Abstract
The aim of this study was to investigate the relationship between the promoter methylation in five cancer-associated genes and clinicopathologic features for identification of molecular markers of tumor metastatic potential and hormone therapy response efficiency in breast cancer. The methylation levels in paraffin-embedded tumor tissues, plasma, and blood cells from 151 sporadic breast cancer patients and blood samples of 50 controls were evaluated by quantitative multiplex methylation-specific polymerase chain reaction. DNA methylation of RAS-association domain family member 1 (RASSF1A), estrogen receptor 1 (ESR1), cadherin 1, type 1, E-cadherin (CDH1), TIMP metallopeptidase inhibitor 3 (TIMP3) and spleen tyrosine kinase (SYK) genes was detected in the tumors of 124, 19, 15, 15, and 6 patients with mean levels of 48.45%, 3.81%, 2.36%, 27.55%, and 10.81%, respectively. Plasma samples exhibited methylation in the same genes in 25, 10, 15, 17, and 3 patients with levels of 22.54%, 17.20%, 22.87%, 31.93%, and 27.42%, respectively. Cumulative methylation results confirmed different spectra in tumor and plasma samples. Simultaneous methylation in tumors and plasma were shown in less than 17% of patients. RASSF1A methylation levels in tumor samples statistically differ according to tumor size (P = .029), estrogen receptor (ER) and progesterone receptor (PR) status (P = .000 and P = .004), and immunohistochemical subtype (P = .000). Moreover, the positive correlation was found between RASSF1A methylation levels and percentage of cancer cells expressing ER and PR. The direct relationship between RASSF1A promoter methylation and expression of ER could aid the prognosis of hormonal therapy response.
Introduction
Breast cancer is the most common cancer in women worldwide. According to data published by the International Agency for Research on Cancer, in 2008, 1,383,000 breast cancer patients were newly diagnosed and 458,000 breast cancer-related deaths occurred [1]. More than 25% of breast cancer patients develop metastatic disease that is mostly incurable and for which there are only palliative therapeutic options [2]. Clinicopathologic characteristics such as tumor size, lymph node (LN) status, invasion of vessels, and hormone receptor status play important roles in metastasis risk [3]. However, the results of a recent multicenter study found differences in clinicopathologic features between patients with and without primary metastases, and for metastasis risk, the lobular histology and luminal B positivity in T1 primary metastatic breast cancer were determined [2].
Similar to other cancer types, breast tumorigenesis is characterized by the progressive accumulation of genetic and epigenetic changes in many genes that regulate cell proliferation and differentiation. Therefore, molecular characterization of tumor tissues allows determination of novel cancer markers including those predicting metastatic potential and therapy response.
Epigenetic abnormalities in neoplastic cells, such as hypermethylation and hypomethylation of DNA, altered patterns of histone modification, and remodeled chromatin structure, result in the modified expression of many essential genes. A well-categorized epigenetic change is hypermethylation of tumor-suppressor promoters that led to inappropriate transcription silencing of these genes [4]. The tumor suppressor gene RAS-association domain family member 1 (RASSF1A) encodes a member of the group of RAS effectors that regulates cell proliferation, apoptosis, and microtubule stability. Hypermethylation of RASSF1A was found in a substantial percentage of various primary tumors [5]. Epigenetic inhibition of RASSF1A is considered to be an early cancer biomarker; however, this phenomenon is extended from primary to metastatic tumors during tumor progression [6]. Moreover, in invasive breast cancers, significantly higher RASSF1A methylation levels were shown compared with in situ carcinomas [7]. These results indicate the possible association of RASSF1A silencing with metastasis. Other studies reported higher frequencies of methylation in RASSF1A alone or in combination with HIN-1 in estrogen receptor (ER)-positive cases compared with ER-negative cases [8,9]. Moreover, a recent in vitro study revealed that RASSF1A inhibits ERα expression and function [product of estrogen receptor 1 (ESR1) gene]; thereby, it plays a key role in suppressing transformation of mammary epithelial cells and ERα-positive breast cancer initiation [10]. In addition to the potential RASSF1A-mediated epigenetic regulation of ESR1, mild or moderate DNA methylation of the ESR1 promoter alone was observed in breast tumorigenesis, indicating the possible influence of epigenetic processes on hormonal therapy response [11,12]. In tumorigenesis, there are numerous changes in the cadherin-catenin adhesion complexes, including the cell adhesion protein E-cadherin encoded by cadherin 1, type 1, E-cadherin (CDH1). In primary breast cancer, the heterogeneous loss of E-cadherin expression corresponding with variable patterns of promoter methylation was observed in the early stages before cell invasion [13]. CDH1 hypermethylation with loss of protein expression was found in both ductal and lobular breast carcinomas; however, no significant correlation was observed between E-cadherin expression and the CDH1 promoter methylation profile [14]. The tissue inhibitors of metalloproteinase (TIMPs) prevent degradation of the extracellular matrix by the metalloproteinases. TIMP metallopeptidase inhibitor 3 (TIMP3) is a matrix-bound protein regulating matrix composition that affects tumor growth, angiogenesis, invasion, and metastasis. TIMP3 promoter methylation was observed in 21% to 27% of breast cancer patients and in invasive ductal carcinomas that were associated with high tumor grading and LN metastasis [15,16]. The spleen tyrosine kinase (SYK) is an intracellular receptor protein kinase involved in cell proliferation, differentiation, and phagocytosis and plays a suppressive function in breast cancer progression and metastasis [17]. The frequencies of SYK promoter hypermethylation at different stages of breast cancer indicate its occurrence shortly before the development of the invasion phenotype [18]. The objective of the present study was to determine the association of the promoter methylation profiles of five genes related to invasion and metastasis with breast cancer clinicopathologic features to identify useful molecular markers indicating the metastatic potential of tumors and patient response to hormonal therapy.
