Abstract
Molecular magnetic resonance imaging (MRI) of tumors improves the specificity of MRI by using targeted probes conjugated to contrast-generating metals. The limitation of this approach is in the identification of a target molecule present in sufficient concentration for visualization and the development of a labeling reagent that can penetrate tumor tissue with the fast kinetics required for use in a clinical setting. The receptor protein tyrosine phosphatase PTPµ is a transmembrane protein that is continuously proteolyzed in the tumor microenvironment to generate a high concentration of extracellular fragment that can be recognized by the SBK2 probe. We conjugated the SBK2 peptide to a gadolinium chelate [SBK2-Tris-(Gd-DOTA)3] to test whether the SBK2 probe could be developed as an MR molecular imaging probe. When intravenously injected into mice bearing flank tumors of human glioma cells, SBK2-Tris-(Gd-DOTA)3 labeled the tumors within 5 minutes with a high level of contrast for up to 2 hours post-injection. The contrast enhancement of SBK2-Tris-(Gd-DOTA)3 was significantly higher than that observed with a current MRI macrocyclic gadolinium chelate (Gadoteridol, ProHance) alone or a scrambled control. These results demonstrate that SBK2-Tris-(Gd-DOTA)3 labeling of the PTPµ extracellular fragment is a more specific MR molecular imaging probe than ProHance or a scrambled control. Consequently, the SBK2 probe may be more useful than the current gold standard reagent for MRI to identify tumors and to co-register tumor borders during surgical resection.
Introduction
Magnetic resonance imaging (MRI) offers excellent spatial resolution while providing basic anatomic detail when used to image tumors. MRI is currently the preferred method of tumor imaging before surgery. Yet MRI is limited to delineating only the gross anatomy of tumors. In the case of brain tumors, MRI has a limited ability to highlight the complex and irregular shapes of tumors and lacks biologic specificity, as the technology images areas of increased blood-brain barrier permeability but not necessarily the full extent of the tumor itself [1]. To improve on the specificity of MRI, molecular MRI has been introduced to label proteins in the area of interest using probes that are conjugated to contrast-generating metals, such as iron oxide or gadolinium chelates [2]. These modifications have their limitations as detection levels of metal ions are in the micromolar range, whereas target proteins tend to be expressed in the nanomolar range, resulting in a disconnect between the quantity of label and labeling agent able to penetrate into a tumor [2]. Amplification methods have been employed to overcome this limitation, including exogenous enzymatic amplification paradigms [3].
An alternative target is one that is biologically amplified by continuous endogenous enzymatic processing and deposition and is therefore found in high concentration in the tumor microenvironment. The receptor protein tyrosine phosphatase PTPµ is an example of a transmembrane protein that is proteolyzed in the tumor microenvironment, but not in normal tissue, to yield an extracellular fragment and a membrane-free intracellular fragment [4,5]. The proteolyzed extracellular fragment of PTPµ is retained in the tumor vicinity and is enriched in the tumor microenvironment of the dispersing tumor cell population typical of glioblastoma multiforme (GBM) [6]. GBM is a devastating cancer of the brain because of the aggressively dispersive/invasive GBM cells [7]. A fluorescent probe that binds to the PTPµ extracellular fragment labels both the main GBM tumor mass in the brain [4] and greater than 99% of the dispersing cells up to 3.5 mm away from the main tumor [6]. The fluorescent probe, named SBK2, is a highly sensitive marker that could possibly be used during neurosurgical resection. Fluorescence imaging cannot, however, be used for brain tumor imaging in patients before surgery because of the limited depth of penetration of the fluorescent light signal.
To test whether the SBK2 probe could be developed as a diagnostic imaging tool, we conjugated it to a gadolinium chelate [SBK2-Tris-(Gd-DOTA)3] to generate an MR-detectable SBK2 probe. We compared the ability of SBK2-Tris-(Gd-DOTA)3 to function as a contrast agent compared to a current MRI macrocyclic gadolinium chelate (Gadoteridol, ProHance) alone or to a scrambled probe linked to gadolinium [scrambled Tris-(Gd-DOTA)3]. When intravenously injected into mice bearing flank tumors of human glioma cells, SBK2-Tris-(Gd-DOTA)3 labeled the tumors within 5 minutes with a high level of contrast persisting for 2 hours post-injection. The contrast enhancement of SBK2-Tris-(Gd-DOTA)3 was significantly higher than that observed with ProHance alone. These results demonstrate that SBK2-Tris-(Gd-DOTA)3 labeling of PTPµ extracellular fragment retained in the tumor microenvironment is a more specific MR molecular imaging probe than a nonspecific gadolinium chelate and may be a useful tool for diagnosis of the extent of tumor dispersal before surgery and co-registering GBM tumor borders during surgical resection.
Materials and Methods
All reagents were used without further purification unless otherwise stated. Benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate, 1-hydroxybenzotriazole hydrate, 2-chlorotritylchloride resin, and all of the Fmoc-protected amino acids were purchased from Chem-Impex International, Inc (Wood Dale, IL). Anhydrous N, N-diisopropylethyl amine and N, N-dimethylformamide were purchased from Alfa Aesar (Ward Hill, MA). Trifluoroacetic acid was purchased from Oakwood Products, Inc (West Columbia, SC). The detailed synthesis of maleimido-Tris-propargyl and azido-(Gd-DOTA) is the subject of a forthcoming publication.
Synthesis and Characterization of SBK2-Tris-(Gd-DOTA)3 and Scrambled Tris-(Gd-DOTA)3
The SBK2 peptide was conjugated to Gd-DOTA using an increased molar ratio of Gd-DOTA monoamide to peptide to generate an MR-visible probe [SBK2-Tris-(Gd-DOTA)3]. A scrambled version of the SBK2 peptide was used to generate a nontargeted control agent [scrambled Tris-(Gd-DOTA)3]. Peptide SBK2 with an N-terminal cysteine (C-GEGDDFNWEQVNTLTKPTSD) was synthesized using standard solid-phase peptide synthesis. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF; Autoflex Speed, Bruker) mass spectra (m/z, M+) were given as follows: 2355.52 (observed) and 2355.00 (calculated). Scrambled peptide (C-GFTQPETGTDNDLWSVDNEK) was synthesized by the same method [MALDI-TOF (m/z, M+): 2355.56 (observed); 2355.00 (calculated)]. SBK2 was conjugated to maleimido-Tris-propargyl, then subsequently to azido-(Gd-DOTA). The reaction was traced by MALDI-TOF until Gd-DOTA was fully attached [MALDI-TOF (m/z, M+): 4664.87 (observed); 4664.62 (calculated); Inductively coupled plasma optical emission spectroscopy (ICP-OES) analysis for Gd3+ content: 9.56% (observed); 10.1% (calculated)]. Scrambled Tris-(Gd-DOTA)3 was synthesized by the same method with a yield of 68% [MALDI-TOF (m/z, M+): 4664.75 (observed); 4664.62 (calculated); ICP (Gd3+ content): 9.68% (observed); 10.1% (calculated)].
