Abstract
BACKGROUND: Methylated Septin 9 (mSEPT9) in plasma has recently been suggested as a screening marker for colorectal cancer (CRC) with variable sensitivity. We aimed to determine the usefulness of plasma mSEPT9 for screening CRC and gastric cancer (GC) and its diagnostic role in postoperative CRC patients. METHODS: A total of 350 peripheral blood samples from 101 CRC patients, 153 GC patients, and 96 healthy persons were collected. In addition, we obtained 35 follow-up blood samples from 27 CRC patients after curative radical surgery. Plasma mSEPT9, serum carcinoembryonic antigen (CEA), and serum CA19-9 were evaluated with clinicopathologic features. RESULTS: The sensitivity of plasma mSEPT9 was 36.6% for detecting CRC and 17.7% for detecting GC, and the specificity was 90.6%. During follow-up periods, mSEPT9 showed negative conversion in eight of nine CRC patients (88.9%) whose plasma mSEPT9 had been positive before radical surgery. The patients with plasma mSEPT9 had a tendency of presence of distant metastasis and lower disease-free survival in both CRC and GC. In GC patients, plasma mSEPT9 was more frequently observed in intestinal (23.5%) and mixed type (40.0%) than diffuse type (7.3%; P =.009). Combined analysis of mSEPT9, CEA, and CA19-9 increased the sensitivity for diagnosing GC to 32.7% (P = .002). CONCLUSION: Considering the high incidence of plasma mSEPT9 in intestinal or mixed type GCs similar to CRCs, GC should be examined through the plasma mSEPT9 screening test. In addition, plasma mSEPT9 is proposed as a follow-up marker in CRC patients, but further validation is required.
Introduction
Colorectal cancer (CRC) is one of the most common cancers and is a leading cause of cancer-related deaths worldwide [1]. In spite of the advances in the treatment, an advanced stage of CRC at the time of diagnosis is still associated with a very unfavorable prognosis. Screening tests decrease CRC-related mortalities by detecting tumors at earlier stages, thereby improving patient prognosis [2,3]. Details of CRC screening policies vary in different countries, but it is generally agreed that there is convincing evidence for the usefulness of testing fecal occult blood or performing colonoscopy to detect early-stage cancer and adenomatous polyps in patients aged ≥50 years [4–6]. Fecal occult blood testing and fecal immunochemical tests have a few advantages including easier methods and inexpensiveness [7] but with low sensitivity and specificity. Flexible sigmoidoscopy and colonoscopy demonstrate high sensitivity and specificity in preventive and therapeutic procedures to remove adenomatous polyps or early cancers [6]. However, it is expensive and presents risks such as perforation or bleeding. Therefore, the recommendations for incorporating invasive procedures into screening policies for CRC vary between countries [4,5,8], and moreover, not all adults receive screening tests despite their recommendation by national guidelines [9,10]. Only 64.2% of adults were reported to be recommended for screening in the United States [10], and the rate was lower in Korea at 35.5% [9]. To maximize the number of individuals who participate in screening and reduce colon cancer deaths and the cost of health care [11,12], studies targeted on more sensitive and noninvasive methods such as blood testing to screen CRC are being investigated.
Because CRC is known to develop from the accumulation of genetic and epigenetic changes in colonic epithelial cells, molecular markers targeting genetic and epigenetic alterations have been evaluated in tumor tissues and peripheral blood. Differences in the methylation status of tumor and nontumor DNA were evaluated for various genes [13], and methylated Septin 9 (mSEPT9) DNA has been proposed as a screening marker for detecting CRC in peripheral blood and tumor tissue [12,14–16]. SEPT9 is a member of the septin family involved in cytokinesis and cytoskeletal organization [17]. Alterations in SEPT9 have been linked to multiple cancers, but the role of SEPT9 in tumorigenesis is still elusive [18]. Instead, several studies have evaluated mSEPT9 in human plasma as a screening marker for CRC [12,14,15]. The sensitivity and specificity of plasma mSEPT9 for detecting CRC varied from 50% to 90% and from 88% to 91%, respectively, depending on the detection methods. Plasma mSEPT9 is suggested to be a screening tool in standard-risk individuals unable or unwilling to receive standard CRC screening tests. However, only limited data regarding CRC screening with mSEPT9 have been analyzed, especially in Asians [19], although some studies have reported differences in tumor DNA methylation between different ethnicities [18,20,21].
Considering that mSEPT9 have been reported to present in other organ cancers [18,22], mSEPT9 in blood sample is suggested not to be specific for CRC. Among various organs, gastric cancer (GC) and small intestinal cancer are morphologically similar to CRC. However, the incidence of small intestinal cancer is very low and we cannot easily meet it at daily practice. GC is the fourth most common malignancy and more common in developing countries and in Asian countries [1]. Intestinal type of GC has very similar histopathology to CRC, and in addition, they share some molecular characteristics including microsatellite instability, hypermethylation of tumor suppressor genes, and some mutations [23,24]. However, mSEPT9 in blood samples of GC patients has not been reported yet.