Materials and Methods
Patients
A total of 151 paraffin-embedded tumor tissue samples and matched 151 peripheral blood samples from nonfamilial breast cancer patients and blood samples of 50 healthy controls were obtained from the Department of Pathology and Department of Senology at hospitals in Bratislava, Slovakia. This study was approved by Ethics Committee of the University Hospital in Bratislava, and written informed consent was obtained from all patients and controls. Relevant clinical and pathologic data were retrieved from the patients' clinical records, and tumors were characterized according to the primary tumor, regional lymph nodes, distant metastasis (TNM) classification. The age of patients ranged from 23 to 91 years (mean, 61.2 ± 10.8 years) at the time of breast cancer diagnosis. Typing was performed according to the current World Health Organization (WHO) classification for breast neoplasms (Table 1). No preoperative radiotherapy or chemotherapy had been performed in any of the cases. Controls included 25 individuals of <50 years and 25 individuals of >50 years who had no signs and symptoms of cancer or other serious diseases.
Table 1.
RASSF1A Methylation Levels in Different Clinical and Histopathologic Categories in Breast Cancer Patients.
| N | RASSF1A Methylation in Tumor Samples | RASSF1A Methylation in Plasma Samples | |||||
| Mean | Median | P Value | Mean | Median | P Value | ||
| 151 | |||||||
| Age | .773 | .669 | |||||
| ≤50 | 17 | 37.9 | 44.4 | 1.8 | 0 | ||
| >50 | 134 | 40.0 | 40.5 | 4.0 | 0 | ||
| Histologic type | .864 | .660 | |||||
| DIC | 131 | 40.1 | 40.6 | 3.8 | 0 | ||
| LIC | 10 | 40.1 | 49.1 | 5.2 | 0 | ||
| Others | 10 | 35.1 | 32.5 | 1.2 | 0 | ||
| Tumor size | .029 | .643 | |||||
| ≤20 | 90 | 41.9 | 42.6 | 4.1 | 0 | ||
| >20 ≤ 50 | 47 | 32.1 | 31.5 | 3.9 | 0 | ||
| >50 | 4 | 64.7 | 64.2 | 0 | 0 | ||
| Histologic grading | .668 | .102 | |||||
| 1 | 36 | 40.6 | 43.2 | 2.5 | 0 | ||
| 2 | 75 | 41.3 | 42.3 | 5.1 | 0 | ||
| 3 | 38 | 36.3 | 35.8 | 2.4 | 0 | ||
| LN status | .718 | .297 | |||||
| 0 | 86 | 42.2 | 43.2 | 3.7 | 0 | ||
| 1 | 36 | 36.5 | 36.8 | 2.2 | 0 | ||
| 2 | 11 | 36.1 | 40.4 | 13.4 | 0 | ||
| 3 | 6 | 43.1 | 49.6 | 0 | 0 | ||
| TNM staging | .066 | .271 | |||||
| I | 80 | 43.4 | 43.2 | 4.3 | 0 | ||
| II | 50 | 31.2 | 28.8 | 1.5 | 0 | ||
| III | 8 | 46.7 | 56.9 | 15.8 | 0 | ||
| IV | 1 | 65.9 | 65.9 | 0 | 0 | ||
| ER status | .000 | .266 | |||||
| Negative | 21 | 18.0 | 3.7 | 2.1 | 0 | ||
| Positive | 129 | 43.6 | 44.4 | 4.0 | 0 | ||
| PR status | .004 | .863 | |||||
| Negative | 33 | 27.4 | 22.4 | 5.1 | 0 | ||
| Positive | 117 | 43.5 | 44.4 | 3.4 | 0 | ||
| HER2 expression | .069 | .419 | |||||
| Negative | 128 | 38.2 | 39.3 | 3.3 | 0 | ||
| Positive | 22 | 50.8 | 43.8 | 6.7 | 0 | ||
| IHC subtypes | .000 | .252 | |||||
| ER+/PR+ HER2- | 114 | 42.2 | 44.2 | 3.3 | 0 | ||
| ER+/PR+ HER2+ | 16 | 51.0 | 46.2 | 9.2 | 0 | ||
| ER- PR- HER2+ | 7 | 43.1 | 42.3 | 0 | 0 | ||
| ER- PR- HER2- | 13 | 5.8 | 0 | 3.4 | 0 | ||
DIC indicates ductal invasive carcinomas; LIC, lobular invasive carcinomas; Others, tubular, micropapillar invasive, cribriform invasive, or mucinous breast carcinomas; LN status, lymph node status; ER status, estrogen receptor status; PR status, progesterone receptor status; IHC subtypes, immunohistochemical subtypes.