The gadolinium content was measured by ICP-OES (Agilent 730 Axial ICP-OES; Agilent Technologies, Wilmington, DE). Relaxation times were measured at 60 MHz (1.5 T) using a Bruker Minispec Relaxometer at 37°C. T1 was measured with an inversion recovery pulse sequence. T2 was measured using a Carr-Purcell-Meiboom-Gill sequence with 500 echoes collected.
Cell Culture and Flank Tumor Implants
The human LN-229 glioma cell line was purchased from American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium supplemented with 5% FBS. Cells were infected with lentivirus encoding green fluorescent protein (GFP), diluted in BD Matrigel Matrix (BD Biosciences, Franklin Lakes, NJ), and injected into both flanks of female nude athymic mice (NCr-nu/+, NCr-nu/nu, 20–25 g each) as previously described [4]. Approximately 1.4 x 106 cells were implanted per flank. To correlate tumor position with GFP fluorescence, mice were imaged using the Maestro FLEX In Vivo Imaging System as previously described [4]. Tumor area was approximately 0.5 cm2, as measured from GFP fluorescence images acquired with the Maestro just before MRI.
Molecular Imaging of Tumors with MRI
The MRI study was performed using a Bruker Biospec 7-T MRI scanner (Bruker Corp, Billerica, MA) with a 72-mm diameter cylindrical radio frequency coil. Mice bearing LN-229 flank tumors were imaged at 2 to 5 weeks after tumor implant. Mice were anesthetized with a 2% isoflurane-oxygen mixture in an isoflurane induction chamber. Tail veins were catheterized with a 30-gauge needle connected to a 1.2-m-long small diameter Tygon tubing filled with heparinized saline. The animals were moved into a magnet and kept under inhalation anesthesia with 1.5% isoflurane-oxygen through a nose cone. A respiratory sensor connected to a monitoring system (SA Instruments, Stony Brook, NY) was placed on the abdomen to monitor rate and depth of respiration. The body temperature was maintained at 37°C by blowing hot air into the magnet through a feedback control system. A group of three mice was used for each agent per experiment. Sagittal section images were acquired with a localizing sequence to identify the tumor location, followed by a fat suppression three-dimensional (3D) FLASH sequence [repetition time (TR) = 8.5 ms, echo time (TE) = 2.6 ms, average = 3, 30° flip angle, in-plane field of view (FOV) = 10 cm, 25-mm slab thickness] and a two-dimensional (2D) T 1-weighted spin echo sequence (TR = 500 ms, TE = 8.1 ms, average = 2, in-plane FOV = 3 cm, 1.2-mm slice thickness) before injection. After pre-injection baseline MRI acquisition, ProHance, the targeted SBK2 agent, or the nontargeted scrambled control was injected at a dose of 0.03 or 0.1 mmol Gd/kg by flushing with 60 µl of heparinized saline. Images were then acquired using the same sequences at 13 different time points after the injection for up to 120 minutes.
Image Processing and Analysis
Image analysis was performed by using Bruker ParaVision 4.0 imaging software. Regions of interest were drawn over the whole tumor in the 2D imaging plane and average signal intensity was measured. Contrast-to-noise ratios (CNRs) in the tumor were calculated at each time point and averaged from different mice (n = 3) using the following equation: CNR = (St - Sm)/(δn), where St and Sm denote the signal in tumor and adjacent thigh muscle and δn is the SD of noise estimated from the background air. The P values were calculated using a two-tailed Student's t test, assuming statistical significance at P < .05. Color-coded display of relative contrast intensity in tumor was processed by SPIN (signal processing in NMR) software (MRI Institute for Biomedical Research, Detroit, MI).
Biodistribution Study
Mice from the MRI study were sacrificed at 7 days post-injection. The organ and tissue samples, including the brain, heart, kidneys, liver, lungs, spleen, skin, muscle, femur, and tumor, were resected and weighed. The tissue samples were liquified with nitric acid and prepared for ICP-OES measurement as previously described [8]. The average gadolinium content in each sample was measured by ICP-OES (n = 3 per contrast agent). The Gd content was calculated as the percentage of injected dose per gram of organ/tissue (% ID/g).
Histologic Analysis of Tumor Tissue Sections
LN-229-GFP flank tumors were excised and fixed with 4% paraformaldehyde in PEM buffer (80 mM Pipes, 5 mM EGTA, 1 mM magnesium chloride, 3% sucrose), pH 7.4, at 72 hours post-MR molecular imaging to allow for clearance of contrast agents. Tumors were embedded in Tissue Freezing Medium (Electron Microscopy Sciences, Hatfield, PA) in a dry ice ethanol slurry and cryosectioned at 7-µm intervals.
For probe labeling, tissue sections were blocked with 2% goat serum in phosphate-buffered saline for 20 minutes at room temperature, then incubated with SBK2-Texas Red or scrambled Texas Red probes [4,5] diluted to 1 µMin block buffer for 1 hour at room temperature in dark conditions. The sections were rinsed with phosphate-buffered saline, coverslipped with Citifluor Antifadent Mounting Medium (Electron Microscopy Sciences), and imaged immediately at x20 magnification on a Leica DMI 6000 B inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) using a Retiga EXi camera (QImaging, Surrey, BC) and Metamorph software (Molecular Devices, Downington, PA). Images were acquired across the entire tissue using bright-field and fluorescence optics for fluorescein (GFP) and Texas Red (probe). The resultant images were tiled and flattened to form a single composite image using Metamorph software. A human specific Vimentin antibody (clone SP20; Thermo Fisher Scientific, Fremont, CA) was used to detect the implanted LN-229 tumor cells and imaged as above.
