Abstract
PURPOSE: There is accumulating evidence that galanin receptors (GALRs) may be tumor suppressors in head and neck squamous cell carcinoma (HNSCC). Promoter methylation status and gene expression were assessed in a large panel of primary tumors, based on the hypothesis that CpG hypermethylation might silence the galanin gene. EXPERIMENTAL DESIGN: Galanin expression was examined using reverse transcription-polymerase chain reaction (PCR). The methylation status of the galanin promoter was studied using bisulfate sequencing and methylation-specific PCR. UM-SCC-54 was stably transfected to express galanin. RESULTS: Galanin expression was absent in 3/12 (25.0%) UM-SCC cell lines, whereas three nonmalignant cell lines had stable expression. Galanin methylation was found in 24/100 (24.0%) cases. HNSCC tumor specimens was significantly correlated with the GALR1 methylation status (P = 1.88E-06). The presence of galanin promoter hypermethylation was statistically correlated with a decrease in disease-free survival (log-rank test, P = 6.02E-05). A multivariate logistic regression analysis showed that methylation of galanin and methylation of the gene pair galanin and GALR1 had an odds ratio for recurrence of 8.95 [95% confidence interval (CI), 2.29–35.03] and 23.84 (95% CI, 2.74–207.17), respectively. UM-SCC-54 cells that are GALR1-proficient but have hypermethylated galanin exhibited suppressed cell proliferation following exogenous expression of galanin. CONCLUSIONS: Association of frequent promoter hypermethylation and gene silencing with poor survival, combined with growth suppression of HNSCC cells after forced gene expression, supports the hypothesis that galanin acts as a tumor suppressor. These data suggest that galanin and GALR1 are potential therapeutic targets and prognostic factors.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most frequent type of cancer and affects ∼500,000 patients per year worldwide [1]. Various factors are linked to the development of HNSCC; sex, tobacco smoking, alcohol consumption, and human papillomavirus (HPV) infection are major risk factors for the disease [2]. The prognosis for patients with HNSCC is still poor today despite tremendous technical advances in surgical treatment, radiotherapy, and chemotherapy. The development of novel therapies for treating HNSCC is therefore urgently required [3]. Current progress in molecular biology should make it possible to create new strategies for cancer, such as molecular targeting therapy. Approximately 30% of all pharmaceuticals on the market today exert their therapeutic effect by interacting with a G protein. coupled receptor (GPCR) [4]. Several studies have suggested that neuropeptides function as tumor suppressor genes and possess potent antitumor activity in an autocrine and/or paracrine manner [5,6]. Galanin is typically thought of as a neuropeptide because it widely expressed in the central and peripheral nervous system and, most abundantly, in the hypothalamus, where it may serve in the regulation of anterior pituitary hormones [7–9]. Galanin binding to the extracellular regions of the GPCRs galanin receptor 1 (GALR1), GALR2, and GALR3 activates the receptors and stimulates numerous signal transduction and integration pathways [10,11]. Galanin is involved in the regulation of many physiological functions, such as feeding, metabolism, and body weight control [12].
Our studies and those of others support a growth regulatory function for galanin and its receptors in epithelial cancer [13–15]. Antibody blockade of GALR1 enhances proliferation of HNSCC cells [13], and galanin and GALR1 induce a marked and prolonged extracellular signal-regulated kinase (ERK)1/2 activation, up-regulation of p27Kip1 and p57Kip2, down-regulation of cyclin D1, and consequent inhibition of cell proliferation [14]. Furthermore, squamous cell cancers and cell lines that fail to express GALR1 exhibit hypermethylation of CpG islands in the promoter region of this gene [15].
This study shows the first evidence that loss of galanin expression is associated with hypermethylation of the promoter region and that expression can be restored after treatment with the demethylating agent 5-azacytidine and the histone deacetylase (HDAC) inhibitor trichostatin A (TSA). Moreover, assessment of tumor specimens confirmed that hypermethylation is common in patients' tumors, with associated GALR1 methylation status and decreased disease-free survival (DFS). Finally, the restoration of galanin expression in HNSCC cells resulted in the inhibition of colony formation in response to galanin stimulation, thereby supporting the hypothesis that this plays a tumor suppressive role in HNSCC.