The present study aimed to investigate the usefulness of mSEPT9 as a diagnostic marker in Korean gastrointestinal cancer patients and to validate the results in healthy controls by performing real-time polymerase chain reaction (PCR) assays with plasma samples from CRC and GC patients and healthy controls. Subsequently, we analyzed the correlation between clinicopathologic characteristics and plasma mSEPT9 in CRC and GC patients, compared the clinical implication of plasma mSEPT9 with serum carcinoembryonic antigen (CEA) and CA19-9 concentration, and evaluated plasma mSEPT9 as a follow-up marker of CRC.
Materials and Methods
Plasma Specimens
To investigate mSEPT9, a total of 254 gastrointestinal cancer patients and 96 anonymous healthy volunteers were enrolled in this study. The cancer patients (101 CRC patients and 153 GC patients) whose diagnosis of CRC or GC was confirmed by endoscopic and histopathologic examination were enrolled. The plasma specimens were collected from the CRC and GC patients before surgical treatment, chemotherapy, or radiation therapy at Seoul National University Bundang Hospital between March 2011 and May 2012. We obtained the pretreatment cell-free plasma samples by collecting remnants of routine blood samples. Using this technique, 35 follow-up specimens were obtained from 27 CRC patients who were treated with curative radical surgery. Clinical and pathologic data for the CRC and GC patients were recorded from the electronic medical record system and histopathologic review. Cancer staging was based on the seventh edition of the Cancer Staging Manual of the American Joint Committee on Cancer [25]. Histologic differentiation was graded on the basis of the World Health Organization (WHO) classification [26]. Follow-up information, including cancer recurrence and the time interval between the date of surgical resection and recurrence, was collected. The patients lost to follow-up and deaths from any cause other than CRC or GC were regarded as censored data for the survival analysis. The mean follow-up period was 518 days (range, 492–543 days) in CRC patients and 413 days (range, 397–460 days) in GC patients. For the disease-free controls, 96 anonymous healthy volunteers were enrolled from the same institute from November 2011 to May 2012. The control subjects did not have any neoplasms in their colorectum or stomach or other organs. This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital. Written consent was obtained from the participants.
Specimen Preparation
Blood samples were collected from remnants of routine blood samples of two 9-ml K2-EDTA bottles. The specimens were processed and stored within 24 hours of collection. Whole-blood specimens were kept at 2 to 8°C before centrifugation and held at 15 to 30°C for approximately 30 minutes immediately before centrifugation. The blood samples were centrifuged in the blood tubes at room temperature for 10 minutes at 1500g and recentrifuged at 1500g for 10 minutes in 15-ml tubes. The buffy coat layer was not to be disturbed while transferring the plasma. The plasma specimens were stored at -70°C
before the analysis. At least 4 ml of plasma was collected for testing.
DNA Extraction
DNA was extracted by using the Abbott m2000sp Sample Preparation System (Abbott Molecular, Abbott Park, IL). The patient specimens and one each of the positive and negative controls included in the Abbott RealTime mS9 Colorectal Cancer Assay Kit (Abbott Molecular) were processed with the Abbott m2000sp. The nucleic acids were eluted by magnetic particle technology to capture the nucleic acids, and unbound sample components were removed by washing.
Bisulfite Conversion and Bisulfite Purification
Bisulfite modification was performed by Abbott Bisulfite Modification Kit according to the manufacturer's protocol. Briefly, 210 µl of bisulfite mix and 38 µl of DNA protect buffer provided in the bisulfite modification kit were mixed with 100 µl of DNA specimen. The tubes were transferred to a thermal cycler and amplified with the following conditions: 99°C for 5 minutes, 60°C for 25 minutes, 99°C for 5 minutes, 60°C for 85 minutes, 99°C for 5 minutes, and 60°C for 2 hours and 55 minutes, followed by holding at 20°C. Bisulfite purification was performed using an Abbott m2000sp System with Abbott Low Residual Volume Tubes.
Real-time PCR for the Detection of mSEPT9
The Abbott RealTime mS9 Colorectal Cancer Assay System, which consisted of an Abbott RealTime mS9 Colorectal Cancer Amplification Reagent Kit and an Abbott RealTime mS9 Colorectal Cancer Control Kit, was used according to the manufacturer's protocol. A PCR master mix was created with the amplification reagent kit. Activation reagent, oligonucleotide reagent, and AmpliTaq gold enzyme were mixed, and 50 µl of the master mix and 30 µl of the purified product were placed on a 96-well optical reaction plate on the Abbott m2000rt (Abbott Molecular) for real-time amplification. The test was performed in triplicate using the contiguous three wells across the row on the same plate. The endogenous human β-actin gene was used as the internal control for sample extraction, bisulfite modification, and amplification efficiency. Specimens whose cycle number of internal control exceeds the established specification range by mS9 Colorectal Cancer Assay m2000rt software were considered as inhibited and retested starting with sample preparation if possible. The positive and negative controls provided by the manufacturer were processed along with the samples from the DNA extraction stage. The results were reported when both controls were valid. Samples were reported as positive when the internal controls were positive and when at least one of three samples was positive.
