Abstract
Previous studies addressing the relationship between gene regulation and inbreeding depression did not allow for discerning the changes that alleviate the depression from those that generate it. We directly addressed this question by analyzing changes in gene expression, using Affymetrix 2.0 arrays in Drosophila melanogaster inbred sublines differing in their magnitudes of inbreeding depression relative to the expression in an outbred control. The total number of arrays analyzed was 27, with 9,133 probe sets showing a significant signal of expression. We found that for those genes differentially expressed between inbred and outbred sublines, most of them showed a pattern of expression consistent with a protective role against inbreeding effects. The observed increase in depression was presumably related to an inability of the genome to do the appropriate expression adjustments. Expression changes detected in our study showed a clear specificity of RNA-splicing and energy derivation functions. Thus, it appears that most of the observed changes in gene expression associated with inbreeding may occur predominantly to alleviate inbreeding depression, i.e., as a protection against the effects of inbreeding.
Keywords: Microarrays, inbreeding depression, gene regulation, transcription
Genetic models of inbreeding have been traditionally focused on inbreeding depression, the most important phenotypic consequence of inbreeding, as well as on the effects and population dynamics of genes that would be involved in its causation.1 In recent times, however, the advent of large-scale platforms to monitor gene expression across the whole genome has made it possible to have a more complete and detailed view of the transcriptomics of inbreeding. The analysis of the changes in expression of thousands of genes may shed light on different possible patterns of transcriptomic responses to the genomic stress originated by inbreeding. The expression changes observed in some genes may have a direct or indirect role in the causation of inbreeding depression for fitness. However, expression changes in other genes might be the reflection of transcriptomic responses directed to compensate or alleviate the negative consequences of inbreeding.2-4 We made a gene expression analysis of inbreeding in Drosophila melanogaster males using the Affymetrix Drosophila Genechip Array 2.0 and aimed to distinguish the changes that alleviate the depression from those that generate it.5 We started with four pairs (lines) of individuals and founded 55 full-sib mating inbred sublines with each. In generation eight, the sublines showed a great variation in pupa productivity (and thus inbreeding depression), despite having the same genealogical inbreeding coefficient (around 0.7). We then took the three most and the three least depressed sublines in each line on the basis of their pupa productivity, and measured their gene expression along with that in outbred samples from the same base population. Thus we could compare the magnitude of gene expression for samples of outbred (control) flies and for samples of highly inbred flies differing in their level of inbreeding depression for fitness. This made it possible to simultaneously measure inbreeding-related differences (by comparing all inbred samples with outbred controls) and depression-related differences (by comparing the most with the least depressed inbred sublines).
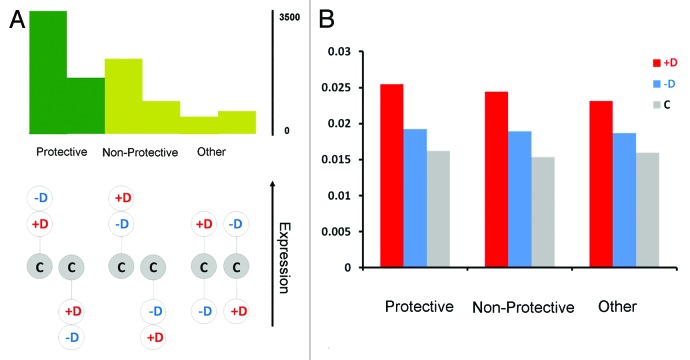
The outcome of the comparisons allowed for the identification of two basic patterns of expression. First, a pattern of transcripts was determined, with intermediate expression for the least depressed samples (Fig. 1A, mid panel). This pattern could in principle correspond to expression changes contributing to depression, as changes were larger in the most depressed samples than in the least depressed ones. Second, a pattern of transcripts in which the most depressed samples, rather than the least depressed ones, were intermediate, i.e., the sublines making the largest changes from outbred controls suffered the least depression, as would be expected in the case of functional adjustments against depression (Fig. 1A, left panel). Our basic finding was that most transcripts (62%) showed this second, “protective” pattern, both for upregulated and downregulated genes, and moreover, that the proportion of protective changes increased for transcripts showing stronger inbreeding or depression effects. The remaining third expression pattern, with the outbred controls in the middle (Fig. 1A, right panel), was represented by a minority (12%) of genes. In summary, changes in expression were consistent with Drosophila having genes or gene networks that are activated in the presence of inbreeding and deliver large changes in transcription across the genome. However, the greatest changes in inbreeding depression did not seem to be generally associated with the greatest changes in gene expression. On the contrary, for most transcripts the largest changes in expression corresponded to the least depressed sublines, consistent with a role of expression changes directed toward a protection against inbreeding effects.