Materials and Methods
Tumor Samples and Cell Lines
Tumor specimens were obtained at surgery from 100 primary HNSCC samples. All patients were treated at the Department of Otolaryngology, Hamamatsu University School of Medicine. All patients provided written informed consent under a protocol approved by the Institutional Review Boards at the Hamamatsu University School of Medicine. Clinical information including age, sex, tumor site, smoking status, alcohol exposure, tumor size, lymph node status, and stage grouping were obtained from the clinical records. The average age was 63.9 years (range, 39–90), and the male/female ratio was 78:22. The primary tumor was located in the oral cavity (n = 34), the hypopharynx (n = 24), the larynx (n = 20), the oropharynx (n = 11), and the paranasal cavity (n = 11). DNA and cDNA from 12 UM-SCC cell lines were established from patients at the University of Michigan [16]. The HPV16 E6/E7-transformed oral keratinocyte cell line HOK16B was a gift from Dr No Hee Park of the University of California, Los Angeles School of Dentistry [17].
Bisulfite Modification and Methylation-Specific Polymerase Chain Reaction Analysis
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Bisulfite modification of genomic DNA was carried out as described earlier [15]. Bisulfite-treated DNA was polymerase chain reaction (PCR) amplified with two pairs of methylation-specific PCR (MSP) primers designed for the promoter region of the galanin gene. The MSP primers for methylated DNA were MSP-galanin-F (5′-TGA CGC GAT TTC GGG CGG TT-3′) and MSP-galanin-R (5′-TAT CCG CCG CCC GAT ATA AC-3′). For unmethylated DNA amplification, the following unmethylated MSP (UMSP) primers were used: UMSP-galanin-F (5′-TGA TGT GAT TTT GGG TGG TT-3′) and UMSP-galanin-R (5′-TAT CCA CCA CCC AAT ATA AC-3′). MSP/UMSP was performed with 25 ng of DNA solution containing 1x PCR buffer, 10x Mg, 10 mM deoxyribonucleotide triphosphate (dNTP), 3% DMSO, 1U Fast Start Polymerase (Roche, Penzberg, Germany), and 10 µM of each primer for a total volume of 25 µl. The PCR conditions were 94°C for 5 minutes; 38 cycles at 94°C for 30 seconds, 58.0°C for 30 seconds, and 72°C for 40 seconds; and a final extension at 72°C for 5 minutes. The 82-bp PCR products were separated by electrophoresis through a 9% acrylamide gel, stained with ethidium bromide. Previously described primers and conditions were used to analyze the methylation status of the GALR1 gene [15], p16 gene [18], RASSF1A gene [19], MGMT gene [20], DAPK gene [20], and DCC gene [21].
Bisulfite Sequencing Analysis for Galanin
Bisulfite-treated DNA was amplified by bisulfite sequencing PCR with a pair of primers that were specific for modified upper strand DNA without any CpG sites in their primers. The primer set was 5′-GTT TAT GAT GGT TTA GAG GA-3′ (forward) and 5′-ACC AAA ACC CTA ACA AAA CC-3′ (reverse). The PCR conditions were 94°C for 5 minutes; 40 cycles at 94°C for 30 seconds, 55.0°C for 30 seconds, and 72°C for 40 seconds; and a final extension at 72°C for 5 minutes. The PCR products were 430 bp long. Bisulfite sequencing analysis was carried out as described previously [15].
RNA Extraction and Quantitative Reverse Transcription-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and treated with RNase-Free DNase (Qiagen). The cDNA was generated from DNase-treated total RNA using random primers (Invitrogen, Carlsbad, CA) with SuperScript II Reverse Transcriptase (Invitrogen). Primers were designed as follows: galanin forward, 5′-AAA CGA GGC TGG ACC CTG AAC-3′; galanin reverse, 5′-ATT CTT GTC GCT GAA TGA CCT GTG-3′, GALR1 forward, 5′-TGCACCTTCGTCTTCGGCTAC-3′; GALR1 reverse, 5′-CAC CAG AAC TGT CTG TGC AGT CTT-3′, GALR2 forward, 5′-CGA CCT GTG TTT CAT CCT GTG-3′; GALR2 reverse, 5′-GGT AGC GGA TGG CCA GAT A-3′; GALR3 forward, 5′-CTG ATG CCC AGA ACA TTT CAC TG-3′; GALR3 reverse, 5′-TGT GCC CAG CAG GAA GAT TAG G-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GCA CCG TCA AGG CTG AGA AC-3′; GAPDH reverse, 5′-TGG TGA AGA CGC CAG TGG A-3′. Quantitative reverse transcription (RT)-PCR was performed with the TaKaRa Thermal Cycler Dice Real Time System TP800 (TaKaRa, Tokyo, Japan). For each PCR evaluation, 2 µl of diluted cDNA, 12.5 µl of SYBR Premix Ex Taq Perfect Real Time (TaKaRa), and 0.5 µl of the primers were added to a final volume of 25 µl. The thermal cycler conditions were one cycle of denaturing at 95°C for 10 seconds, followed by 45 cycles of denaturing at 95°C for 5 seconds and annealing/extension at 60°C for 30 seconds. Analysis was performed with Thermal Cycler Dice Real Time System TP800 Software Ver. 1.03A (TaKaRa) following the manufacturer's instructions. The mRNA expression of the target genes was normalized to GAPDH mRNA expression for comparisons between samples.