Measurement of CEA and CA19-9
Serum CEA and CA19-9 concentrations were analyzed using an electrochemiluminescence immunoassay kit on the Modular Analytics E170 module (Roche Diagnostics Corp, Indianapolis, IN) according to the manufacturer's instructions.
Statistical Analysis
The sensitivity and specificity of plasma mSEPT9 were compared using receiver operating curve with the area under the curve. The correlation between clinicopathologic features and mSEPT9 was analyzed using the Fisher exact method or χ2 test. The mean values for age, CEA, CA19-9, and tumor size were compared according to mSEPT9 status, using the nonparametric Mann-Whitney test. The Kaplan-Meier method with the log-rank test was performed for the univariate analysis of the effect of mSEPT9 for predicting disease-free survival (DFS). Multivariate analysis was performed using the Cox proportional hazards model to determine hazard ratios. Statistical analysis was performed with SPSS Standard version 17.0 (SPSS Inc, Chicago, IL). All of the P values were two-sided, and P < .05 was considered to be statistically significant.
Results
Diagnostic Performance of mSEPT9 in Gastrointestinal Cancer Patients
Specimens from 254 gastrointestinal cancer patients and 96 healthy controls were tested for mSEPT9. mSEPT9 was identified in the pretreatment plasma of 64 of 254 gastrointestinal cancer patients (25.2%), consisting of 37 CRC and 27 GC patients. Therefore, overall sensitivity was 36.6% [95% confidence interval (CI), 27.8–46.8%] in CRC and overall sensitivity was 17.7% (95% CI, 12.0–24.6%) in GC (P = .001). mSEPT9 was detected in 9 (9.4%) of the 96 control specimens collected from the healthy blood donors. The test had an overall specificity of 90.6% (95% CI, 82.9–95.6%). By comparing the results from the cancer patients and healthy controls, the positive detection rate of mSEPT9 was higher in the CRC patients (P < .001) and the GC patients (P = .05).
Clinicopathologic Characteristics and mSEPT9
The clinical features of the CRC and GC patients according to mSEPT9 were shown in Table 1. The positive rate of mSEPT9 was 30.8% for stage 0/I, 36.7% for stage II, 25.0% for stage III, and 64.7% for stage IV CRC patients, with borderline statistical significance (P = .051). The detection of mSEPT9 was significantly higher in the CRC patients with synchronous distant metastasis than in those without metastasis (P = .008). There was a tendency for higher CEA concentration in the mSEPT9-positive CRC group (44.69 ng/ml vs 3.31 ng/ml, P = .024). Of a total of 101 CRC patients, 15 were treated with preoperative chemoradiation therapy and then radical surgery and 3 were treated with chemotherapy only. Thus, the pathologic characteristics were compared according to mSEPT9 in 83 CRC patients (Table 2). Tumor size was significantly larger in the mSEPT9-positive CRC patients than in the mSEPT9-negative patients (P = .004). The CRC patients with lymphatic or perineural invasion did not show higher mSEPT9 positivity than those without invasion (P > .05), but the patients with venous invasion showed a higher percentage of mSEPT9 detection than the patients without invasion (63.6% vs 30.6%, P = .044). Among the other findings, mSEPT9 positivity was significantly higher in CRCs with peritumoral abscess (75.0%, P = .004).
Table 1.
Clinical Features of a Total of 101 CRC Patients and 153 GC Patients according to mSEPT9 Status.
