Abstract
Hox proteins are a prominent class of transcription factors that specify cell and tissue identities in animal embryos. In sharp contrast to tissue-specifically expressed transcription factors, which coordinate regulatory pathways leading to the differentiation of a selected tissue, Hox proteins are active in many different cell types but are nonetheless able to differentially regulate gene expression in a context-dependent manner. This particular feature makes Hox proteins ideal candidates for elucidating the mechanisms employed by transcription factors to achieve tissue-specific functions in multi-cellular organisms. Here we discuss how the recent genome-wide identification and characterization of Hox cis-regulatory elements has provided insight concerning the molecular mechanisms underlying the high spatiotemporal specificity of Hox proteins. In particular, it was shown that Hox transcriptional outputs depend on the cell-type specific interplay of the different Hox proteins with co-regulatory factors as well as with epigenetic modifiers. Based on these observations it becomes clear that cell-type specific approaches are required for dissecting the tissue-specific Hox regulatory code. Identification and comparative analysis of Hox cis-regulatory elements driving target gene expression in different cell types in combination with analyses on how cofactors, epigenetic modifiers and protein-protein interactions mediate context-dependent Hox function will elucidate the mechanistic basis of tissue-specific gene regulation.
Keywords: Drosophila, Hox, homeodomain, tissue-specific, transcription, gene regulation, genome-wide, cofactors
Hox Proteins: Broadly Expressed Yet Highly Cell-Type Specific Regulators of Gene Expression
Hox genes encode evolutionarily conserved and essential transcription factors (TFs) expressed in precise domains along the anterior-posterior axis of animal embryos where they regulate segment morphogenesis.1 Besides their role in patterning the early embryo, Hox TFs control organogenesis in later developmental stages2 and are critical for the maintenance of tissue homeostasis in adult organisms.3 Hox proteins regulate cellular and tissue identities in a cell-autonomous manner by binding to DNA sequences in Hox response elements (HREs), thereby activating or repressing downstream targets.4,5 Despite their rather broad expression Hox TFs execute their regulatory function in a highly context-dependent manner.6-8 However, the molecular mechanisms underlying the spatiotemporal specificity of Hox TFs have remained a long-standing question.
Hox TFs are characterized by the presence of a homeodomain, a 60 amino acids DNA-binding domain. The Hox homeodomain recognizes DNA consensus sequences containing an -ATTA- core,9,10 thus Hox TFs show overlapping in vitro binding behaviors.11,12 Nonetheless, different Hox proteins exhibit very diverse in vivo binding preferences13 and execute distinct regulatory functions,5 showing that the loose DNA recognition properties of the homeodomain are not sufficient to confer specificity to the Hox proteins.12,14,15
Combinatorial assembly of TF and transcriptional co-regulator complexes on shared cis-regulatory elements is a widespread mechanism driving context-dependent transcriptional responses. Along these lines, it has been proposed that HREs integrate multiple inputs and that the high in vivo specificity of Hox proteins is achieved in cooperation with cofactors and collaborators.13 Well-established examples of Hox cofactors include members of the PBC and Meis protein families, i.e., Extradenticle (Exd) and Homothorax (Hth) in Drosophila.16 These cofactors improve Hox TF selectivity and permit the differential regulation of target genes through cooperative complex formation.13,17 As demonstrated by a recent study, complex formation between Drosophila Hox TFs and their cofactor Exd has profound effects on the DNA recognition properties of Hox proteins and in particular it reveals novel DNA binding specificities.18 Strikingly, the Exd-Hox complex derived specificity is unique for each one of the closely related Hox TFs tested in the study,18 highlighting the crucial role of protein-protein interactions in fine-tuning the in vivo selectivity of different Hox TFs. On the other hand, several TFs coordinate the regulatory activity of Hox proteins without the requirement of complex formation by acting as collaborators.19 Examples include the collaboration between Ubx and Smads for repressing spalt in the Drosophila haltere,19 the collaboration of Ubx and Abd-A with Sloppy paired for repressing Distal-less in the Drosophila abdomen,20 and the regulation of reaper expression in the anterior part of the maxillary segment in Drosophila embryos by the combinatorial activity of the Hox TF Deformed (Dfd) and eight other TFs.21
Combinatorial Input on Hox-Regulated Enhancers Defines the Cell-Type Specificity of Hox Proteins
As suggested by studies on selected developmental enhancers,19-21 the integration of a combinatorial transcriptional input on HREs is most likely a frequently employed mechanism for regulating Hox TF activity in vivo. Nonetheless, the information obtained by dissecting a small number of HREs is not sufficient for globally unravelling the regulatory code underlying the spatio-temporal precision of Hox TFs. An in depth analysis of Hox cis-regulatory elements on the genome-wide level is undoubtedly essential in order to understand the mechanistic basis of how Hox TFs regulate target gene expression. In recent years, the wide-spread use of genome-wide chromatin-profiling methods such as chromatin immunoprecipitation (ChIP) followed by microarray analysis (ChIP-on-Chip) or coupled to massively parallel sequencing (ChIP-Seq) has resulted in mapping of the in vivo binding profiles of several TFs, among them a number of Hox proteins.22-26 These studies allowed for the first time the genome-wide identification and characterization of Hox-bound cis-regulatory elements.