Figure 1. (A) Patterns of expression observed for the 9133 probes analyzed in the experiment. In the protective pattern, the least depressed sublines (–D) have a more extreme expression than the most depressed sublines (+D) relative to the outbred control expression (C). In the non-protective pattern, the most depressed sublines have more extreme expression than the least depressed sublines. The remaining pattern (other) refers to probes where the outbred expression is intermediate between the two inbred ones. The histograms represent the number of probes showing each particular expression pattern. (B) Coefficient of variation of expression among sublines for each of the three patterns of expression.
Two observations support the existence of gene regulation networks responding to inbreeding. First, genes showing protective expression patterns had more defined functional annotations, with higher levels of enrichment in particular categories than genes expressed in the other patterns. The list of protectively upregulated genes was significantly enriched in transcription, RNA splicing and protein folding functions while genes downregulated included respiration and energy derivation terms, which suggests measures to save energy. Second, the expression levels tended to be less variable in the least than in the most depressed sublines (Fig. 1B). The higher variation in expression between the most depressed sublines suggests an erratic behavior resulting from defective regulation of gene networks, a potential cause of inbreeding depression.6,7 This erratic behavior was consistent with defects in transcription occurring at random and being thus different in different lines and sublines, as would be the case if the depression was caused by the fixation of rare deleterious recessive genes, as considered by current population genetic models. The more regular and stable changes in the least depressed sublines were consistent with functional responses aimed to alleviate this depression.
Some of the genes showing the greatest upregulation in the least depressed sublines already had well-known cytoprotective functions. This was the case of Sec61γ8 and that of the Hsp70-related gene Dmel\CG2918.9 Hps70 is a family of heat-shock proteins part of the cell’s machinery for protein folding that play an important role in the protection of cells against stress. However, it is not clear if the observed large-scale change in transcription was a specific response to inbreeding or a general response to situations of poor body condition or decreased fitness, either due to inbreeding or any other stress source. It is difficult to evaluate a priori the likelihood of these two alternatives, because the precise biological mechanisms causing inbreeding depression are not completely understood.10,11 It has recently been proposed that DNA methylation could be involved12 but our results do not allow us to address this matter, due to present limitations in the details of gene functional annotations. A Flybase search for the ontology term “DNA methylation” found only five genes described in Drosophila melanogaster [CG34356, Ipod, Mt2, CG14478 and Su(var)3–9 ], of which only the latter two were expressed in our experiment and only the last one showed an (upregulated) protective-consistent response. It could be argued that an evolved specific pathway to alleviate inbreeding effects would be more likely in populations experiencing frequent demographic bottlenecks and facing high levels of inbreeding. In contrast, in species with large effective population sizes, such as Drosophila melanogaster, it would be more reasonable to expect a general transcriptomic response to poor body condition. However, in that case it is puzzling first, that the lists of genes with protective patterns found in our experiment were not particularly enriched in stress-related functional terms; and second, the limited overlap between our list of protectively-behaving genes and those of genes described as responding to stress in this species. When we examined the genes under the Gene Ontology (GO) term “response to stress” in Flybase, we found (Fig. 2) that the proportion of protective expressions in our experiment was even slightly lower than in the whole list of expressed genes. The situation was similar for other broad categories often considered as functionally related to response to stress, as “immune response” and “aging.”11,13 However, other terms such as “response to oxidative stress,” “cellular response to stress” and “cellular response to starvation” included more protective profiles, in proportion, than the general list.