Reactivation of Galanin Expression
Cultures were incubated either for 48 hours with 5-azacytidine (15 and 30 µg/ml; Sigma, St Louis, MO), an inhibitor of DNA methyltransferase, for 24 hours with 300 nM TSA (Sigma), an inhibitor of HDAC, or for 48 hours with 5-azacytidine followed by 24 hours with TSA. The medium was then removed and cultures were maintained in standard Dulbecco's modified Eagle's medium, which was replaced every other day [15].
Plasmid Constructs and Growth Suppression Analysis
The UM-SCC-54 cell line, which does not express galanin due to hypermethylation but does express GALR1, GALR2, and GALR3, was used in colony-forming assays. Using Lipofectamine Reagent (Invitrogen), 8 µg of pCMV-SPORT6-preprogalanin mammalian expression construct or pCMV-SPORT6-empty construct and pTRE-2hyg (Clontech, Palo Alto, CA) were used to transfect each 6-cm dish containing 4 x 105 cells seeded 48 hours before transfection. The cells were maintained in Dulbecco's modified Eagle's medium with hygromycin B (180 µg/ml) (Invitrogen) 48 hours after transfection. The surviving colonies were counted 14 days later, after staining with crystal violet (Fisher, Pittsburgh, PA). All experiments were repeated at least three times. Media from pCMV-SPORT6-preprogalanin and pCMV-SPORT6-empty-transfected cells were assessed using a galanin ELISA Kit (Peninsula Labs, San Carlos, CA) according to the manufacturer's directions.
Statistical Analysis
Statistical analyses of the associations between variables were performed by Fisher exact test. Comparisons and tests of the colony formation assay were made by Student's t test. The disease-free interval was measured from the date of the treatment to the date when locoregional recurrence or distant metastasis was diagnosed. DFS probabilities were estimated by the Kaplan-Meier method, and the log-rank test was applied to assess the significance of differences among actuarial survival curves. A multivariate logistic regression analysis was used to identify the predictive value of the prognostic factors, including the age, sex, smoking status, alcohol intake, stage grouping, and methylated genes [22,23]. A significant difference was identified when the probability was less than .05. The statistical analysis was done with the StatMate IV software package (ATMS Co, Ltd, Tokyo, Japan).