Analysis of the newly identified HREs revealed several features that appear to be crucial for the cell-type specific regulatory functions of Hox proteins in vivo. In our recent study,22 genomic regions bound by the Hox TF Dfd in stage 9–12 Drosophila embryos were identified by ChIP-Seq. A subsequent analysis of the architectural features of a number of Dfd regulated enhancers led to the conclusion that motif composition and short distance spacing of TF binding sites is critical for transcriptional regulation by Dfd in vivo. Importantly, the analysis of gene classes associated with enhancers displaying different motif compositions and motif pair associations showed that architectural features of Dfd CRMs are sufficient to predict target gene function and expression patterns with high accuracy.22 Furthermore, a comparison of regulatory regions bound by Dfd22 and Ubx,27 two Hox proteins specifying the morphology of different segments in the Drosophila embryo, revealed that in contrast to their loose DNA binding specificity in vitro Dfd and Ubx bind to non-overlapping genomic regions in vivo. Analysis of Dfd and Ubx HREs indicated that regulatory specificity/selectivity is encoded by distinct combinations of co-occurring TF binding motifs, highlighting once again the essential role of co-regulatory TFs. Along the same lines, computational analysis of genomic regions bound by Ubx in Drosophila haltere and T3 leg imaginal discs revealed the presence of distinct TF motifs in each data set.25 Interestingly these TFs are locally expressed in either of the two analyzed tissues and therefore they are expected to function as Ubx co-regulators in the respective tissues.25 Nevertheless, this hypothesis remains to be experimentally addressed. In yet another genome-wide study, the in silico analysis of Ubx bound genomic regions in Drosophila wing imaginal discs26 showed that even though no specific Ubx consensus motif was enriched in Ubx HREs, binding motifs for other TFs were over-represented in these sequences, suggesting that these TFs assist Ubx in identifying target HREs and/or in regulating target gene expression.26 Collectively these studies point out that the restricted spatiotemporal availability of Hox co-regulators dictates the highly precise regulatory function of Hox TFs.
Identification and Analysis of HREs from Isolated Cell-Types is Essential for Unraveling the Hox Regulatory Code
As increasing evidence indicates that the cellular context is an essential determinant of the regulatory output of Hox TFs, it is becoming clear that cell-type specific approaches are required in order to identify all relevant aspects of the interplay of Hox TFs with cis-regulatory elements and co-regulatory factors. Genome-wide data obtained from whole embryos or whole tissues (i.e., imaginal discs) represent an averaged signal originating from the mixture of heterogeneous cell types. In such experimental setups, TF-DNA interactions taking place in less frequently encountered cell-types will most likely not be detected, as they will be diluted by signals arising from abundant cell types. Recently, different methods have been presented for isolating pure populations of nuclei from selected cell-types using organisms ranging from plants to animals.28-30 These methods include affinity purification of nuclei tagged in selected cell types28,30 and fluorescent activated cell sorting of nuclei that have been fluorescently labeled for a nuclear protein expressed in the cell type of interest.29 Both approaches were shown to be suitable for genome-wide experiments aiming at identifying TF-bound genomic regions, obtaining gene expression profiles and analyzing epigenetic modifications. The combination of these nuclear sorting approaches with techniques successfully employed to amplify ChIP signals from limited amounts of starting material31 will permit cell-type specific genome-wide analyses of Hox-DNA interactions even using less-abundant cell-types. Using isolated nuclei and genome-wide methods to identify cis-regulatory enhancer modules bound by Hox TFs is a promising approach for identifying the mechanisms that confer specificity to Hox proteins in the different cellular contexts.
As indicated by our comparative analysis,22 the genomic regions bound by different Hox TFs show little if any overlap, in sharp contrast to their highly similar in vitro binding properties. Nevertheless, the two data sets used for this analysis were generated using different methods, namely ChIP-Seq for Dfd-bound regions and ChIP-Chip for Ubx-bound regions and embryos of not entirely identical developmental stages, 4–9 h old and 3–8 h old embryos respectively.22,27 A comparison of cis-regulatory elements bound by different Hox proteins in the same cell-type and identified under comparable conditions will provide more detailed insight concerning how the involvement of different co-regulatory TFs properties affects Hox cell-type and segment specific activity.
