MAP kinase PMK-1 is required for defense against the bacterial pathogen Y. pestis.
- 30 ••.Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, Kamanzi O, Matsumoto K, Hisamoto N, Kim DH. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000892. doi: 10.1371/journal.pgen.1000892. The authors identify ATF-7, a conserved transcription factor, and demonstrate that it functions downstream of the p38 MAP kinase PMK-1 to regulate the defense response toward the bacterial pathogen P. aeruginosa.
- 31.Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32 ••.Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011;7:e1002074. doi: 10.1371/journal.ppat.1002074. In this study, the authors use a transcriptional profiling approach to show that C. elegans mounts a pathogen-specific immune response toward the human fungal pathogen C. albicans, which involves the specific downregulation of antibacterial immune effectors.
- 33 ••.Youngman MJ, Rogers ZN, Kim DH. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002082. doi: 10.1371/journal.pgen.1002082. The authors show that a decline in the activity of the p38 MAP kinase PMK-1 underlies the increased susceptibility to bacterial infection that occurs in aging nematodes.
- 34 ••.Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. The authors demonstrate that S. aureus induces a robust immune response in C. elegans. They also show that the features of S. aureus infection are different from those caused by P. aeruginosa.
- 35.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36 •.Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, Ewbank JJ. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE. 2011;6:e19055. doi: 10.1371/journal.pone.0019055. The authors use new methods to conduct a comparative study of the transcriptional response to bacterial and fungal pathogens in C. elegans.
- 37 •.Marsh EK, van den Berg MCW, May RC. A two-gene balance regulates Salmonella typhimurium tolerance in the nematode Caenorhabditis elegans. PLoS ONE. 2011;6:e16839. doi: 10.1371/journal.pone.0016839. The authors demonstrate that C. elegans mutated for the antimicrobial peptide LYS-7 are more susceptible to infection with the human fungal pathogen Cryptococcus neoformans, but are resistant to the bacterial pathogen Salmonella Typhimurium.
- 38.Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 39 ••.Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. The authors use a reverse genetic screen to identify the transcription factor zip-2. They demonstrate that it is required for defense against P. aeruginosa infection in a manner that is independent of the p38 MAP kinase PMK-1.
- 40.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 42.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 43.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Yang J, Shi J, Gong Y-N, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 46.Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo Z-Q, Vance RE. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ting JP-Y, Williams KL. The CATERPILLER family: an ancient family of immune/apoptotic proteins. Clin Immunol. 2005;115:33–37. doi: 10.1016/j.clim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 49.Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere G, Ip WE, Fracchia S, Hennessy E, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–549. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50 ••.Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 2011;332:729–732. doi: 10.1126/science.1203411. This study shows that sensory neurons suppress innate immune responses by negatively regulating a noncanonical unfolded protein response pathway in C. elegans.
- 51 ••.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. The authors demonstrate that p38 MAP kinase PMK-1-mediated defenses require compensatory activation of the unfolded protein response by the X-box binding protein XBP-1.
- 52.Bischof LJ, Kao C-Y, Los FCO, Gonzalez MR, Shen Z, Briggs SP, van der Goot FG, Aroian RV. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog. 2008;4:e1000176. doi: 10.1371/journal.ppat.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53 ••.Kao C-Y, Los FCO, Huffman DL, Wachi S, Kloft N, Husmann M, Karabrahimi V, Schwartz J-L, Bellier A, Ha C, et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011;7:e1001314. doi: 10.1371/journal.ppat.1001314. The authors perform a whole genome RNA interference screen for genes that when knocked down, cause C. elegans to be hypersusceptible to a bacterial pore-forming toxin. They demonstrate that two pathways mediated by p38 MAP kinase PMK-1 and JNK MAP kinase are important for defense against this toxin.
- 54.Richardson CE, Kinkel S, Kim DH. Physiological IRE-1-XBP-1 and PEK-1 Signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. 2011;7:e1002391. doi: 10.1371/journal.pgen.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy KC, Hunter RC, Bhatla N, Newman DK, Kim DH. Caenorhabditis elegans NPR-1-mediated behaviors are suppressed in the presence of mucoid bacteria. Proc Natl Acad Sci U S A. 2011;108:12887–12892. doi: 10.1073/pnas.1108265108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anyanful A, Easley KA, Benian GM, Kalman D. Conditioning protects C. elegans from lethal effects of enteropathogenic E. coli by activating genes that regulate lifespan and innate immunity. Cell Host Microbe. 2009;5:450–462. doi: 10.1016/j.chom.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aballay A. Neural regulation of immunity: role of NPR-1 in pathogen avoidance and regulation of innate immunity. Cell Cycle. 2009;8:966–969. doi: 10.4161/cc.8.7.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Powell J, Kim D, Ausubel F. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci U S A. 2009;106:2782–2787. doi: 10.1073/pnas.0813048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial–pathogen interactions. Proc Natl Acad Sci U S A. 2008;105:17469–17474. doi: 10.1073/pnas.0809527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh V, Aballay A. Regulation of DAF-16-mediated innate immunity in Caenorhabditis elegans. J Biol Chem. 2009;284:35580–35587. doi: 10.1074/jbc.M109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 64.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 65 ••.van der Hoeven R, McCallum KC, Cruz CR, Garsin DA. Ce-Duox1/BLI-3 Generated Reactive Oxygen Species Trigger Protective SKN-1 Activity via p38 MAPK Signaling during Infection in C. elegans. PLoS Pathog. 2011;7:e1002453. doi: 10.1371/journal.ppat.1002453. The authors demonstrate that the conserved transcription factor SKN-1 protects C. elegans from reactive oxygen species that are produced by the host during infection with P. aeruginosa.
- 66.Chávez V, Mohri-Shiomi A, Garsin DA. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun. 2009;77:4983–4989. doi: 10.1128/IAI.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol. 2009;4:527–533. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]