| mSEPT9 | P Value | ||||
| Positive | Negative | Total | |||
| 101 CRCs | |||||
| Age | 65.76 ± 11.25 | 62.34 ± 10.97 | 63.59 ± 11.14 | NS | |
| Gender | |||||
| Male | 17 (32.7%) | 35 (67.3%) | 52 | NS | |
| Female | 20 (40.8%) | 29 (59.2%) | 49 | ||
| TNM stage | |||||
| 0/I | 8 (30.8%) | 18 (69.2%) | 26 | ||
| II | 11 (36.7%) | 19 (63.3%) | 30 | .051 | |
| III | 7 (25.0%) | 21 (75.0%) | 28 | ||
| IV | 11 (64.7%) | 6 (35.3%) | 17 | ||
| Distant metastasis | |||||
| Absent | 26 (31.0%) | 58 (69.0%) | 84 | .008 | |
| Present | 11 (64.7%) | 6 (35.3%) | 17 | ||
| CEA concentration | 44.69 ± 133.65 | 3.31 ± 6.23 | 18.21 ± 82.09 | .024 | |
| Preoperative CCRT | NS | ||||
| Not done | 31 (36.0%) | 55 (64.0%) | 86 | ||
| Done | 6 (40.0%) | 9 (60.0%) | 15 | ||
| Location | NS | ||||
| Right colon | 6 (28.6%) | 15 (71.4%) | 21 | ||
| Left colon | 31 (38.8%) | 49 (61.2%) | 80 | ||
| Total | 37 (36.6%) | 64 (63.4%) | 101 | ||
| 153 GCs | |||||
| Age | 65.30 ± 13.03 | 60.45 ± 13.34 | 61.31 ± 13.37 | NS | |
| Gender | NS | ||||
| Male | 19 (20.9%) | 72 (79.1%) | 91 | ||
| Female | 8 (12.9%) | 54 (87.1%) | 62 | ||
| TNM stage | .016 | ||||
| I | 6 (11.8%) | 45 (88.2%) | 51 | ||
| II | 7 (15.9%) | 37 (84.1%) | 44 | ||
| III | 6 (15.0%) | 34 (85.0%) | 40 | ||
| IV | 8 (44.4%) | 10 (55.6%) | 18 | ||
| Distant metastasis | .004 | ||||
| Absent | 19 (14.1%) | 116 (85.9%) | 135 | ||
| Present | 8 (44.4%) | 10 (55.6%) | 18 | ||
| CEA concentration | 5.46 ± 9.89 | 3.95 ± 10.52 | 4.21 ± 10.40 | NS | |
| CA19-9 concentration | 22.22 ± 44.38 | 28.11 ± 106.47 | 27.08 ± 98.38 | NS | |
| Location of tumor | NS | ||||
| Lower third | 19 (20.0%) | 76 (80.0%) | 95 | ||
| Middle third | 4 (10.8%) | 33 (89.2%) | 37 | ||
| Upper third | 4 (19.0%) | 17 (81.0%) | 21 | ||
| Total | 27 (17.6%) | 126 (82.4%) | 153 | ||
CCRT indicates chemoradiation therapy; NS, not significant.
Table 2.
Pathologic Characteristics according to mSEPT9 Status in 83 CRC Patients and 138 GC Patients with Pathologic Examination of Radical Surgery Specimens.
| mSEPT9 | P Value | |||
| Positive | Negative | Total | ||
| CRC | ||||
| Histologic grade | NS | |||
| WD | 3 (42.9%) | 4 (57.1%) | 7 | |
| MD | 24 (34.8%) | 45 (65.2%) | 69 | |
| PD | 1 (33.3%) | 2 (66.7%) | 3 | |
| Mucinous | 1 (25.0%) | 3 (75.0%) | 4 | |
| Tumor size | 5.75 ± 2.73 | 3.99 ± 1.92 | 4.59 ± 2.37 | .004 |
| Depth of invasion | NS | |||
| pTis/T1 | 4 (28.6%) | 10 (71.4%) | 14 | |
| pT2 | 4 (25.0%) | 12 (75.0%) | 16 | |
| pT3 | 14 (35.9%) | 25 (64.1%) | 39 | |
| pT4 | 7 (50.0%) | 7 (50.0%) | 14 | |
| LN metastasis | NS | |||
| pN0 | 19 (35.8%) | 34 (64.2%) | 53 | |
| pN1 | 6 (28.6%) | 15 (71.4%) | 21 | |
| pN2 | 4 (44.4%) | 5 (55.6%) | 9 | |
| Lymphatic invasion | NS | |||
| Absent | 20 (31.7%) | 43 (68.3%) | 63 | |
| Present | 9 (45.0%) | 11 (55.0%) | 20 | |
| Venous invasion | .044 | |||
| Absent | 22 (30.6%) | 50 (69.4%) | 72 | |
| Present | 7 (63.6%) | 4 (36.4%) | 11 | |
| Perineural invasion | NS | |||
| Absent | 19 (30.2%) | 44 (69.8%) | 63 | |
| Present | 10 (50.0%) | 10 (50.0%) | 20 | |
| Tumor deposits | NS | |||
| Absent | 22 (34.4%) | 42 (65.6%) | 64 | |
| Present | 7 (36.8%) | 12 (63.2%) | 19 | |
| Preexisting adenoma | NS | |||
| Absent | 26 (40.0%) | 39 (60.0%) | 65 | |
| Present | 3 (16.7%) | 15 (83.3%) | 18 | |
| Other findings | .004 | |||
| Absent | 19 (30.2%) | 44 (69.8%) | 63 | |
| Mucin production | 1 (12.5%) | 7 (87.5%) | 8 | |
| Peritumoral abscess | 9 (75.0%) | 3 (25.0%) | 12 | |