P < .05 was regarded as statistically significant (in bold). LN status was categorized according to the number of cancer cell-positive nodes as 0, 1, 2, and 3 with none, 1 to 3, 4 to 10, and >10 of positive LNs, respectively. ER or PR status was considered as positive in cases with ≥1% of positively responding cells. HER2 expression was regarded as positive, if the intensity of IHC reaction was 3+ in 30% of tumor cells or with fluorescence in situ hybridization proven HER2 gene amplification in cases with ambiguous IHC positive at 2+ intensity reaction. According to ER, PR, and HER2 expression, four IHC subtypes were recognized, luminal A and B (ER+ and/or PR+ HER- and ER+ and/or PR+ HER2+), HER2 overexpression positive (ER- PR- HER2+), and triple negative (ER- PR- HER2-). Tumor sizes are shown in millimeters.
DNA Extraction and Sodium Bisulfite Modification
Blood samples of patients and controls were collected in EDTA-treated tubes and centrifuged at 1000g for 10 minutes at room temperature within 2 hours of venepuncture. Then, supernatants were collected and centrifuged at 1000g for 10 minutes at room temperature to prevent cellular DNA contamination. Plasma samples were stored at -70°C until further processing. Cell-free DNA from plasma samples was isolated using a QIAamp DSP Virus Kit (Qiagen, Hilden, Germany), DNA from paraffin-embedded tumor tissues was isolated by the MagneSil Genomic, Fixed Tissue System (Promega, Madison, WI), and genomic DNA from peripheral blood was obtained using a FlexiGene DNA Kit (Qiagen) according to the manual instructions. DNA concentrations were measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Bremen, Germany). Tumor DNA (2 µg), cell-free DNA (2 µg), and genomic DNA (1 µg) were modified by sodium bisulfite treatment according to the protocols of the EpiTect Bisulfite Kit (Qiagen) and CpGenome DNA Modification Kit (Chemicon, Billerica, MA), respectively. DNA was stored at -18°C until use.
Quantitative Multiplex Methylation-Specific Polymerase Chain Reaction Analysis
For quantitative evaluation of promoter methylation, the two-color modification of quantitative multiplex methylation-specific polymerase chain reaction (QM-MSP) technology was used [19]. QM-MSP was performed in two sequential polymerase chain reaction (PCR) reactions. In the first step, co-amplification of three and two gene loci (RASSF1A, CDH1, SYK and ESR1, TIMP3) was performed using three and two pairs of methylation-independent external primers, respectively. Multiplex PCRs were performed in 30-µl volumes containing 30 to 60 ng of modified DNA, 15 µl of 2x QIAGEN Multiplex PCR Master Mix (Qiagen), and aliquots of six/four primers at a final concentration of 0.2 µM. PCR conditions were 95°C for 15 minutes, 35 cycles at 94°C for 30 seconds, 62/56°C for 60 seconds, hybridization at 72°C for 90 seconds, and final extension at 72°C for 10 minutes. In the second step (quantitative real-time PCR), 1 µl of the first reaction PCR product was used at a dilution of up to 1:102 in a duplex reaction with both pairs of primers and specific TaqMan probes for methylated and unmethylated DNA substrates for each gene. PCRs were performed in 15-µl volumes containing 7.5 µl of Maxima Probe qPCR Master Mix (2x; Fermentas, Amherst, NY), methylation- and unmethylation-specific primers for RASSF1A, ESR1, CDH1, TIMP3, or SYK gene at a final concentration of 0.3 µM, and methylation- and unmethylation-specific TaqMan probes at concentrations ranging from 0.1 to 0.27 µM. The reaction conditions were 50°C for 2 minutes, 95°C for 10 minutes, followed by 35 cycles at 95°C for 15 seconds and 60°C for 30 to 90 seconds with final extension at 72°C for 30 seconds. For quantitative PCR, a CFX96TM Real-Time PCR System (Bio-Rad, Hercules, CA) was used. Standard curve establishment and evaluation of quantitative analysis of DNA methylation were performed as previously described [20]. The relative amount of methylation (%) was calculated in each sample according to the formula [M/(U + M)] x 100. Concentrations of methylated (M) and unmethylated (U) portions were determined from simultaneously amplified standard curves for each gene. Methylation levels up to 0.5% were considered to be the background of this sensitive quantitative method. The cumulative methylation index (CMI) was calculated as the sum of percentage methylation for all evaluated genes. For all five genes, CMI of 500 was the maximum value of methylation. Primers and TaqMan probes are summarized in the supplementary material (Table W1) [21].