Results
Synthesis and Characterization of SBK2-Tris-(Gd-DOTA)3 and Scrambled Tris-(Gd-DOTA)3
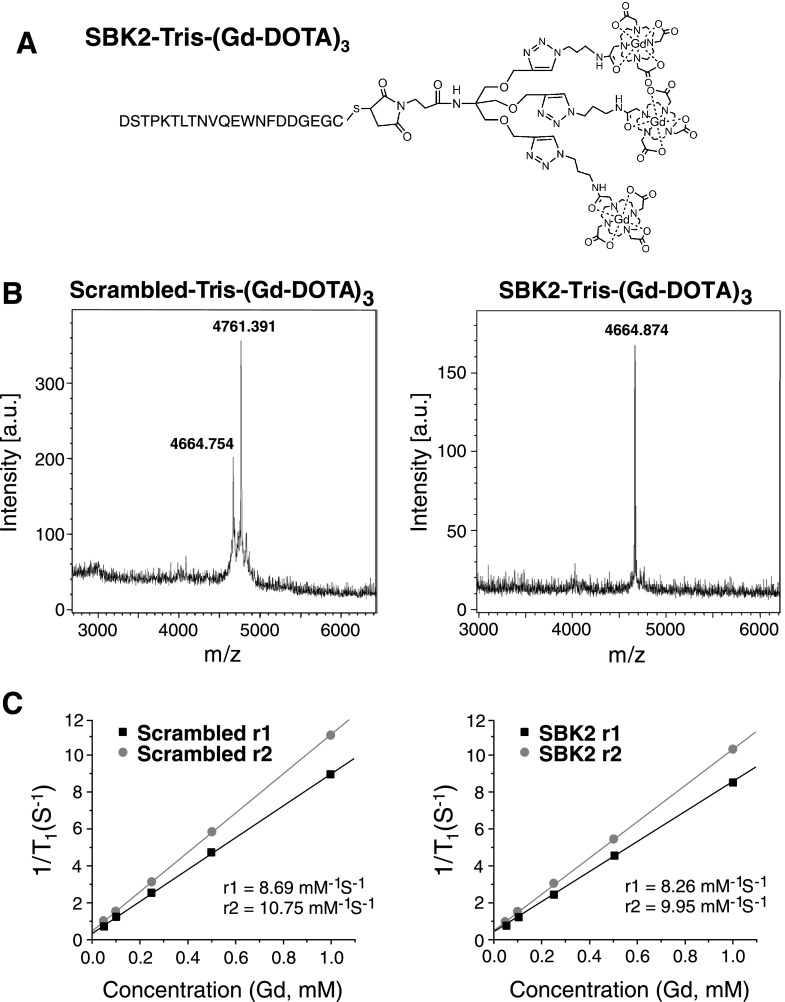
The SBK2 peptide was conjugated to Gd-DOTA using an increased molar ratio of Gd-DOTA monoamide to peptide to generate an MR-visible probe [SBK2-Tris-(Gd-DOTA)3] for effective targeted contrast enhancement (Figure 1A). The final product had good water solubility. A scrambled version of the SBK2 peptide was used to generate a nontargeted control agent [scrambled Tris-(Gd-DOTA)3]. The final products were characterized by MALDI-TOF (Figure 1B) and ICP-OES and with a Bruker Minispec Relaxometer. Both targeted and control contrast agents had similar relaxivity (Figure 1C).
Figure 1.
Structure and characterization of PTPµ targeted and control contrast agents. (A) The structure of the targeted contrast agent SBK2-Tris-(Gd-DOTA)3 is shown. (B) MALDI-TOF mass spectra characterization of the nontargeted control [scrambled Tris-(Gd-DOTA)3] and targeted contrast agent [SBK2-Tris-(Gd-DOTA)3]. (C) Plots of relaxivity versus the concentration of gadolinium (mM) of the contrast agents scrambled Tris-(Gd-DOTA)3 and SBK2-Tris-(Gd-DOTA)3.
MR Tumor Molecular Imaging with SBK2-Tris-(Gd-DOTA)3
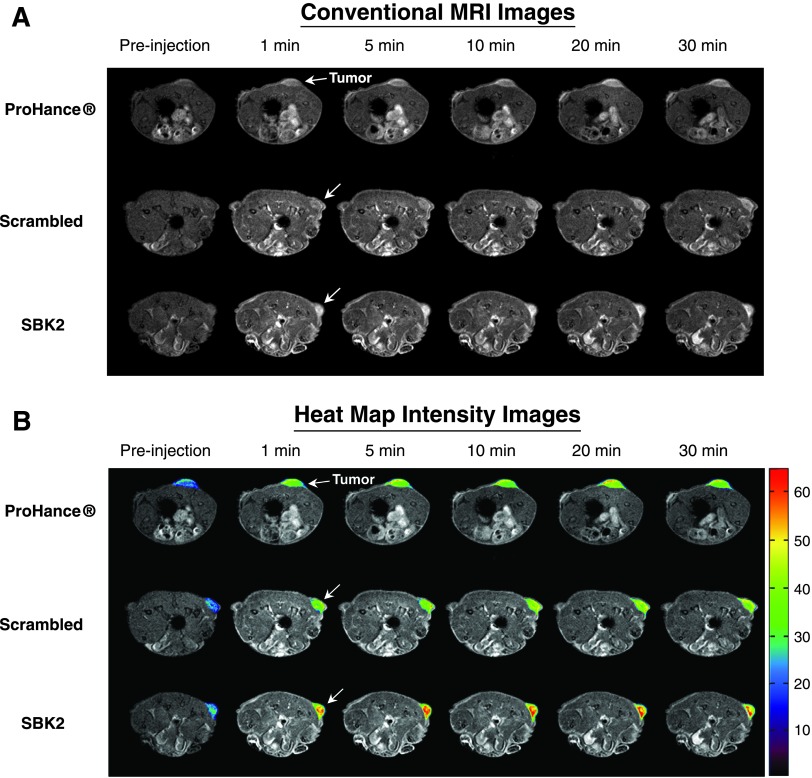
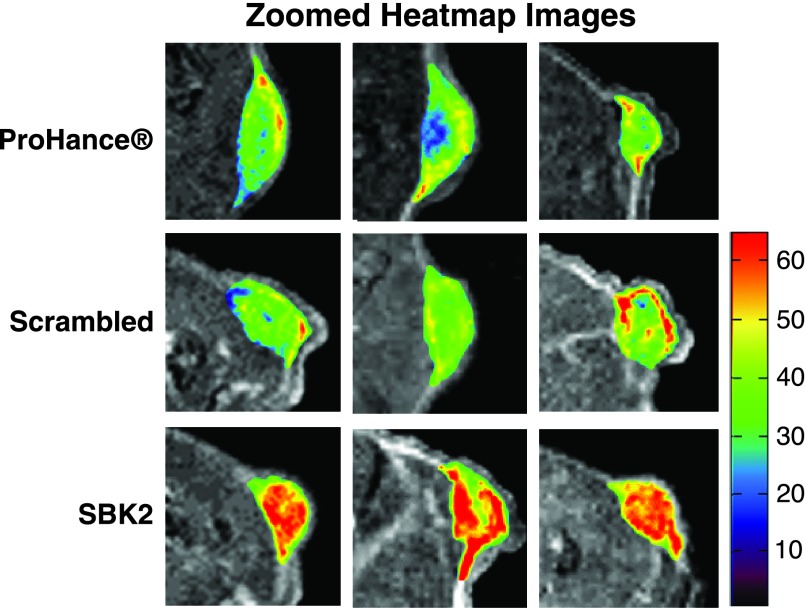
Athymic (nu/nu) mice bearing LN-229 flank tumors (2–5 weeks post-implantation) were used for MRI studies. 2D axial T 1-weighted gradient echo images of mice were acquired before injection of contrast agents, then at 1-, 5-, 10-, 15-, 20-, 25-, and 30-minute intervals following intravenous injection of either ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 administered at a dose of 0.1 mmol/kg (Figure 2). Representative examples of 2D axial images are shown in Figure 2A for each contrast agent, with tumors indicated by white arrows. In comparison with the nontargeted contrast agents, the targeted SBK2-Tris-(Gd-DOTA)3 agent resulted in greater enhancement of tumors that was maintained for a longer period of time (Figure 2A). This improved tumor enhancement is better illustrated with heat map intensity overlays applied to the tumor regions (Figure 2B). The color key indicates the relative enhancement level from lowest (purple) to highest (red). The tumor enhancement generated by the SBK2-Tris-(Gd-DOTA)3 was greater than that observed for the scrambled or ProHance agents (Figure 2B). Digitally magnified MRI and heat map intensity images show greater detail of the