Results
MSP, Bisulfite Sequencing Analysis, and Quantitative RT-PCR for Galanin
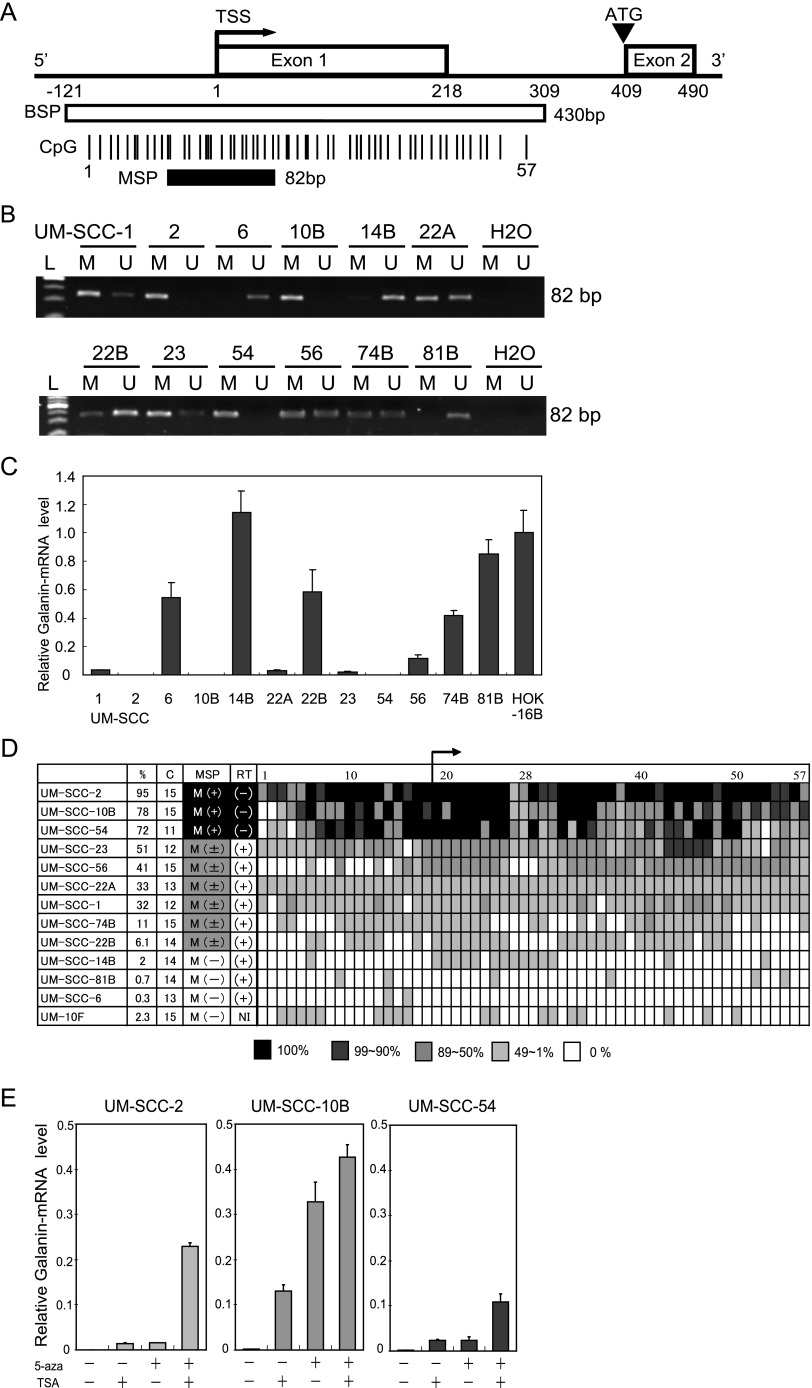
The guanine and cytosine content in the first 500 bp, including the first exon and intron, is 79% and is much higher than the overall guanine and cytosine content of the galanin gene (59%) [24]. Figure 1A indicates the individual CpG sites as vertical lines and numbered from 1 to 57. CpG sites 1 to 18 are located upstream of the transcription start site (TSS) and 19 to 57 are located downstream of TSS. A bent arrow represents the TSS. An arrowhead represents the translation start site (ATG). The nucleotide sequence of the fragment was used for methylation analysis. The first exon of galanin is in the untranslated region (Figure 1A).
Figure 1.
Diagrammatic representation of the galanin gene and its proximal promoter, galanin methylation analysis using theMSP assay, the bisulfite sequencing analysis, and galanin expression by quantitative RT-PCR in HNSCC cell lines. (A) Galanin CpG sites within expanded views of the promoter region relative to the TSS. The white box encloses the 430-bp region that includes the TSS and the 57 individual CpG sites that were examined for frequency of methylation. The region analyzed using MSP is shown by the black box (82 bp). The TSS is represented by a bent arrow. The ATG start codon is represented by an arrowhead. (B) Representative examples of MSP of galanin in UM-SCC cell lines, showing samples that are fully methylated (UM-SCC-2, UM-SCC-10B, and UM-SCC-54), partially methylated (UM-SCC-1, UM-SCC-22A, UM-SCC-22B, UM-SCC-23, UM-SCC-56, and UM-SCC-74B), or unmethylated (UM-SCC-6, UM-SCC-14B, and UM-SCC-81B). (C) The relative mRNA expression as assessed by quantitative RT-PCR of galanin in 12 UM-SCC cell lines. The housekeeping gene GAPDH was run as a control for RNA integrity. (D) Summary of bisulfite sequencing analysis of galanin in UM-SCC cell lines. The numbers at the top row indicate the CpG dinucleotide (labeled 1–57) in the amplicon. The cell line number is shown on the left. The next columns show the rate of methylation of CpG dinucleotides (%) and the number of clones (C) analyzed in each sample. In the column labeled MSP, M(+) indicates positive for the galanin-methylated form by MSP; M(±) indicates positive for the galanin-methylated and unmethylated form by MSP; M(-) indicates negative for the galanin-methylated form by MSP. The column labeled RT indicates galanin expression by quantitative RT-PCR: (+), positive expression of galanin; (-), negative expression of galanin; NI, no information. (E) The effect of treatment with 5-azacytidine or TSA or both on galanin expression in three cell lines with densely methylated galanin is shown using quantitative RT-PCR. The controls were cells treated similarly but without 5-azacytidine or TSA.