Interplay of Hox TFs with Epigenetic Modifiers
Epigenetic characteristics including chromatin structure and histone modifications on enhancer modules strongly correlate with spatiotemporal enhancer activity, as shown by genome-wide experiments using cell lines of variable origins and isolated Drosophila embryonic mesoderm cells.29,32,33 Assuming that epigenetic changes resulting in an open chromatin conformation generally potentiate accessibility of DNA to TFs, the observed tissue-specific epigenetic marks are interpreted as a means to control the selective occupancy of TFs in different cellular contexts. As shown in our study, epigenetic regulators bind to Hox enhancers on a genome wide level, suggesting that their interaction with Hox TFs is crucial for the regulation of Hox target gene expression.22 For instance, the Hox protein Dfd shares a substantial number of target genomic regions with the transcriptional coactivator dCBP.22 dCBP bears intrinsic acetyltransferase activity and induces histone acetylation and subsequent open chromatin conformation in the vicinity of its binding,34 thus active histone marks i.e., H3K27ac were expected to be enriched at Dfd/dCBP bound genomic regions. When we analyzed the genome-wide epigenetic data generated by the modENCODE consortium using Drosophila embryos,27 we were not able to detect enrichment for acetylated histone variants. Nevertheless, as the combinatorial binding of Dfd and dCBP takes place only in a small embryonic segment where Dfd is expressed, it is most probably impossible to extract information concerning the epigenetic marks at these loci from data generated using whole embryos. A cell-type specific analysis of histone modifications is required for clarifying whether dCBP-mediated histone acetylation is encountered at Dfd/dCBP bound loci. Generally, the identification of epigenetic marks at genomic regions bound by Hox TFs in different cell-types will determine to what extent chromatin accessibility determines the binding of Hox TFs to their target HREs in the different cellular contexts.
The most obvious explanation for the joint binding of epigenetic regulators and Hox proteins at overlapping genomic regions22 is that the presence of these histone-modifying enzymes is a prerequisite for inducing open chromatin conformation required for the binding of Hox TFs to DNA. But is this the only plausible explanation? Interestingly, in agreement with the reported activity of histone acetyltransferases to acetylate TFs in addition to histones,35 we showed that dCBP acetylates Glial cells missing (Gcm), a co-regulatory TF of Dfd and that the interaction of Gcm and CBP is required for gene expression driven by a given Dfd/Gcm regulated HRE.22 TF acetylation is a commonly used mechanism for modifying/enhancing transcriptional regulatory activity as shown for a number of TFs among which Gcma,36 Myocardin,37 EWS-FLI138 and GATA-1.39 Therefore the interplay of Hox TFs and epigenetic modifiers might mediate the local recruitment of such enzymes in order to modulate the transcriptional activity of Hox co-regulators or even of the Hox TFs themselves. Further experiments are definitely required in order to validate this hypothesis and to address whether post-translational modifications of Hox co-regulators mediated by epigenetic regulators are a commonly observed theme and not restricted to the example of Gcm in the Drosophila embryo.
In addition to the traditional view that epigenetic patterning of enhancers precedes TF binding, a more complex model was proposed recently for the establishment of lineage-specific epigenetic marks (reviewed in40). As shown by a number of independent studies, TFs as well as transcriptional co-regulators are often involved in establishing cell-type specific epigenetic signatures by binding at enhancer modules and mediating the recruitment of chromatin modifying enzymes.40 It would be worthwhile to investigate whether Hox proteins and/or their co-regulatory TFs employ this strategy in order to initiate cell-type specific transcriptional responses.