tumor enhancement achieved with each contrast agent for three separate tumors per agent (Figure 3). The entire tumor was enhanced over adjacent tissues with all of the contrast agents evaluated. ProHance and scrambled Tris-(Gd-DOTA)3 resulted in enhanced contrast of the tumor edge compared with the tumor center. This heterogeneous pattern is not due to necrosis because the LN-229 tumors grow slowly and do not exhibit necrosis in the time frame examined here. In comparison, the SBK2-Tris-(Gd-DOTA)3 resulted in the most intense tumor enhancement, particularly in the tumor center. These results suggest that active targeting of the tumor with SBK2-Tris-(Gd-DOTA)3 achieved greater penetration and contrast of the entire tumor.
Figure 2.
The SBK2 targeted contrast agent greatly improves the enhancement of LN-229 tumors. (A) Representative T 1-weighted axial 2D gradient images of LN-229 flank tumor-bearing mice before (pre-injection) and at 1, 5, 10, 20, and 30 minutes after the intravenous injection of ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 at 0.1mmol Gd/kg [n = 7 tumors from four mice for ProHance; n = 11 tumors from six mice for both SBK2-Tris-(Gd-DOTA)3 and SBK2-Tris-(Gd-DOTA)3]. White arrow indicates tumor. (B) Axial 2D gradient images shown in A with heat map overlays on the tumor to indicate contrast intensity. Color-coded key indicates relative level of contrast intensity with red being the most intense.
Figure 3.
Digitally magnified axial 2D gradient images of tumors with heat map intensity overlays. Zoomed views of 5-minute T 1-weighted axial 2D gradient images of LN-229 flank tumor-bearing mice illustrate details of the heat map intensity overlay in the tumors. Three representative examples are shown for the ProHance control, scrambled Tris-(Gd-DOTA)3 (scrambled), and SBK2-Tris-(Gd-DOTA)3 (SBK2) probes. Color-coded key indicates relative level of contrast intensity with red being the most intense.
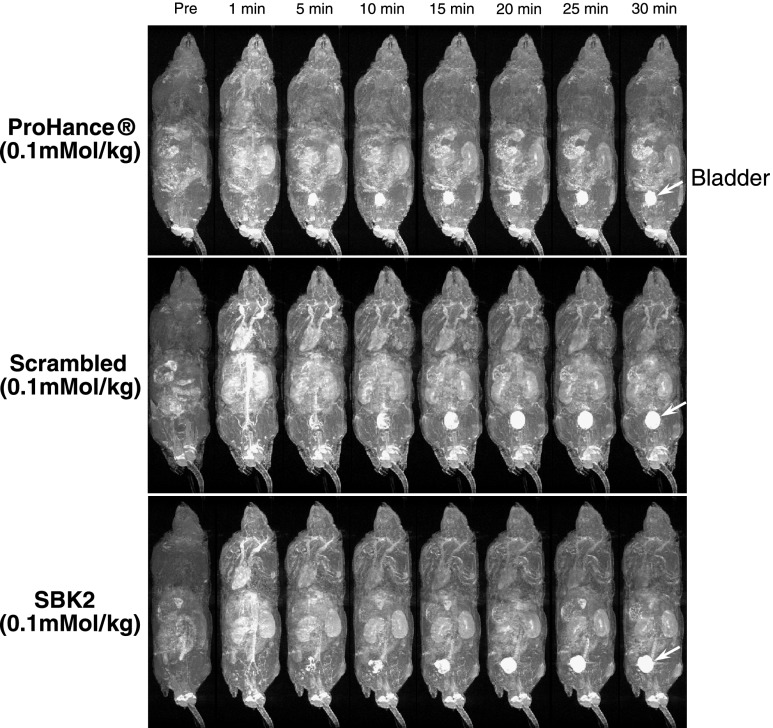
To examine the biodistribution of each contrast agent, 3D longitudinal T 1-weighted gradient echo images were also acquired for each mouse (Figure 4). Contrast enhancement was observed in kidneys and other major organs at the earliest time points but was significantly decreased at later time points. Alternatively, enhancement of the urinary bladder (white arrows in Figure 4) increased over time with all labeling agents, suggesting that the unbound agents were being cleared through renal filtration.