Each sample was tested with both sets of primers (MSP and UMSP) for galanin. These primers were designed on the basis of the bisulfite sequencing data. The forward primers for MSP and UMSP contained 11 to 14 CpG dinucleotides. The reverse primers for MSP and UMSP included 24 to 26 CpG dinucleotides. The MSP assay for galanin correlated with the expression status. All cell lines (UM-SCC-2, UM-SCC- 10B, and UM-SCC-54) with loss of expression showed only methylated alleles. Cell lines with apparently normal galanin mRNA levels showed only unmethylated alleles (UM-SCC-6, UM-SCC-14B, and UM-SCC-81B) or both alleles were detected (UM-SCC-1, UM-SCC-22A, UM-SCC-22B, UM-SCC-23, UM-SCC-56, and UM-SCC-74B). This study analyzed 12 UM-SCC cell lines for the expression of galanin by using quantitative RT-PCR and found that 3 of 12 (25.0%) cell lines (UM-SCC-2, UM-SCC-10B, and UM-SCC-54) had loss of galanin mRNA expression (Figure 1, B and C).
Sodium bisulfate DNA sequencing was carried out to assess the percentage of CpG alleles that are methylated in the galanin 5′ region. The percentage of methylated alleles for each of the 57 different CpG sites are shown in Figure 1D for 12 HNSCC cell lines and one control culture, UM-10F fibroblasts. Bisulfite sequencing was used to more precisely map and determine the density of methylation throughout the galanin promoter. Bisulfite modification of the cell line's DNA and PCR amplification of the galanin promoter region were performed and at least 11 clones were sequenced for each of the cell lines. Quantitative RT-PCR showed that UM-SCC-2, UM-SCC-10B, and UM-SCC-54 had loss of galanin expression (ΔΔCT value, 3.7E.0.5 + 1.9E -0.5, 0.0011 + 0.0001, 0.0010 + 0.0004) and had heavy methylation levels in 57 CpG sites (average, 95.3%, 78.0%, and 71.8%). UM-SCC-1, UM-SCC-6, UM-SCC-14B, UM-SCC-22A, UM-SCC-22B, UM-SCC-23, UM-SCC-56, UM-SCC-74B, and UM-SCC-81B had stable galanin expression (ΔΔCT value, 0.0205 + 0.0027 to 1.1447 + 0.1479) and moderate or low methylation levels ranging from 0.3% to 65.5%. The fibroblast cell line UM-10F had a much lower methylation level (2.3%), in comparison to UM-SCC-10B (78.0%). The concordance between mRNA expression levels and aberrant methylation of galanin was almost complete (Figure 1, C and D).