Protein-Protein Interactions as Determinants of Hox TF Regulatory Specificity
The assembly of transcriptional regulatory complexes on target enhancer elements is a key aspect of transcriptional regulation. After the binding of the first TFs at DNA sequences, further recruitment of regulatory proteins determines the regulatory output, namely activation or repression of transcription and maintenance of the respective transcriptional state. The ability of Hox proteins to bind frequently encountered DNA sequences offers them the possibility to interact with a vast number of genomic regions. Interestingly, even though different Hox proteins are highly similar with respect to their homeodomains, they display pronounced variability concerning other protein domains and amino acid sequences.13 Thus, amino acid sequences and protein structures outside the homeodomain very likely mediate protein-protein interactions that subsequently refine the binding of Hox TFs to target HREs. Concerning the extensively studied Hox/PBC heterodimers, a hexapeptide motif upstream of the homeodomain has long been described to mediate the interaction of the two proteins (summarized in13). Interestingly, a recent study demonstrated that the picture is probably more complicated, as the hexapeptide seems to be at a certain extent dispensable for the in vivo Hox/PBC interaction, while additional cofactors as well as paralog-specific amino acid sequences are implicated in the protein-protein interaction, resulting in high functional plasticity.41 Considering the indispensable role of co-regulators in regulating Hox transcriptional activity, identification of Hox protein domains mediating intermolecular interactions with co-regulatory TFs will improve our understanding of how Hox proteins achieve functional specificity. Importantly, as it has been recently suggested by structure-function studies of different Hox proteins, the role of each protein domain should not be considered individually but rather as one of the components of a multifunctional unit that collectively defines Hox-DNA interactions and regulatory activity.42
Hox Proteins: Ideal Models for Revealing the Mechanisms of Cell-Type Specific Transcriptional Regulation
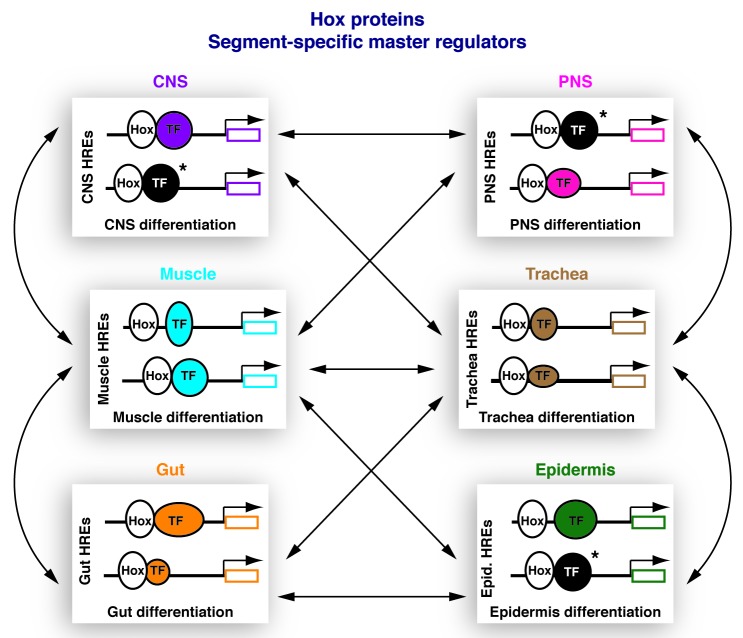
Even though cell-type specific transcriptional regulation of gene expression is a fundamental aspect of multi-cellularity, the molecular mechanisms conferring tissue specificity to TFs remain largely unknown. In many cases, the expression of TFs coordinating the differentiation of a given tissue is restricted to the population of precursor cells that will eventually lead to the formation of this tissue. For example, the TF Twist, which drives somatic muscle specification, is only expressed in mesoderm and muscle precursor cells in the Drosophila embryo.43 In sharp contrast to TFs that are tissue-specifically expressed, Hox TFs are active in large embryonic segments containing multiple cell types and giving rise to several different tissues and body structures. This particular feature of Hox TFs makes them ideal candidates for understanding the mechanistic basis of cell- and tissue-specificity. A comparative analysis of the mechanisms employed by a Hox TF for regulating target gene expression in different cell types will allow the identification of general regulatory mechanisms used in more than one cell types and will additionally reveal cell-type specific mechanisms driving target gene regulation solely in a defined cellular context (Fig. 1). As discussed above, these molecular mechanisms may include recruitment of cell-type specific transcriptional co-regulators, interaction with distinct epigenetic modifiers and discrepancy in DNA-binding properties emerging from differential protein domain usage and protein-protein interactions. Furthermore, a comparative analysis of Hox regulatory networks in different cell-types within an embryonic segment will offer valuable insight concerning how gene expression is coordinated in different tissue types in order to ensure the proper implementation of complex biological programs.
Figure 1. Model: Hox transcription factors function as segment-specific master regulators executing cell-type specific functions. Central nervous system (CNS), peripheral nervous system (PNS), muscle, trachea, gut and epidermis are shown as selected examples of tissues where Hox proteins differentially regulate target gene expression. In the different tissue-types, Hox proteins bind to HREs and promote expression of tissue-specific target genes. Spatially restricted TFs (color-coded depending on the different tissues) fine-tune the regulatory activities of Hox proteins in the different cellular contexts. Co-regulatory TFs active in more than one cell types are shown in black and are marked by an asterisk. Tissue-specific epigenetic modifications implicated in transcriptional regulation are omitted for simplicity reasons. Arrows indicate the interactions between transcriptional regulatory networks active in different tissues, resulting in the coordination of complex processes, necessary for embryonic development and tissue homeostasis.