| Total | 29 | 54 | 83 | |
| GC | ||||
| Lauren classification | .009 | |||
| Intestinal | 12 (23.5%) | 39 (76.5%) | 51 | |
| Diffuse | 6 (7.3%) | 76 (92.7%) | 82 | |
| Mixed | 2 (40.0%) | 3 (60.0%) | 5 | |
| Tumor size | 5.54 ± 3.63 | 4.37 ± 2.88 | 4.54 ± 3.02 | NS |
| Depth of invasion | NS | |||
| pT1 | 6 (12.2%) | 43 (87.7%) | 49 | |
| pT2 | 1 (4.5%) | 21 (95.5%) | 22 | |
| pT3 | 7 (20.0%) | 28 (80.0%) | 35 | |
| pT4 | 6 (18.8%) | 26 (81.3%) | 32 | |
| LN metastasis | NS | |||
| pN0 | 9 (14.1%) | 55 (85.9%) | 64 | |
| pN1 | 3 (14.3%) | 18 (85.7%) | 21 | |
| pN2 | 1 (4.5%) | 21 (95.5%) | 22 | |
| pN3 | 7 (22.6%) | 24 (77.4%) | 31 | |
| Lymphatic invasion | NS | |||
| Absent | 10 (14.9%) | 57 (85.1%) | 67 | |
| Present | 10 (14.1%) | 61 (85.9%) | 71 | |
| Venous invasion | NS | |||
| Absent | 16 (14.0%) | 98 (86.0%) | 114 | |
| Present | 4 (16.7%) | 20 (83.3%) | 24 | |
| Perineural invasion | NS | |||
| Absent | 13 (17.1%) | 63 (82.9%) | 76 | |
| Present | 7 (11.3%) | 55 (88.7%) | 62 | |
| Tumor border | .001 | |||
| Expanding | 11 (33.3%) | 22 (66.7%) | 33 | |
| Infiltrative | 9 (8.6%) | 96 (91.4%) | 105 | |
| Preexisting adenoma | NS | |||
| Absent | 18 (13.8%) | 112 (86.2%) | 130 | |
| Present | 2 (25.0%) | 6 (75.0%) | 8 | |
| Total | 20 | 118 | 138 | |
WD indicates well differentiated; MD, moderately differentiated; PD, poorly differentiated; NS, not significant.
In the GC patients, mSEPT9 was also associated with advanced stage (P = .016) and distant metastasis (P = .004; Table 1). In contrast to CRC group, CEA concentrations of the mSEPT9-positive GC group were similar to the mSEPT9-negative GC group (P > .05). Of a total of 153 GC patients, radical surgery was not performed in 15 patients. Thus, pathologic characteristics were compared according to mSEPT9 in 138 GC patients (Table 2). Interestingly, mSEPT9 detection was more frequently found in intestinal type (23.5%) and mixed type (40.0%) than diffuse type (7.3%, P = .009). Among the other findings, mSEPT9 positivity was significantly higher in GCs with expanding border (P = .001).
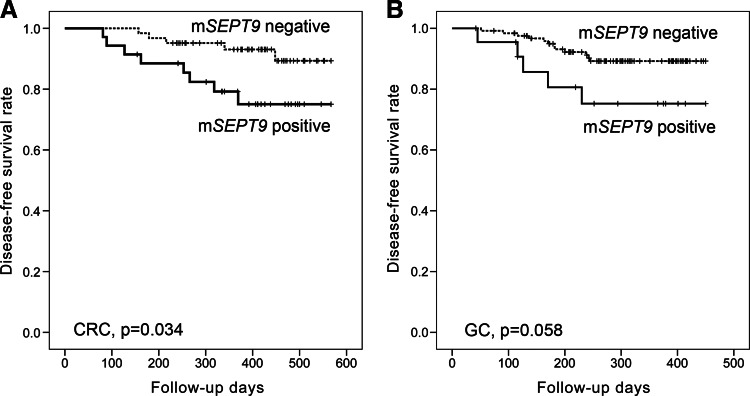
The results of the Kaplan-Meier univariate analysis showed that mSEPT9 positivity was significantly associated with lower DFS in the CRC patients (P = .034; Figure 1A). The mSEPT9-positive CRC cases had estimated mean DFS duration of 480 days (95% CI, 426–534) compared with the 539 days (95% CI, 516–563) in the mSEPT9-negative CRC cases. However, mSEPT9 showed borderline prognostic significance in the GC patients (P = .058; Figure 1B). mSEPT9 was not an independent prognostic marker in the CRC or GC patients by Cox regression analysis (P > .05; data not shown).
Figure 1.
Kaplan-Meier univariate survival curves according to mSEPT9 status in (A) CRC patients and (B) GC patients.
mSEPT9 as a Follow-Up Marker
Thirty-five postoperative samples (one to four samples in each patient) from 27 CRC patients treated with curative radical surgery were tested for mSEPT9 (Table 3). Postoperative specimens were obtained 118 days after surgery on average (71–232 days). Among 27 CRC patients with follow-up plasma samples, 24 (88.9%) patients showed negative mSEPT9 after curative radical surgery irrespective of the pretreatment results. Sixteen patients (61.5%) showed negative results for mSEPT9 before and after surgery. Of the nine patients who were found with mSEPT9 before surgery, negative conversion was observed in eight patients (88.9%).