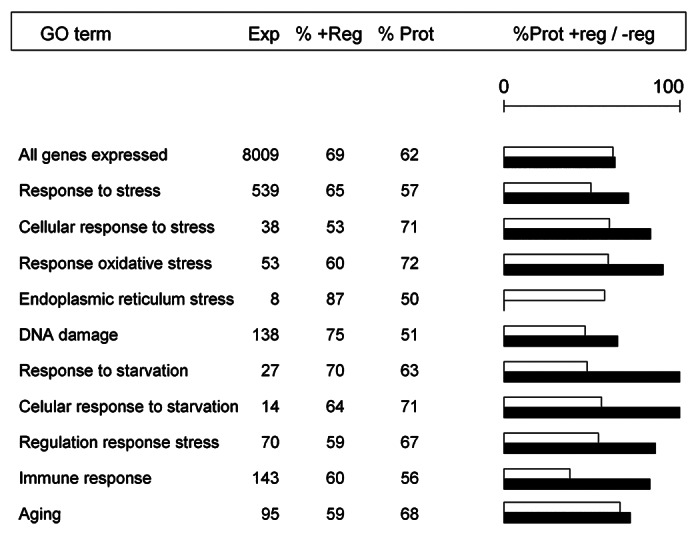
Figure 2. Number of genes corresponding to different Gene Ontology (GO) terms. Exp, number of genes with probes expressed in the experiment; %+Reg, proportion of these genes upregulated in the inbred lines; %Prot, proportion of these genes showing protective-consistent expression patterns. White and black bars represent the proportions of protective patterns among the up- and the downregulated genes in the term.
A more detailed analysis of these results provides an intriguing result: the increased frequency of protective profiles relative to that observed in the general gene list occurred almost exclusively in the downregulated genes (with the exception of the term “Endoplasmic reticulum stress,” although this class consisted of only 8 expressed genes). Thus, many genes related with stress responses were, somewhat unexpectedly, downregulated in the least depressed sublines. One possible interpretation is that genes in these GO terms really tend to be involved in responses against the sudden exposure to stress sources. However, changes in these genes could be counterproductive if the stress source is maintained for a long-term, as would be the case in a situation of chronic stress such as that likely caused by inbreeding depression. The transcription analyses of Sørensen, et al.,14 using Drosophila melanogaster lines selected to withstand different kinds of stressful conditions, resulted in lists of differentially expressed genes that did not coincide with our lists of protective-like genes more than expected by chance. This can also be consistent with the above interpretation, because the stressful stimuli used in that experiment tended to be acute rather than chronic. In addition, not all transcriptomic responses to stress need to be protective. For example, Girardot, et al.15 found a lack of correlation between the direction of genes’ transcriptional responses to oxidative stress and the effects of their mutations on resistance to that stress, which suggests the coexistence of protective and deleterious changes in gene expression.
In any case, the interpretation of changes in the expression of individual genes in terms of functional descriptions as broad as “stress” may not be realistic. The effects of genes on complex phenotypic traits could heavily depend on physiological context, or, as suggested by Sarup, et al.,16 on genetic context. In a review of published transcription analyses of inbreeding in Drosophila populations, Sarup, et al. found that the lists of genes responding to inbreeding had little overlap when they corresponded to populations with different genetic backgrounds, whereas this overlap was considerable when the populations` backgrounds were similar. This supports the view that the regulation of transcription occurs in closely integrated networks of genes17 and that considering genetic backgrounds is required to understand the behavior of most genes.18 Several studies found that the quantitative genetics of transcription is very complex, involving transgressive segregation, epistasis, and evolved regulatory strategies to limit or to exploit gene expression noise.6,19 In addition, and as illustrated above by the limited number of genes ascribed at present to the GO category “DNA methylation,” the enterprise of gene annotation is far from finished. Even in this model species, there are many genes without annotation, and many ontology terms without genes. Moreover, genes included in the same term can influence the same biological process in opposite directions, or it could even be too simplistic to consider that genes affect a given character always in the same direction.