Statistical Analysis
For statistical analyses, SPSS statistics 15.0 was applied, with P < .05 regarded as statistically significant. Normally distributed data were tested by Pearson correlations, Student's t tests, or analysis of variance with Bonferroni or Tamhane tests for multiple comparisons, depending on homogeneity of variance. For non-normally distributed data, Spearman correlations, nonparametric Mann-Whitney U or Kruskal-Wallis H tests were used. Normality of distribution was assessed by Kolmogorov-Smirnoff tests. All tests were two-tailed. Categorical data were tested by Chi square.
Results
DNA Methylation in Tumor and Plasma Samples
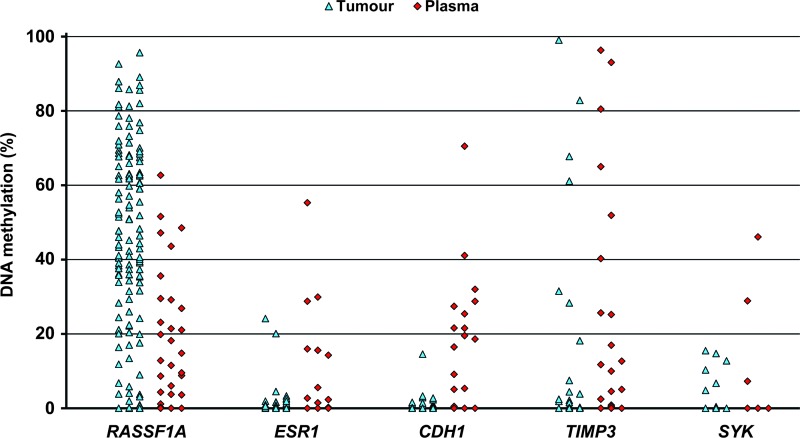
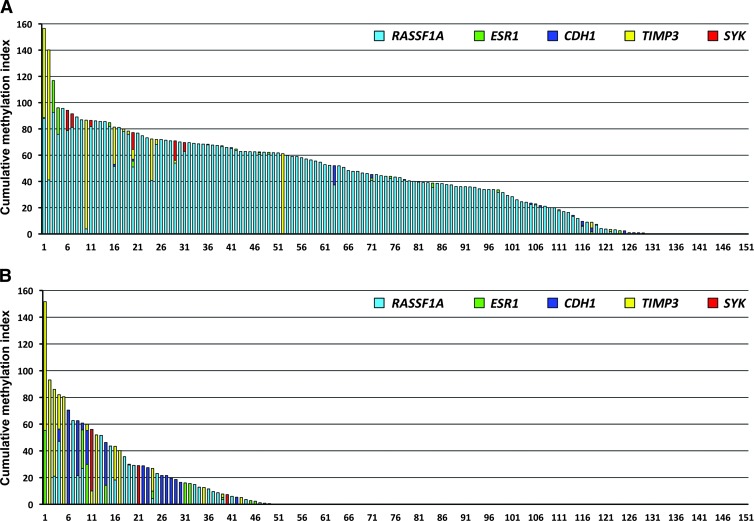
Quantitative analyses of DNA methylation were performed in paraffin-embedded tumor tissues, blood cells, and plasma samples from 151 breast cancer patients. Of these, 129 patients (85.4%) demonstrated different levels of methylation in at least one of the evaluated promoters in their tumors: in 124, 19, 15, 15, and 6 patients, mean methylation levels of 48.45%, 3.81%, 2.36%, 27.55%, and 10.81% were found in RASSF1A, ESR1, CDH1, TIMP3, and SYK, respectively. In plasma samples, RASSF1A methylation was observed at a markedly lower frequency of 22.54% in 25 patients. ESR1, CDH1, TIMP3, and SYK were methylated in 10, 15, 17, and 3 patients at 17.20%, 22.87%, 31.93%, and 27.42%, respectively. DNA methylation levels of five evaluated genes in tumor and plasma samples are graphically depicted in Figure 1. Simultaneously methylated promoters in both tumor and plasma samples were found in 25, 1, 2, 4, and 1 patients in RASSF1A, ESR1, CDH1, TIMP3, and SYK genes, respectively (Table 2). Low levels of TIMP3 methylation (0.53–5.15%) were detected in the genomic DNA of four breast cancer patients. Of 50 healthy controls, methylation levels of >0.5% in the CDH1 promoter was observed in the genomic DNA of one person alone. However, in the control plasma samples, rare methylation events in CDH1 were found in one person (1.67%), TIMP3 in two (16.52% and 20.40%), and ESR1 in two (7.12% and 16.72%). The number of methylated genes was determined in individual patients. Of 129 patients with any methylation in tumor tissues, 91, 30, and 7 samples were methylated in one, two, and three evaluated genes, respectively. One patient manifested promoter methylation in all five genes; however, the CMI was only 77.14. Of 49 patients with methylation in plasma, 37, 7, and 5 samples were methylated in one, two, and three genes, respectively (Figure 2). The cumulative methylation levels for the five evaluated genes were significantly higher in the tumors and plasma than in the genomic DNA of the same patients. In the methylation-positive tumor and plasma samples, the mean CMIs were 50.52 and 32.96, respectively, compared with no methylation in genomic DNA except for four patients with TIMP3 methylation. However, a similar range of cumulative methylation in tumor and plasma DNA (0.74–156.57 and 0.51–151.62) was observed. In the majority of tumors, the substantial portion of CMI was represented by RASSF1A methylation when compared with more frequent methylation of other genes in plasma samples.