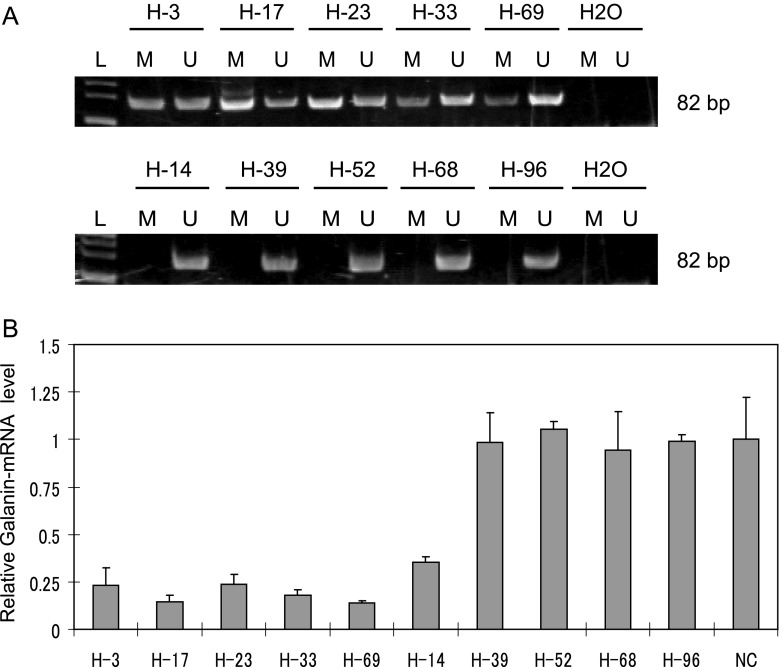
Head and neck tumors were examined for galanin methylation, showing methylated samples (H-3, H-17, H-23, H-33, and H-69) and unmethylated samples (H-14, H-39, H-52, H-68, and H-96) (Figure 2A). mRNA was isolated from frozen tissue to determine whether galanin methylation in tumor specimens was also associated with gene silencing. Galanin expression analyzed by quantitative RT-PCR showed that five tumors, H-3, H-17, H-23, H-33, and H-69 with strong methylation signals, exhibited little mRNA expression. In contrast, four of five (H-39, H-52, H-68, and H-96) tumors without galanin promoter methylation exhibited relatively robust mRNA expression (Figure 2B).
Figure 2.
Galanin methylation analysis using the MSP assay and galanin expression by quantitative RT-PCR in HNSCC tumor samples. (A) Representative examples of MSP of galanin in primary tumors from Hamamatsu University Hospital, showing samples that are methylated (H-3, H-17, H-23, H-33, and H-69) or unmethylated (H-14, H-39, H-52, H-68, and H-96). (B) Relative galanin mRNA expression as assessed by quantitative RT-PCR in 10 tumor specimens that were also evaluated for galanin promoter methylation. Specimens H-3, H-17, H-23, H-33, and H-69 exhibited promoter methylation, but H-14, H-39, H-52, H-68, and H-96 did not.
Galanin Expression after 5-Azacytidine and TSA Treatment
Three cell lines (UM-SCC-2, UM-SCC-10B, and UM-SCC-54) that showed absence of galanin expression and hypermethylation were cultured with 5-azacytidine alone, TSA alone, or 5-azacytidine plus TSA. This facilitated assessing the effect of demethylation induced by 5-azacytidine and an inhibitor of HDAC by TSA. Figure 1E shows that all three cell lines demonstrated restored galanin expression that correlates in association with demethylation and histone acetylation. The results show that galanin expression was upregulated after 5-azacytidine and TSA treatment in comparison to before treatment in the heavily methylated cell lines.
Quantitative RT-PCR for GALRs and Tumor Suppressor Activity of Galanin
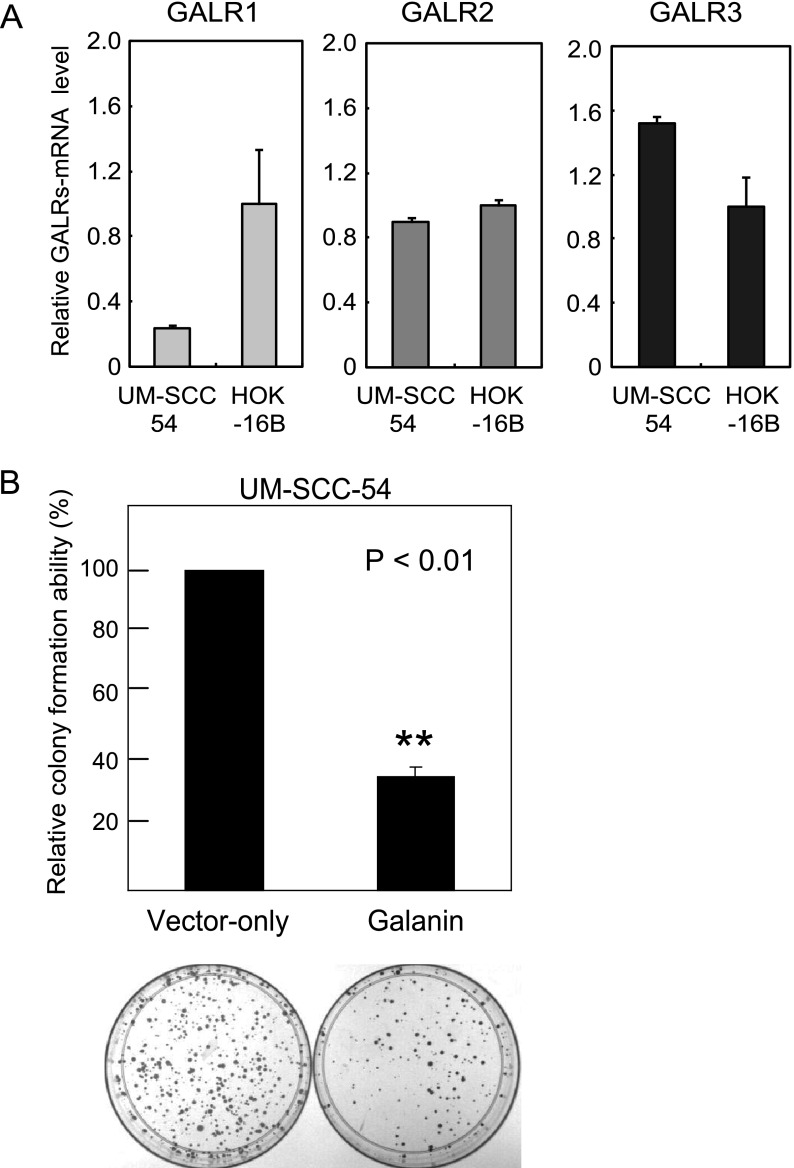
The expression of GALR1, GALR2, and GALR3 was examined in UM-SCC-54 and HOK-16B by quantitative RT-PCR. UM-SCC-54 did not express galanin mRNA exhibiting hypermethylation of the promoter region (Figure 1C). UM-SCC-54 had stable expression in GALR1 (ΔΔCT value, 0.2365 + 0.0129), GALR2 (ΔΔCT value, 0.8950 + 0.0299), and GALR3 (ΔΔCT value, 1.5210 + 0.0407; Figure 3A).