Acknowledgments
The work discussed in this article was funded by the Deutsche Forschungsgemeinschaft (DFG: LO 844/3-2).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/22939
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Hombría JC, Lovegrove B. Beyond homeosis--HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–76. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 3.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–76. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 4.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- 5.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 6.Akam M. Hox genes: from master genes to micromanagers. Curr Biol. 1998;8:R676–8. doi: 10.1016/S0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- 7.Lohmann I, McGinnis W. Hox Genes: it’s all a matter of context. Curr Biol. 2002;12:R514–6. doi: 10.1016/S0960-9822(02)01025-4. [DOI] [PubMed] [Google Scholar]
- 8.Tour E, Hittinger CT, McGinnis W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development. 2005;132:5271–81. doi: 10.1242/dev.02138. [DOI] [PubMed] [Google Scholar]
- 9.McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308:428–33. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- 10.Scott MP, Tamkun JW, Hartzell GW., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 11.Ekker SC, von Kessler DP, Beachy PA. Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J. 1992;11:4059–72. doi: 10.1002/j.1460-2075.1992.tb05499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 1994;13:3551–60. doi: 10.1002/j.1460-2075.1994.tb06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desplan C, Theis J, O’Farrell PH. The sequence specificity of homeodomain-DNA interaction. Cell. 1988;54:1081–90. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekker SC, Young KE, von Kessler DP, Beachy PA. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 1991;10:1179–86. doi: 10.1002/j.1460-2075.1991.tb08058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–9. doi: 10.1016/S0959-437X(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 18.Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–82. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh CM, Carroll SB. Collaboration between Smads and a Hox protein in target gene repression. Development. 2007;134:3585–92. doi: 10.1242/dev.009522. [DOI] [PubMed] [Google Scholar]
- 20.Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–9. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- 21.Stöbe P, Stein MA, Habring-Müller A, Bezdan D, Fuchs AL, Hueber SD, et al. Multifactorial regulation of a hox target gene. PLoS Genet. 2009;5:e1000412. doi: 10.1371/journal.pgen.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorge S, Ha N, Polychronidou M, Friedrich J, Bezdan D, Kaspar P, et al. The cis-regulatory code of Hox function in Drosophila. EMBO J. 2012;31:3323–33. doi: 10.1038/emboj.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–98. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choo SW, White R, Russell S. Genome-wide analysis of the binding of the Hox protein Ultrabithorax and the Hox cofactor Homothorax in Drosophila. PLoS One. 2011;6:e14778. doi: 10.1371/journal.pone.0014778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slattery M, Ma L, Négre N, White KP, Mann RS. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS One. 2011;6:e14686. doi: 10.1371/journal.pone.0014686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal P, Habib F, Yelagandula R, Shashidhara LS. Genome-level identification of targets of Hox protein Ultrabithorax in Drosophila: novel mechanisms for target selection. Sci Rep. 2011;1:205. doi: 10.1038/srep00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–77. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–56. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 30.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shankaranarayanan P, Mendoza-Parra MA, Walia M, Wang L, Li N, Trindade LM, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods. 2011;8:565–7. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 32.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 33.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–3. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 35.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- 36.Chang CW, Chuang HC, Yu C, Yao TP, Chen H. Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol Cell Biol. 2005;25:8401–14. doi: 10.1128/MCB.25.19.8401-8414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao D, Wang C, Tang R, Chen H, Zhang Z, Tatsuguchi M, et al. Acetylation of myocardin is required for the activation of cardiac and smooth muscle genes. J Biol Chem. 2012;287:38495–504. doi: 10.1074/jbc.M112.353649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlottmann S, Erkizan HV, Barber-Rotenberg JS, Knights C, Cheema A, Uren A, et al. Acetylation Increases EWS-FLI1 DNA Binding and Transcriptional Activity. Front Oncol. 2012;2:107. doi: 10.3389/fonc.2012.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–8. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–30. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudry B, Remacle S, Delfini MC, Rezsohazy R, Graba Y, Merabet S. Hox proteins display a common and ancestral ability to diversify their interaction mode with the PBC class cofactors. PLoS Biol. 2012;10:e1001351. doi: 10.1371/journal.pbio.1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merabet S, Sambrani N, Pradel J, Graba Y. Regulation of Hox activity: insights from protein motifs. Adv Exp Med Biol. 2010;689:3–16. doi: 10.1007/978-1-4419-6673-5_1. [DOI] [PubMed] [Google Scholar]
- 43.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–4. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]