Figure 4.
Distribution and clearance of control or PTPµ targeted contrast agents following intravenous administration in xenograft mice. Representative T 1-weighted longitudinal 3D maximum intensity projection images of mice bearing LN-229 flank tumors before (Pre) and at 1, 5, 10, 15, 20, 25, and 30 minutes after intravenous injection of ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 contrast agents at 0.1 mmol Gd/kg in nude mice [n = 4 for ProHance; n = 6 for both scrambled Tris-(Gd-DOTA)3 and SBK2-Tris-(Gd-DOTA)3]. Arrow indicates bladder localization.
Quantitative Tumor Signal Analysis
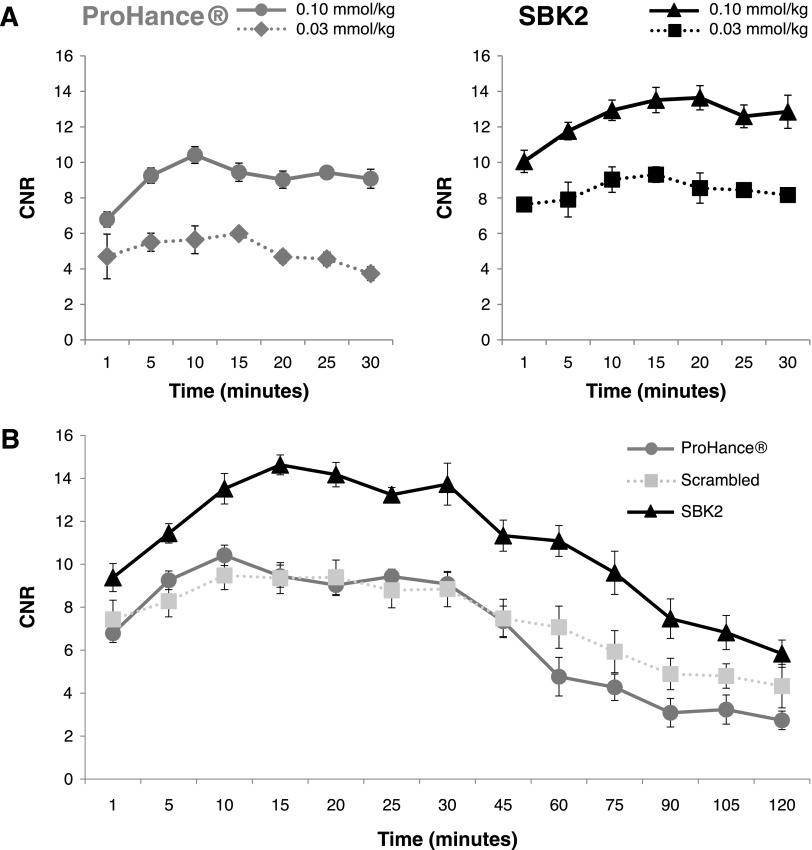
Comparison of tumor enhancement was done using quantitative signal analysis. Regions of interest were drawn over the tumor and adjacent muscle tissue for each mouse, and the CNR was determined at each time point following injection of contrast agents. Dose-response tumor signal analysis was performed for ProHance or SBK2-Tris-(Gd-DOTA)3 administered at 0.03 mmol/kg or 0.1 mmol/kg (Figure 5A). The results indicate that the tumor contrast achieved with SBK2-Tris-(Gd-DOTA)3 at 0.1 mmol/kg was significantly greater over the entire 30-minute period than that achieved with ProHance at 0.1 mmol/kg (P < .05). Similar results were observed when comparing SBK2-Tris-(Gd-DOTA)3 administered at 0.03 mmol/kg with ProHance (0.03 mmol/kg) at 10 to 30 minutes post-injection (P < .05). Of interest, the lowest dose of SBK2-Tris-(Gd-DOTA)3 (0.03 mmol/kg) resulted in tumor contrast that was similar to the contrast achieved with the highest dose of ProHance (0.1 mmol/kg). These results suggest that the targeted SBK2-Tris-(Gd-DOTA)3 is superior for tumor enhancement.
Figure 5.
Quantitation of LN-229 flank tumor enhancement following intravenous administration of control or targeted contrast agents in xenograft mice. (A) Dose-response plots of LN-229 flank tumor CNR with ProHance or SBK2-Tris-(Gd-DOTA)3 administered at 0.03 mmol Gd/kg (n = 3 per condition) or 0.1 mmol Gd/kg [n = 7 tumors from four mice for ProHance; n = 11 tumors from six mice for SBK2-Tris-(Gd-DOTA)3]. (B) Comparison of LN-229 flank tumor CNR over a 2-hour period following intravenous administration of ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 contrast agents (0.1 mmol Gd/kg; n = 6 tumors from three mice per condition). Data shown as means ± SEM.
To evaluate the dynamics of tumor enhancement over a longer period of time, quantitative signal analysis was performed in tumors over a 2-hour period following injection of contrast agents (Figure 5B). The targeted agent SBK2-Tris-(Gd-DOTA)3 showed improved tumor CNR when compared with the nontargeted scrambled Tris-(Gd-DOTA)3 or ProHance. SBK2-Tris-(Gd-DOTA)3 resulted in an approximate 55% increase in tumor CNR over scrambled Tris-(Gd-DOTA)3 or ProHance at 15 to 45 minutes post-injection (P < .001). At 60 to 120 minutes post-injection, the ProHance cleared more rapidly than SBK2-Tris-(Gd-DOTA)3 resulting in greater than 110% increase in SBK2-Tris-(Gd-DOTA)3 tumor CNR compared with ProHance (P < .001). The SBK2-Tris-(Gd-DOTA)3 tumor CNR was approximately 53% greater than the scrambled Tris-(Gd-DOTA)3 tumor CNR at 60 to 90 minutes (P < .001). The larger size of the scrambled Tris-(Gd-DOTA)3 probe in comparison with ProHance may be partially responsible for its slower clearance rate between 60 and 120 minutes.