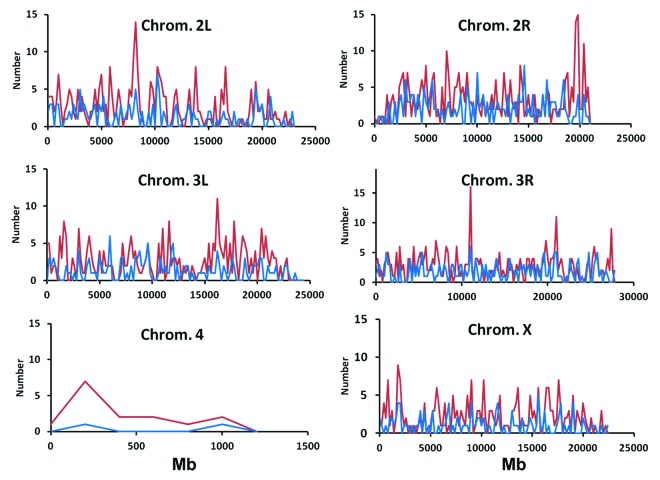
Our results change the “detrimental paradigm” of inbreeding depression, where expression changes are usually interpreted as fitness damaging. We suggest that many changes in gene expression, widespread throughout all chromosomes of the Drosophila genome (Fig. 3), are in fact compensatory adjustments to keep organisms functional. Analyzing the nature of these adjustments could greatly increase our understanding of inbreeding depression, as well as have practical applications in the fields of animal and plant breeding and the genetic management of endangered species. The main strategy proposed to improve the fitness of inbred populations considered by the current models of depression is the purging of deleterious mutations, and in the case of small populations this purging would work only for mutations of large effect. Our experiment found evidence of genetic effects protecting against inbreeding depression, which could facilitate the eventual adaptation of endangered species to living in small populations. The quantification of both the adaptive and detrimental variations in populations could be the key in evaluating their genetic health20 and in the application of conservation plans in the future.
Figure 3. Distribution of probes differentially expressed in inbred vs. outbred sublines with a False Discovery Rate (FDR) of 0.01 over the physical map of the Drosophila chromosomes (scale in Megabases, Mb). Probes with protective-consistent pattern are shown in red and those with non-protective pattern are shown in blue.
Acknowledgments
This work was funded by Ministerio de Ciencia y Tecnología (CGL2009–13278-C02), Xunta de Galicia (IN825B 2009/6–0 and Grupos de Referencia Competitiva, 2010/80) and Fondos Feder: unha maneira de facer Europa.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/22559
References
- 1.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–96. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 2.Kristensen TN, Dahlgaard J, Loeschcke V. Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evol Ecol Res. 2002;4:1209–16. [Google Scholar]
- 3.Kristensen TN, Pedersen KS, Vermeulen CJ, Loeschcke V. Research on inbreeding in the 'omic' era. Trends Ecol Evol. 2010;25:44–52. doi: 10.1016/j.tree.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Leimu R, Kloss L, Fischer M. Inbreeding alters activities of the stress-related enzymes chitinases and β-1,3-glucanases. PLoS One. 2012;7:e42326. doi: 10.1371/journal.pone.0042326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia C, Ávila V, Quesada H, Caballero A. Gene-expression changes caused by inbreeding protect against inbreeding depression in Drosophila. Genetics. 2012;192:161–72. doi: 10.1534/genetics.112.142687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–23. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Lemos B, Araripe LO, Fontanillas P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci U S A. 2008;105:14471–6. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arsham AM, Neufeld TP. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS One. 2009;4:e6068. doi: 10.1371/journal.pone.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker’s guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–8. doi: 10.1379/1466-1268(1996)001<0023:AHSGTT>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–9. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paige KN. The functional genomics of inbreeding depression: A new approach to an old problem. BioScience. 2010;60:267–77. doi: 10.1525/bio.2010.60.4.5. [DOI] [Google Scholar]
- 12.Vergeer P, Wagemaker NC, Ouborg NJ. Evidence for an epigenetic role in inbreeding depression. Biol Lett. 2012;8:798–801. doi: 10.1098/rsbl.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:7663–8. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sørensen JG, Nielsen MM, Loeschcke V. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. J Evol Biol. 2007;20:1624–36. doi: 10.1111/j.1420-9101.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- 15.Girardot F, Monnier V, Tricoire H. Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics. 2004;5:74. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarup P, Sørensen JG, Kristensen TN, Hoffmann AA, Loeschcke V, Paige KN, et al. Candidate genes detected in transcriptome studies are strongly dependent on genetic background. PLoS One. 2011;6:e15644. doi: 10.1371/journal.pone.0015644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–3. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 19.Chalancon G, Ravarani CNJ, Balaji S, Martinez-Arias A, Aravind L, Jothi R, et al. Interplay between gene expression noise and regulatory network architecture. Trends Genet. 2012;28:221–32. doi: 10.1016/j.tig.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohn MH, Murphy WJ, Ostrander EA, Wayne RK. Genomics and conservation genetics. Trends Ecol Evol. 2006;21:629–37. doi: 10.1016/j.tree.2006.08.001. [DOI] [PubMed] [Google Scholar]