Table 3.
mSEPT9 Status in Follow-Up Plasma Samples of 27 CRC Patients.
| mSEPT9 | No. of Patients | CRC Recurrence during Follow-Up | |
| Preoperative Samples | Postoperative Samples | ||
| Negative | Negative | 16 | 3 |
| Negative | Positive | 2 | 1 |
| Positive | Negative | 8 | 0 |
| Positive | Positive | 1 | 0 |
| Total | 27 | 4 | |
CRC recurrence was diagnosed in 4 of 27 patients (Table 3), of whom 1 patient was diagnosed as recurred cancer by plasma mSEPT9. In this patient, two serial plasma samples showed mSEPT9 positivity at 128 and 215 days after the first surgery (Figure W1), but clinical impression was abscess at anastomosis site by computed tomography (CT) imaging. Serum CEA concentration of the patient was 2.9, 4.2, and 12.2 ng/ml at -1, 128, and 215 days, respectively, thus with borderline clinical significance. Recurrence was confirmed by intraoperative frozen tissue examination and radical resection of the recurred cancer was performed. Negative conversion of mSEPT9 was found after the second surgery for recurrent cancer (at 348 days after the first surgery). mSEPT9 was also found in the primary and recurrent CRC tissue samples from this patient.
Serum CEA and CA19-9 Concentrations
No serum biomarker currently exists for initial diagnosis of CRC or GC in daily practice, but in 2010, the AJCC Cancer Staging, Seventh Edition has accepted serum tumor marker CEA as site-specific prognostic factor in CRC and CEA and CA19-9 as prognostic factors in GC [25]. Thus, concentrations of CEA in CRC patients and CEA and CA19-9 in GC patients were additionally evaluated. A serum concentration higher than 5 ng/ml was defined as CEA positive and a serum concentration higher than 27 U/ml as CA19-9 positive [27]. Positive CEA was found in 21 (20.8%) of the 101 CRC patients, which were correlated with advanced stage and shorter DFS (Table W1). CEA positivity was found in 14 (9.2%) and CA19-9 positivity was found in 22 (14.4%) of 153 GC patients, which were also correlated with advanced stage and shorter DFS (Table W2).
To increase the sensitivity for the diagnosis of CRC or GC, we performed combined analysis of mSEPT9 and serum markers. CEA positivity and/or plasma mSEPT9 was observed in 44 (43.6%) of 101 CRC patients; thus, the sensitivity of the combined analysis was similar to the sensitivity of mSEPT9 for CRC diagnosis (P > .05). In contrast to CRC patients, the sensitivity of combined analysis of CEA, CA19-9, and/or mSEPT9 (50 of 153 patients, 32.7%) was significantly higher than any single marker (P = .002 in mSEPT9; P < .001 in CEA and CA19-9).
Discussion
In the present study, mSEPT9, which is being evaluated as a noninvasive plasma screening marker of CRC, was tested in Korean CRC patients. Overall sensitivity was 36.6% (95% CI, 27.8–46.8%) and overall specificity was 90.6% (95% CI, 82.9–95.6%). In contrast to our results in Korean CRC patients, sensitivities of plasma mSEPT9 in previous studies ranged from 50.0% to 90.0% and specificities ranged from 73.0% to 94.0% as determined by various methods [12,14,28,29]. Thus, the sensitivity of our results was relatively low, but the specificity was high. The possible causes of low frequency of mSEPT9 in the Korean CRC patients may be ethnic variations in the methylation pattern of SEPT9 or the method of detection. Although ethnic variation in the methylation patterns of SEPT9 has not been studied, differences in methylation status between ethnic groups have been published for other genes in prostate or breast cancer patients [18,20]. The screening of CRC with mSEPT9 has been performed mostly in the United States and Germany and showed sensitivities ranging from 50% to 90% with various methods [12,28,29], whereas a small population study in China showed a sensitivity of 75% using MethylLight assay [19]. Therefore, further investigations are required in various ethnics with various methods. To demonstrate the levels of mSEPT9 in CRC tissue in the Korean population, we evaluated mSEPT9 in 17 CRC tissues and 17 matched normal tissues. mSEPT9 was detected in 16 cancer tissues (94.1%) and 5 normal tissues (29.4%; P = .001; data not shown). Therefore, the levels of mSEPT9 in Koreans were higher in cancer tissue than normal tissue, but the detection rate of plasma mSEPT9 was lower than other ethnics. In addition, 25 GC tissues and 25 matched normal tissues were evaluated for mSEPT9. mSEPT9 was detected in 14 cancer tissues (56.0%) and 4 normal tissues (16.0%; P = .002; data not shown), which suggested that mSEPT9 was not specific for CRC diagnosis.