Biodistribution in Tissues
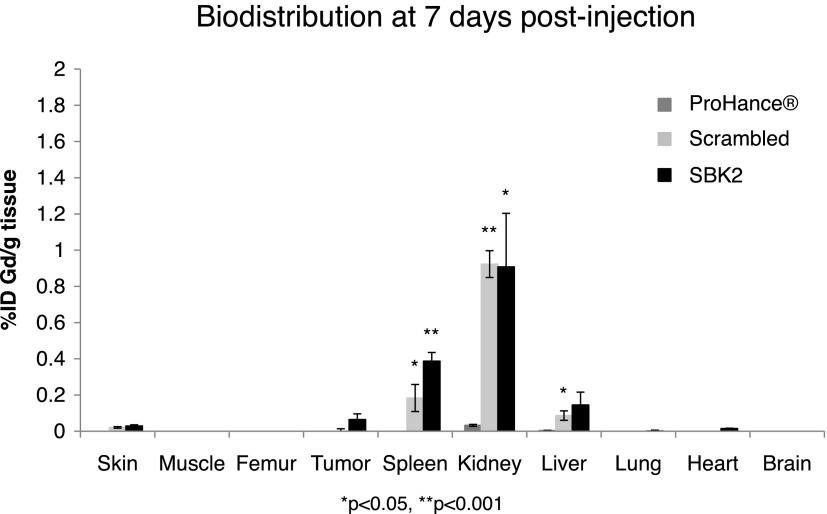
Tissue clearance of the targeted probes is important to avoid the accumulation of toxic gadolinium ions. The biodistribution of gadolinium was examined in the brain, heart, kidneys, liver, lungs, spleen, skin, muscle, femur, and tumor at 7 days post-injection of the ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 (Figure 6). The gadolinium retention of scrambled Tris-(Gd-DOTA)3 and SBK2-Tris-(Gd-DOTA)3 was similar in all tissues. Low residual accumulation of approximately 0.39%, 0.91%, and 0.14% of injected targeted SBK2-Tris-(Gd-DOTA)3 per gram of tissue was detected in spleen, kidney, and liver, while less than 0.1% was detected in all other tissues, including tumor. Accumulation of gadolinium from ProHance was less than 0.1% for all tissues examined.
Figure 6.
Biodistribution of gadolinium in the major organs and tissues of mice at 7 days after intravenous administration of ProHance, scrambled Tris-(Gd-DOTA)3, or SBK2-Tris-(Gd-DOTA)3 contrast agents at a dose of 0.1 mmol Gd/kg in nu/nu athymic mice bearing LN-229 flank tumors (n = 3 mice per condition). Data shown as means ± SEM (*P < .05, **P < .001 compared to ProHance).
Histology/Tumor Binding Specificity
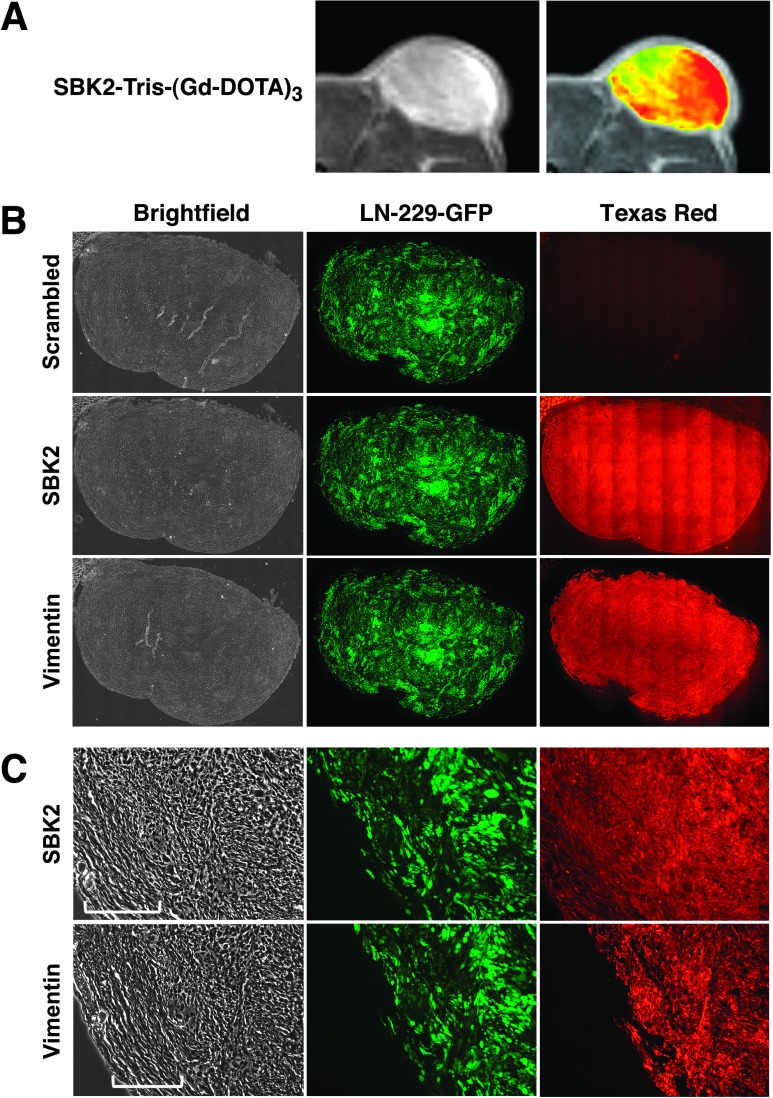
LN-229-GFP flank tumors used for MRI were excised, fixed, and cryosectioned for histologic analysis. Zoomed MR and corresponding heat map images for the tumor are shown (Figure 7A). The heterogeneous signal enhancement in the heat map overlay suggests that the target of the SBK2-Tris-(Gd-DOTA)3 probe was concentrated in the tumor center and at the outer edge of the tumor. The corresponding flank tumor sections were incubated with SBK2-Texas Red or scrambled Texas Red probes [4,5]. SBK2-Texas Red labeled the entire tumor with greatest intensity in the tumor center (Figure 7B). This pattern is similar to that observed in the MR images following labeling with the SBK2-Tris-(Gd-DOTA)3 probe. In contrast, the scrambled Texas Red probe did not label the tumor (Figure 7B). These data indicate that the SBK2 probe recognized the flank tumors both in vitro and in vivo. Anti-human vimentin antibody was used to label all the human cells in the mouse flank. There is co-registration of the GFP-positive and vimentin-positive cells (Figure 7, A and B), demonstrating that all GFP-positive cells in the tumor xenograft are of human origin. The LN-229 flank tumor is encapsulated by GFP-negative cells and matrix, as shown in the magnified bright-field images in Figure 7C. The proteolyzed extracellular fragment of PTPµ is retained both in the main tumor and, to a lower extent, in the encapsulated region, as illustrated by labeling with the SBK2-Texas Red probe (Figure 7C). Similar gradients of PTPµ extracellular fragment, as detected by the SBK2 probe, have been observed previously [4].