Figure 1.
Methylation levels of five genes evaluated in tumor and plasma samples of breast cancer patients.
Table 2.
Frequencies of Breast Cancer Patients with DNA Methylation in Tumor and Plasma Samples.
| Evaluated Genes | Promoter Methylation in Tumor, N (%) | Promoter Methylation in Plasma, N (%) | Promoter Methylation in Both Tumor and Plasma, N (%) |
| RASSF1A | 124 (82.1) | 25 (16.6) | 25 (16.6) |
| ESR1 | 19 (12.8) | 10 (6.7) | 1 (0.7) |
| CDH1 | 15 (9.9) | 15 (10) | 2 (1.3) |
| TIMP3 | 15 (9.9) | 17 (11.5) | 4 (2.7) |
| SYK | 6 (4) | 3 (2) | 1 (0.7) |
Figure 2.
Cumulative DNA methylation levels in breast cancer patients. The results from tumor tissues of 129 patients (A) and plasma samples of 49 patients (B) are shown. The CMI is the sum of percentage methylation for five evaluated genes.
RASSF1A Methylation Levels and Clinicopathologic Categories
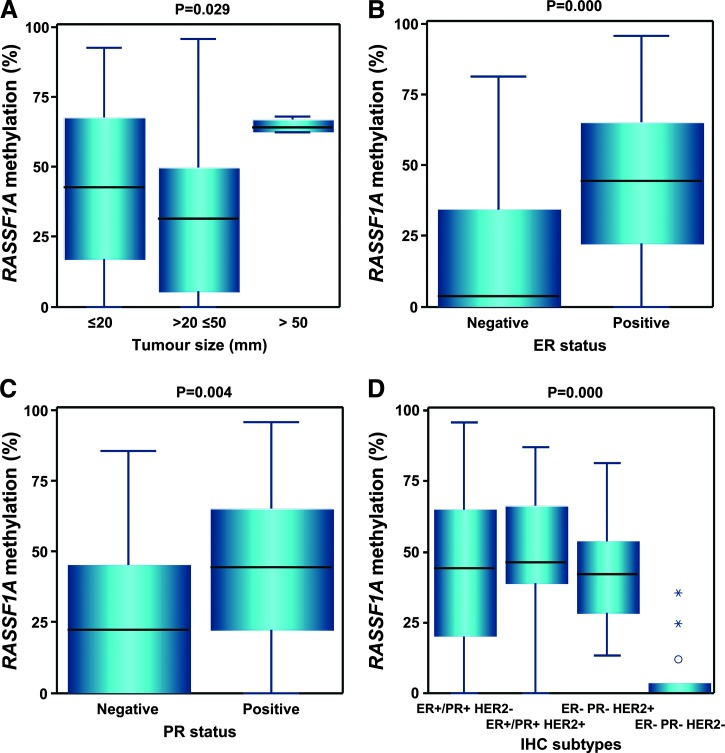
Statistical analysis of the correlation between methylation levels and clinicopathologic features of 151 breast cancer patients was performed for the highly and frequently methylated RASSF1A gene. The evaluated categories were age, histologic type, tumor size, histologic grading, LN status, TNM staging, ER status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) expression, and immunohistochemical (IHC) subtypes. In plasma samples, RASSF1A methylation ranged from 0% to 15.8%, with no significant differences between the subgroups of each clinicopathologic category. The tumor samples of these patients exhibited visibly higher levels of RASSF1A methylation, and statistically significant differences were observed between patients with various tumor size (P = .029), ER-negative and ER-positive status (P = .000), PR-negative and PR-positive status (P = .004), and different IHC subtypes (P = .000; Table 1 and Figure 3). Moreover, there was a positive correlation between RASSF1A methylation levels and the percentage of cancer cells with ER (r = 0.251, P = .002) or PR expression (r = 0.200, P = .014).
Figure 3.
Distribution of RASSF1A methylation levels in four clinical and histopathologic categories of breast cancer patients. Box plots show the significant differences in the subgroups of patients with different tumor sizes (A), negative and positive estrogen or progesterone status (B and C), and different IHC subtypes (D). The length of the boxes is the interquartile range (IQR) that represents values between the 75th and 25th percentiles. Values more than three IQRs from the end of a box are labeled as extreme (*). Values more than 1.5 IQRs but less than 3 IQRs from the end of the box are labeled as outliers (O). The median is depicted by a horizontal line.