In addition to CRCs, we investigated plasma mSEPT9 in 153 GC patients because mSEPT9 was reported in other organ cancers [18,22] and intestinal type GC has similar morphologic and molecular features to CRC. mSEPT9 was infrequently detected in the plasma samples of GCs (17.7%). However, 23.5% of intestinal type and 40.0% of mixed type GCs showed presence of mSEPT9 in the plasma samples, which were similar frequency to CRCs. Furthermore, plasma mSEPT9 was observed in 44.4% (8 of 18) stage IV GCs. Therefore, GCs, especially intestinal/mixed type or stage IV, should be considered in differential diagnosis at the screening test by using plasma mSEPT9. This study is the first to validate a role of plasma mSEPT9 as a noninvasive diagnostic biomarker in large-scaled GC cases.
To validate the specificity of mSEPT9 in CRC patients and to evaluate a possibility of using mSEPT9 as a noninvasive follow-up marker, we investigated plasma mSEPT9 in follow-up samples from 27 CRC patients after curative radical surgery. Twenty-four (88.9%) of 27 patients showed negative mSEPT9 after curative radical surgery and negative conversion was observed in 8 of 9 patients (88.9%), which was similar to the overall specificity (90.6%) of pretreatment plasma mSEPT9. It is suggested that plasma mSEPT9 before and after treatment is specific for CRCs and have potential for a follow-up marker of CRCs. The usefulness of mSEPT9 as a follow-up marker was additionally suggested by the patient in whom recurrent CRC was detected by only plasma mSEPT9 in our cohort. It is the first to demonstrate the negative conversion of plasma mSEPT9 after curative radical surgery and the diagnostic role of mSEPT9 for detecting recurrence. Follow-up results for mSEPT9 in these patients will aid in deciding whether mSEPT9 can be used as a follow-up marker. Further validation is required for the clinical use of plasmamSEPT9 as a follow-up marker of CRCs.
In daily practice, serum CEA and/or CA19-9 concentration is used for screening, prognosis prediction, and follow-up examination in CRC and GC patients. We also evaluated pretreatment serum CEA concentrations in CRC patients and CEA and CA19-9 in GC patients. To increase the sensitivity for the diagnosis of CRC or GC, we performed combined analysis of mSEPT9 and serum markers. The sensitivity of combined analysis was significantly increased in GCs (32.7%; P = .002) but not in CRCs. In contrast to CRCs, multiple markers including plasma mSEPT9 and serum CEA and CA19-9 are suggested to be more useful for diagnosing GC than any single marker.
Despite the several reports evaluating mSEPT9 in CRC patients, its clinicopathologic implication has not been confirmed yet. Neither did in GC patients. Thus, we analyzed whether the detection of mSEPT9 showed any relationship with clinicopathologic characteristics and patient outcome. There was no significant difference in mSEPT9 detection according to depth, lymph node metastasis, and lymphatic or perineural invasion in both CRC and GC. However, those patients with distant metastasis had significantly higher positivity for mSEPT9. Furthermore, the mSEPT9-positive group exhibited lower DFS than the mSEPT9-negative group (P = .034 in CRC; P = .058 in GC). Therefore, mSEPT9 is suggested to be useful for predicting aggressive tumor behavior in CRC and GC patients. In contrast to the present results, septin 9, which is encoded by SEPT9, was identified as a pseudopod-specific protein in various cancer cell lines that is essential for pseudopod protrusion and tumor cell migration and invasion [30]. This discrepancy may be because genetic, epigenetic, and expression status is different according to the various transcripts encoding different isoforms of septin 9, and the relationship between mSETP9 and septin 9 expression is not clearly elucidated due to isoforms of septin 9 [31]. Further study is needed to clarify it.
In conclusion, mSEPT9 was detected at a relatively low frequency in the Korean CRC patients but highly specific. mSEPT9 detection was correlated with clinicopathologic characteristics such as distant metastasis and poor DFS. In addition to CRCs, we revealed that plasma mSEPT9 was also observed in GC patients, especially in intestinal/mixed type or stage IV GC patients. Therefore, we suggest that GC should be considered through the screening test using plasma mSEPT9. Follow-up of mSEPT9-positive patients would reveal the usefulness of mSEPT9 as an early detection marker for recurrence.
Supplementary Material
Footnotes
This study was supported by grant 06-2011-140 from the Seoul National University Bundang Hospital Research Fund and by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0023625).