Figure 7.
Tumor labeling and histology. LN-229-GFP flank tumor MRI and histologic sections show PTPµ probe labeling of tumors. (A) Zoomed grayscale and heat map images are shown by MRI following administration of SBK2-Tris-(Gd-DOTA)3. (B) The corresponding histologic sections were labeled with SBK2-Texas Red or scrambled Texas Red probes, or anti-human vimentin antibody. Images were acquired across the entire tissue using bright-field and fluorescence optics for fluorescein (GFP) and Texas Red (probe and antibody). The resultant images were tiled and flattened to form a single composite image using Metamorph software. Anti-human vimentin antibody recognizes the human glioma tumor cells. There is co-registration of the GFP-positive and vimentin-positive cells in these tumors. (C) Zoomed images of the histologic sections are shown for each type of staining. The bracketed region in C indicates the encapsulated region surrounding the tumor.
Discussion
In this publication, we demonstrate that the SBK2 probe can achieve better tumor contrast enhancement than the nonspecific clinical contrast agent currently in use. The SBK2 probe targets PTPµ molecules enriched in the tumor microenvironment and, in so doing, improves the specificity of tumor enhancement. We previously conjugated the SBK2 peptide to a fluorescent molecule and were able to detect it in GBM cells in vivo using preclinical imaging modalities [4,6]. We now demonstrate that the SBK2 probe can be used for clinical imaging (MRI) in addition to its aforementioned use in preclinical and histologic staining of GBM tumors.
PTPµ is a cell-cell adhesion molecule that mediates adhesion through a homophilic binding mechanism. Adhesive domains within the extracellular segment of PTPµ are critical for mediating homophilic binding [9]. The key adhesive domains are retained in the PTPµ extracellular fragment that accumulates in association with glial brain tumors, specifically GBM, but not in normal brain [4]. The presence of this extracellular adhesive fragment prompted the development of the SBK2 probe to bind homophilically to the fragments to label GBM tumor cells and their microenvironment [4,6]. In this study, we demonstrate that SBK2-Tris-(Gd-DOTA)3 intensely labeled both the tumor core and the edge within 5 minutes (Figures 2 and 3). The signal remained strong for 90 minutes post-injection and provided greater contrast than ProHance for 2 hours after injection (Figure 5). We conclude that the kinetics of SBK2-Tris-(Gd-DOTA)3 labeling of GBM cells in vivo will allow ample time to perform MRI in a clinical setting.
To compare ProHance to the molecular imaging agent SBK2-Tris-(Gd-DOTA)3 and its scrambled control, all agents were dosed on the basis of gadolinium content/kg of animal weight. This experimental paradigm allows direct comparison of the three signals because it should equalize the amount of gadolinium per animal, even though it would reduce the targeting peptide in the SBK2 probe by approximately one third. The SBK2 probe provided enhanced contrast over both ProHance and the scrambled probe, which also contained the three gadolinium groups. The presence of the three gadolinium groups and the higher relaxivity of the SBK2 probe contribute to its ability to function as a better contrast agent than ProHance. The kinetics suggest that the binding of SBK2 to the target has a fast off rate, likely because it binds homophilically to PTPµ fragments in the tumor microenvironment, which is a lower affinity interaction than that observed in high affinity binding such as with antibodies. This is advantageous because it promotes probe clearance from the tumor tissue and body.
The SBK2 probe is specifically and actively targeting GBM cells and is not exploiting the enhanced permeability and retention (EPR) effect to reach the tumor microenvironment. The EPR effect is a means of passively targeting tumor tissue and is currently used to direct lipophilic therapeutic and imaging macromolecules to tumors [10,11]. The SBK2 probe uses an active targeting strategy by homophilically binding to the PTPµ fragment in the GBM tumor microenvironment and, at 4.7 kDa, is too small to be retained through an EPR mechanism [11]. Given that the scrambled probe, which has similar lipophilicity and size to the SBK2 probe, does not target the tumor as efficiently further confirms that EPR is not responsible for the binding of SBK2 probe to the GBM cells.
The SBK2 probe offers several advantages over other MR molecular imaging reagents used to target cell surface proteins (see [12–15]), including a fast time course to achieve peak labeling and rapid clearance from tumor tissue. The time course of peak labeling with the SBK2 probe occurs within 15 minutes (Figure 5), while the SBK2 probe levels decline rapidly in the tumor tissue, typically at 2 hours (Figure 5) with clearance from most tissues of the body by 7 days (Figure 6). Peak binding for other cell surface probes occurs within 1 to 4 hours post-injection [12–15], and in the case of integrin binding RGD liposomes and nanoparticles, the rate of clearance is much slower, beginning 24 to 48 hours after injection [12,15].
The SBK2 probe may also function better than antibodies linked to contrast agents that are currently in development as MR molecular imaging reagents [3,16,17]. As a 4.7-kDa peptide-based probe, SBK2-Tris-(Gd-DOTA)3 is much smaller than an antibody and, therefore, is expected to have superior tissue penetration. Because PTPµ is endogenously processed and continuously deposited in tumor tissue to give rise to the extracellular fragment product, no additional exogenous amplification is required to visualize the probe. An anti-epidermal growth factor receptor (EGFR) antibody developed to visualize GBM cells requires an exogenous amplification step to provide sufficient contrast enhancement in vivo [3]. This added step necessitates that the anti-EGFR antibody be injected 5 hours before the injection of the contrast agent [3], making it less suitable than the SBK2 probe for use in a clinical setting.