Discussion
Quantification of DNA methylation levels in cancer-associated genes contributes to the more complex molecular characterization of tumors required for the development of new diagnostic and therapeutic strategies for cancer patients. High methodical diversity of DNA methylation status evaluation was found in the current literature; therefore, we compared our results with these studies using quantitative methods based on real-time technology. In tumor samples, we found RASSF1A methylation in 82.1% of evaluated breast cancer patients, with a mean level of 48.45% in methylated cases. In other studies, analogous frequencies (68% and 82.5%) but lower means of methylation levels (18.5 ± 4.7% and more than 10%) were found in the majority of patients [21,22]. Similar to our results, previous studies recorded low incidences of ESR1 methylation in breast cancers and weak correlation with low ERα expression levels, indicating a sporadic role of DNA methylation in ER silencing [11,23]. In the present study, 9.9% of patients exhibited CDH1 promoter methylation levels of up to 14.56% in tumor tissues, similar to other groups of patients where low levels of CDH1 methylation frequencies from 5.8% to 22.5% were observed [20,22,24]. Variable TIMP3 methylation levels of 3% to 42% were identified in the samples of nine patients [25], in accordance with our findings; however, to our knowledge, quantitative evaluation of SYK methylation has not yet been performed. Comparison of studies in European, American, and Saudi Arabian females revealed that the highest methylation levels were in RASSF1A similar to our study, confirming the important role of RASSF1A epigenetic silencing in breast cancer regardless of ethnicity [21,23,24].
Circulating cell-free DNA from plasma, serum, or other body fluids seems to be an appropriate biologic material for qualitative or quantitative testing of tumor-specific molecular alterations including DNA methylation. The serum of patients with invasive ductal carcinomas showed significantly higher RASSF1A methylation frequencies compared with control persons [26], as in our study. We found significantly lower frequencies of RASSF1A methylation in plasma samples (16.6%) than in tumors (82.1%); however, all 25 patients with positive findings in plasma had simultaneous methylation in their tumor samples. For ESR1, CDH1, TIMP3, and SYK, rare incidences were observed in both tumor and plasma samples. ESR1 was evaluated in the serum of healthy controls and disease-free breast cancer and metastatic breast cancer patients and revealed no differences in the low levels of ESR1 methylation between these three groups [27]. Our results oppose the above-mentioned hypothesis describing the possible influence of ESR1 epigenetic silencing alone in the strategy of breast cancer therapy. Many researchers have focused on the identification of useful sets of methylated genes to improve diagnosis, prognosis, or therapeutic strategy; therefore, CMI appears to be a useful parameter. The high incidence of RASSF1A methylation in CMI shows the value of this silenced gene in tumor development in our patients. However, in 49 plasma samples, we found a different spectrum in CMI, with visibly higher occurrences of TIMP3 and CDH1 methylation. These results indicate that cell-free DNA could be derived from a degraded cell subpopulation, which is active in invasive and metastatic processes, for example, circulating tumor cells, rather than from products of apoptosis and necrosis in heterogeneous tumor masses [28]; therefore, these DNA samples could be used for metastatic potential testing. However, after the critical evaluation of methodical diversity, variability of results, and limited diagnostic sensitivity and specificity of cell-free DNA alterations in many published studies, we agree that clinical utilization of such DNA requires further studies to assess sample collection, processing, analysis, and measurement of results [29].
In our previous study, no relationship between tumor size and RASSF1A methylation levels was observed [20], but in the presented group of patients we found significantly higher levels in four cases with breast tumors larger than 50 mm, three of which were at an advanced stage of disease. The accumulation of DNA methylation changes could be associated with aggressive phenotype rather than larger size of tumor, because small cancers can also invade and metastasize as a result of higher numbers of molecular changes compared with early-stage cancer. Most importantly, we found a relationship between RASSF1A methylation levels and expression of hormonal receptors. Previous studies showed higher frequency of RASSF1A methylation in breast cancers with ER+ and PR+ status than in ER- and PR- cases [24,30]; however, we observed a positive correlation between RASSF1A methylation levels in tumor tissues and number of cancer cells with positive expression in both ER and PR. Analyses of RASSF1A methylation in four different IHC subtypes showed very low levels in ER- PR- HER2- but not in ER- PR- HER2+. Moreover, in cancers with HER2 overexpression, higher but statistically insignificant differences in methylation levels were observed compared with HER2-negative cancers. These results indicate the possible influence of HER2 on DNA methylation processes.