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figure W1 and are available online at www.transonc.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): an update. Am J Gastroenterol. 2008;103:1541–1549. doi: 10.1111/j.1572-0241.2008.01875.x. [DOI] [PubMed] [Google Scholar]
- 3.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 4.Qaseem A, Denberg TD, Hopkins RH, Jr, Humphrey LL, Levine J, Sweet DE, Shekelle P. Screening for colorectal cancer: a guidance statement from the American College of Physicians. Ann Intern Med. 2012;156:378–386. doi: 10.7326/0003-4819-156-5-201203060-00010. [DOI] [PubMed] [Google Scholar]
- 5.Lee BI, Hong SP, Kim SE, Kim SH, Kim HS, Hong SN, Yang DH, Shin SJ, Lee SH, Park DI, et al. Korean guidelines for colorectal cancer screening and polyp detection. Clin Endosc. 2012;45:25–43. doi: 10.5946/ce.2012.45.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 7.Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288–1296. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 8.The Lancet, author. New guidance for colorectal cancer screening. Lancet. 2012;379:978. doi: 10.1016/S0140-6736(12)60413-9. [DOI] [PubMed] [Google Scholar]
- 9.Lee EH, Lee HY, Choi KS, Jun JK, Park EC, Lee JS. Trends in cancer screening rates among Korean men and women: results from the Korean National Cancer Screening Survey (KNCSS), 2004–2010. Cancer Res Treat. 2011;43:141–147. doi: 10.4143/crt.2011.43.3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frieden TR. Forward: CDC Health Disparities and Inequalities Report—United States, 2011. MMWR Surveill Summ. 2011;60:1–2. [PubMed] [Google Scholar]
- 11.Pignone M, Sox HC. Screening guidelines for colorectal cancer: a twice-told tale. Ann Intern Med. 2008;149:680–682. doi: 10.7326/0003-4819-149-9-200811040-00247. [DOI] [PubMed] [Google Scholar]
- 12.Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, Fang JC, Samowitz WS, Heichman KA. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 14.deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, Grutzmann R, Pilarsky C, Habermann JK, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–1346. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 15.Grutzmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F, Roblick UJ, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toth K, Galamb O, Spisak S, Wichmann B, Sipos F, Valcz G, Leiszter K, Molnar B, Tulassay Z. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res. 2011;17:503–509. doi: 10.1007/s12253-010-9338-7. [DOI] [PubMed] [Google Scholar]
- 17.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Dorsey TH, Terunuma A, Kittles RA, Ambs S, Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q, Chen HY, Bai EQ, Luo YX, Fu RJ, He YS, Jiang J, Wang HQ. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010;202:1–10. doi: 10.1016/j.cancergencyto.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 20.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16:3539–3547. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 21.Nieminen TT, Shoman S, Eissa S, Peltomaki P, Abdel-Rahman WM. Distinct genetic and epigenetic signatures of colorectal cancers according to ethnic origin. Cancer Epidemiol Biomarkers Prev. 2012;21:202–211. doi: 10.1158/1055-9965.EPI-11-0662. [DOI] [PubMed] [Google Scholar]
- 22.Bennett KL, Karpenko M, Lin MT, Claus R, Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D, Plass C. Frequently methylated tumor suppressor genes in head and neck squamous cell carcinoma. Cancer Res. 2008;68:4494–4499. doi: 10.1158/0008-5472.CAN-07-6509. [DOI] [PubMed] [Google Scholar]
- 23.Lee HS, Choi SI, Lee HK, Kim HS, Yang HK, Kang GH, Kim YI, Lee BL, Kim WH. Distinct clinical features and outcomes of gastric cancers with microsatellite instability. Mod Pathol. 2002;15:632–640. doi: 10.1038/modpathol.3880578. [DOI] [PubMed] [Google Scholar]
- 24.Wu M, Semba S, Oue N, Ikehara N, Yasui W, Yokozaki H. BRAF/K-ras mutation, microsatellite instability, and promoter hypermethylation of hMLH1/MGMT in human gastric carcinomas. Gastric Cancer. 2004;7:246–253. doi: 10.1007/s10120-004-0300-9. [DOI] [PubMed] [Google Scholar]
- 25.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer; 2010. pp. 117–126.pp. 143–164. [Google Scholar]
- 26.Bosman FT, Carneiro F, Hruban RH, Theise ND. In: WHO Classification of Tumours of the Digestive System. Bosman FT, Jaffe ES, Lakhani SR, Ohgaki H, editors. France: IARC, Lyon; 2010. pp. 48–58.pp. 134–146. [Google Scholar]
- 27.Lee HS, Lee HE, Park do J, Kim HH, Kim WH, Park KU. Clinical significance of serum and tissue Dickkopf-1 levels in patients with gastric cancer. Clin Chim Acta. 2012;413:1753–1760. doi: 10.1016/j.cca.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272–277. doi: 10.1016/j.cgh.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tänzer M, Balluff B, Distler J, Hale K, Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70:3780–3790. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 31.Connolly D, Yang Z, Castaldi M, Simmons N, Oktay MH, Coniglio S, Fazzari MJ, Verdier-Pinard P, Montagna C. Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression. Breast Cancer Res. 2011;13:R76. doi: 10.1186/bcr2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.