To conclude, the SBK2-Tris-(Gd-DOTA)3 probe is a rapid and specific marker of glioma cells using MRI. This study has established that the targeted SBK2-Tris-(Gd-DOTA)3 probe is effectively delivered to LN-229 tumors for enhanced contrast and can be quickly cleared from the animal. A fluorescent SBK2 probe can cross the blood-brain barrier, even in areas of the brain where it is largely intact, to label most dispersive cells in GBMtumors [6]. Because GBM cells tend to migrate as chains of cells [6,18] or can exist as “cellular sheets” [19], we speculate that the SBK2-Tris-(Gd-DOTA)3 probe could effectively cross the blood-brain barrier and label these dispersive cell chains using MRI technology. Future studies will explore the use of this probe in orthotopic intracranial glioma tumors and test whether the SBK2 probe used with MRI can achieve the high resolution required to detect chains of dispersive GBM cells.
Acknowledgments
The authors thank Xueming Wu for technical assistance with MRI, Scott Howell for assistance with the heat map images, and Cathy Doller for expert technical support with histology. In addition, the authors also thank Jim Basilion, Chris Flask, Jeff Duerk, and Mark Griswold for critical reading of the manuscript.
Footnotes
This research was supported by National Institutes of Health grant R01-NS063971 (S.M.B.-K. and Z.-R.L.). This work was also supported by the Tabitha Yee-May Lou Endowment Fund for Brain Cancer Research, the Case Center for Imaging Research, and the Athymic Animal Core Facility of the Case Comprehensive Cancer Center (P30 CA43703).
References
- 1.Upadhyay N, Waldman AD. Conventional MRI evaluation of gliomas. Br J Radiol. 2011;(84 Spec No. 2):S107–S111. doi: 10.1259/bjr/65711810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kircher MF, Willmann JK. Molecular body imaging: MR imaging, CT, and US. Part I. Principles. Radiology. 2012;263:633–643. doi: 10.1148/radiol.12102394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shazeeb MS, Sotak CH, DeLeo M, III, Bogdanov A., Jr Targeted signal-amplifying enzymes enhance MRI of EGFR expression in an orthotopic model of human glioma. Cancer Res. 2012;71:2230–2239. doi: 10.1158/0008-5472.CAN-10-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, Sloan AE, Miller RH, Basilion JP, Brady-Kalnay SM. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12:305–316. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, Brady-Kalnay SM. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69:6960–6968. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Tan M, Craig SE, Basilion JP, Lu ZR, Wilson DL, Brady-Kalnay SM. Single cell molecular recognition of migrating and invading tumor cells using a targeted fluorescent probe to receptor PTPmu. Int J Cancer. 2012;132:1624–1632. doi: 10.1002/ijc.27838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacob G, Dinca EB. Current data and strategy in glioblastoma multiforme. J Med Life. 2009;2:386–393. [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Burden-Gulley SM, Yu GP, Tan M, Lindner D, Brady-Kalnay SM, Lu ZR. Synthesis and evaluation of a peptide targeted small molecular Gd-DOTA monoamide conjugate for MR molecular imaging of prostate cancer. Bioconjug Chem. 2012;23:1548–1556. doi: 10.1021/bc300009t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aricescu AR, Siebold C, Jones EY. Receptor protein tyrosine phosphatase micro: measuring where to stick. Biochem Soc Trans. 2008;36:167–172. doi: 10.1042/BST0360167. [DOI] [PubMed] [Google Scholar]
- 10.Maeda H. Macromolecular therapeutics in cancer treatment: the EPR effect and beyond. J Control Release. 2012;164:138–144. doi: 10.1016/j.jconrel.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 11.Arias JL. Drug targeting strategies in cancer treatment: an overview. Mini Rev Med Chem. 2011;11:1–17. doi: 10.2174/138955711793564024. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Su B, Meng S, Ju L, Yan L, Ding Y, Song Y, Zhou W, Li H, Tang L, et al. RGD-targeted paramagnetic liposomes for early detection of tumor: in vitro and in vivo studies. Eur J Radiol. 2011;80:598–606. doi: 10.1016/j.ejrad.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 13.Lim EK, Kim HO, Jang E, Park J, Lee K, Suh JS, Huh YM, Haam S. Hyaluronan-modified magnetic nanoclusters for detection of CD44-overexpressing breast cancer by MR imaging. Biomaterials. 2011;32:7941–7950. doi: 10.1016/j.biomaterials.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 14.Meier R, Henning TD, Boddington S, Tavri S, Arora S, Piontek G, Rudelius M, Corot C, Daldrup-Link HE. Breast cancers: MR imaging of folate-receptor expression with the folate-specific nanoparticle P1133. Radiology. 2010;255:527–535. doi: 10.1148/radiol.10090050. [DOI] [PubMed] [Google Scholar]
- 15.Wu PC, Su CH, Cheng FY, Weng JC, Chen JH, Tsai TL, Yeh CS, Su WC, Hwu JR, Tzeng Y, et al. Modularly assembled magnetite nanoparticles enhance in vivo targeting for magnetic resonance cancer imaging. Bioconjug Chem. 2008;19:1972–1979. doi: 10.1021/bc800092w. [DOI] [PubMed] [Google Scholar]
- 16.Quan G, Du X, Huo T, Li X, Wei Z, Cui H, Chang X, Cheng Y, Ye X, Cheng H. Targeted molecular imaging of antigen OC183B2 in ovarian cancers using MR molecular probes. Acad Radiol. 2010;17:1468–1476. doi: 10.1016/j.acra.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Serda RE, Adolphi NL, Bisoffi M, Sillerud LO. Targeting and cellular trafficking of magnetic nanoparticles for prostate cancer imaging. Mol Imaging. 2007;6:277–288. [PubMed] [Google Scholar]
- 18.Burden-Gulley SM, Qutaish MQ, Sullivant KE, Lu H, Wang J, Craig SE, Basilion JP, Wilson DL, Brady-Kalnay SM. Novel cryo-imaging of the glioma tumor microenvironment reveals migration and dispersal pathways in vivid three-dimensional detail. Cancer Res. 2011;71:5932–5940. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger PC, Scheithauer BW, Vogel FS. Surgical Pathology of the Nervous System and Its Coverings. 3rd ed. New York: Wiley; 2002. [Google Scholar]