In normal human breast epithelium, ERα expression is fairly consistent over time, and women with ER overexpression in the normal breast may have increased estrogen sensitivity that is associated with higher breast cancer risk [31]. In previous case-control studies, the incidence of breast epithelial cells expressing ER was higher in breast cancer cases than benign breast disease controls [32]; however, Woolcott and colleagues did not confirm this strong association [33]. ER expression in luminal breast cancers varied from 1% to 100% of positively stained cells, and even patients with 1% of ER-expressing tumor cells experience some clinical benefit from endocrine therapies [34]. Therefore, in addition to ER levels, ER dynamics could play an important role in tumor behavior including therapy response. In a recent study, a mouse model using patient-derived ER+ tumor xenografts was developed for the evaluation of intratumoral hormone and receptor action. The researchers reported that analysis of the ER transcriptome in selected tumors showed notable differences in the ER mechanism of action and downstream-activated signaling networks, in addition to identifying a small set of common estrogen-regulated genes. Mapping of conserved and tumor-unique ER programs can contribute to the development of more personalized therapeutic strategies [35]. Both estrogen signaling and epigenetic modifications, in particular DNA methylation, are involved in the regulation of gene expression in breast cancers. Putnik and colleagues investigated the potential regulatory cross talk between these two pathways in human MCF-7 breast cancer cells. They identified approximately 140 genes that were influenced by both 17β-estradiol and a demethylating agent 5-aza-20-deoxycytidine; however, they did not show a direct molecular interplay of estrogen mediators and epigenetic signaling at the promoters of regulated genes [36]. Furthermore, in a recent study of the RASSF1A tumor-suppressive function in MCF-7 cells, reconstitution of RASSF1A expression decreased ERα levels, followed by reduced expression of Id1 and the E2-responsive genes BCL-2 and C-MYC, up-regulation of p21Cip1/Waf1 induction of cell-cycle arrest and senescence, and inhibition of signaling pathways involved in breast epithelial cell transformation. These findings indicate a central role of RASSF1A in suppressing transformation of human breast epithelial cells in part through ERα inhibition [10]. This hypothesis is supported by the present study, because we observed a positive correlation between RASSF1A methylation levels and percentage of ER- or PR-positively stained cells in contrary to hormone receptor-negative cases with low levels of methylation. In another study of heterogeneity of matched breast primary tumors and metastases, 3 of 10 evaluated patients exhibited no RASSF1A methylation and a hormone-negative phenotype in both types of tissues [37]. Previous work carried out in vitro is convincing; however, detailed studies of cancer-associated changes in the ER mechanism of action in human tumors will enable association of the “ER-dependent pathway patterns” with the effectiveness of hormonal therapy and may help to develop new therapeutic molecules.
To summarize, in the present study, we observed high frequencies of RASSF1A methylation and positive correlation of RASSF1A methylation levels with ER and PR expression in breast cancer patients. Therefore, we speculate that the levels of RASSF1A methylation in ER+ breast cancer patients could be helpful in determining prognosis and hormonal therapy response. Clinical utility of cell-free DNA isolated from plasma for cancer-associated molecular characteristics testing, namely, DNA methylation, is unsatisfactory because of its low detection sensitivity. Furthermore, the specific methylation spectra in plasma samples related to invasive and metastatic processes need to be further evaluated in larger study cohorts.
Supplementary Material
Acknowledgments
We thank Gabriela Gasajova for excellent technical assistance.
Footnotes
This publication is the result of the implementation of the following projects: No. APVV-0076-10 supported by the Slovak Research and Development Agency (60%); No. 26240220058, Research and Development Operational Programme funded by the European Regional Development Fund (20%); Nos 2/0065/10 and 2/0120/13 funded by the Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and the Slovak Academy of Sciences (20%). All authors declare that they have no competing interests.
This article refers to supplementary material, which is designated by Table W1 and is available online at www.transonc.com.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2008;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Barinoff J, Hils R, Bender A, Groß J, Kurz C, Tauchert S, Mann E, Schwidde I, Ipsen B, Sawitzki K, et al. Clinicopathological differences between breast cancer in patients with primary metastatic disease and those without: a multicentre study. Eur J Cancer. 2012;49:305–311. doi: 10.1016/j.ejca.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Clark GM, Sledge GW, Jr, Osborne CK, McGuire WL. Survival from first recurrence: relative importance of prognostic factors in 1, 015 breast cancer patients. J Clin Oncol. 1987;5:55–61. doi: 10.1200/JCO.1987.5.1.55. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 5.Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP. The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol. 2005;20:645–663. doi: 10.14670/HH-20.645. [DOI] [PubMed] [Google Scholar]
- 6.Feng W, Orlandi R, Zhao N, Carcangiu ML, Tagliabue E, Xu J, Bast RC, Jr, Yu Y. Tumor suppressor genes are frequently methylated in lymph node metastases of breast cancers. BMC Cancer. 2010;10:378. doi: 10.1186/1471-2407-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasquali L, Bedeir A, Ringquist S, Styche A, Bhargava R, Trucco G. Quantification of CpG island methylation in progressive breast lesions from normal to invasive carcinoma. Cancer Lett. 2007;257:136–144. doi: 10.1016/j.canlet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Sunami E, Shinozaki M, Sim MS, Nguyen SL, Vu AT, Giuliano AE, Hoon DS. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10:46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res. 2007;9:57. doi: 10.1186/bcr1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaler S, Schmidt M, Schad A, Sleeman JP. RASSF1A inhibits estrogen receptor alpha expression and estrogen-independent signalling: implications for breast cancer development. Oncogene. 2012;31:4912–4922. doi: 10.1038/onc.2011.658. [DOI] [PubMed] [Google Scholar]
- 11.Gaudet MM, Campan M, Figueroa JD, Yang XR, Lissowska J, Peplonska B, Brinton LA, Rimm DL, Laird PW, Garcia-Closas M, et al. DNA hypermethylation of ESR1 and PGR in breast cancer: pathologic and epidemiologic associations. Cancer Epidemiol Biomarkers Prev. 2009;18:3036–3043. doi: 10.1158/1055-9965.EPI-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widschwendter M, Siegmund KD, Müller HM, Fiegl H, Marth C, Müller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 13.Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- 14.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 16.Lui EL, Loo WT, Zhu L, Cheung MN, Chow LW. DNA hypermethylation of TIMP3 gene in invasive breast ductal carcinoma. Biomed Pharmacother. 2005;59:363–365. doi: 10.1016/s0753-3322(05)80079-4. [DOI] [PubMed] [Google Scholar]
- 17.Coopman PJ, Mueller SC. The Syk tyrosine kinase: a new negative regulator in tumor growth and progression. Cancer Lett. 2006;241:159–173. doi: 10.1016/j.canlet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Y, Liu H, Sahin A, Dai JL. Reactivation of SYK expression by inhibition of DNA methylation suppresses breast cancer cell invasiveness. Int J Cancer. 2005;113:654–659. doi: 10.1002/ijc.20628. [DOI] [PubMed] [Google Scholar]
- 19.Swift-Scanlan T, Blackford A, Argani P, Sukumar S, Fackler MJ. Two-color quantitative multiplex methylation-specific PCR. Biotechniques. 2006;40:210–219. doi: 10.2144/000112097. [DOI] [PubMed] [Google Scholar]
- 20.Sebova K, Zmetakova I, Bella V, Kajo K, Stankovicova I, Kajabova V, Krivulcik T, Lasabova Z, Tomka M, Galbavy S, et al. RASSF1A and CDH1 hypermethylation as potential epimarkers in breast cancer. Cancer Biomark. 2011–2012;10:13–26. doi: 10.3233/CBM-2012-0230. [DOI] [PubMed] [Google Scholar]
- 21.Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, Garrett E, Argani P, Sukumar S. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 22.Cho YH, Yazici H, Wu HC, Terry MB, Gonzalez K, Qu M, Dalay N, Santella RM. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- 23.Buhmeida A, Merdad A, Al-Maghrabi J, Al-Thobaiti F, Ata M, Bugis A, Syrjänen K, Abuzenadah A, Chaudhary A, Gari M, et al. RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Res. 2011;31:2975–2981. [PubMed] [Google Scholar]
- 24.Cho YH, Shen J, Gammon MD, Zhang YJ, Wang Q, Gonzalez K, Xu X, Bradshaw PT, Teitelbaum SL, Garbowski G, et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat. 2012;131:197–205. doi: 10.1007/s10549-011-1712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagnon JF, Sanschagrin F, Jacob S, Tremblay AA, Provencher L, Robert J, Morin C, Diorio C. Quantitative DNA methylation analysis of laser capture microdissected formalin-fixed and paraffin-embedded tissues. Exp Mol Pathol. 2010;88:184–189. doi: 10.1016/j.yexmp.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Shin MH, Kweon SS, Park MH, Yoon JH, Lee JS, Choi C, Fackler MJ, Sukumar S. Evaluation of promoter hypermethylation detection in serum as a diagnostic tool for breast carcinoma in Korean women. Gynecol Oncol. 2010;118:176–181. doi: 10.1016/j.ygyno.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 27.Zurita M, Lara PC, del Moral R, Torres B, Linares-Fernández JL, Arrabal SR, Martínez-Galán J, Oliver FJ, Ruiz de Almodóvar JM. Hypermethylated 14-3-3-σ and ESR1 gene promoters in serum as candidate biomarkers for the diagnosis and treatment efficacy of breast cancer metastasis. BMC Cancer. 2010;10:217. doi: 10.1186/1471-2407-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 29.Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker—a critical appraisal of the literature. Clin Chim Acta. 2010;411:1611–1624. doi: 10.1016/j.cca.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, Kim IA, Jung N, Cho NY, Kang GH. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- 31.Khan SA, Yee KA, Kaplan C, Siddiqui JF. Estrogen receptor α expression in normal human breast epithelium is consistent over time. Int J Cancer. 2002;102:334–337. doi: 10.1002/ijc.10737. [DOI] [PubMed] [Google Scholar]
- 32.Khan SA, Rogers MA, Khurana KK, Meguid MM, Numann PJ. Estrogen receptor expression in benign breast epithelium and breast cancer risk. J Natl Cancer Inst. 1998;90:37–42. doi: 10.1093/jnci/90.1.37. [DOI] [PubMed] [Google Scholar]
- 33.Woolcott CG, SenGupta SK, Hanna WM, Aronson KJ. Estrogen and progesterone receptor levels in nonneoplastic breast epithelium of breast cancer cases versus benign breast biopsy controls. BMC Cancer. 2008;8:130. doi: 10.1186/1471-2407-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, Manuel CA, Edgerton SM, Harrell JC, Elias A, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135:415–432. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putnik M, Zhao C, Gustafsson JÅ, Dahlman-Wright K. Global identification of genes regulated by estrogen signaling and demethylation in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2012;426:26–32. doi: 10.1016/j.bbrc.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, Griffin C, Fetting J, Davidson NE, De Marzo AM, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–1946. doi: 10.1158/1078-0432.CCR-07-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.