Figure 3.
Expression of GALRs and growth suppression analysis. (A) Representative examples of quantitative RT-PCR for the GALR expression in UM-SCC-54. (B) Relative colony-forming ability for UM-SCC-54. The vector-transfected number of hygromycin B-resistant colonies was set at 100%. Colonies larger than 1 mm were counted, and the results are presented as the means ± SD (bars) of three separate experiments, each performed in triplicate.
The effect of reexpressing galanin was assessed in the UM-SCC-54 cell line in a colony-forming assay. Wild-type preprogalanin in the pCMV-SPORT6 construct or empty pCMV-SPORT6 construct were transfected into the UM-SCC-54 cell line exhibiting galanin hypermethylation and loss of galanin expression. Reconstitution of galanin expression in UM-SCC-54 suppressed relative colony-forming ability by approximately 65% (P = 7.77E-06; Figure 3B). These results were consistent through three independent experiments. ELISAs confirmed increased concentrations of galanin of 1.96 ± 1.04 ng/ml in the UM-SCC-54 cells transfected with wild-type preprogalanin in the pCMV-SPORT6 construct compared to 0.20 ± 0.18 ng/ml in the empty pCMV-SPORT6 construct-transfected cells.
Clinicopathologic Characteristics
The galanin promoter was methylated in 24 of 100 (24.0%) previously untreated primary tumors tested with the same primers and unmethylated in 76 (76.0%). The methylation of galanin significantly correlated with the GALR1 methylation status (P = 1.88E-06), p16 methylation status (P = .005), DAPK methylation status (P = .001), E-cadherin methylation (P = 1.03E-05), and H-cadherin methylation (P = .0002). The presence of galanin promoter methylation was not associated with any difference in the age of onset, gender, smoking status, tumor site, tumor size, lymph node status, clinical stage, RASSF1A methylation, MGMT methylation, and DCC methylation (Table 1).
Table 1.
Galanin Gene Methylation Status in HNSCC Primary Samples.
| Patient and Tumor Characteristics (n = 100) | Methylation | P Value* | ||
| Present (n = 24) | Absent (n = 76) | |||
| Age | ||||
| 70 and older (29) | 7 | 22 | ||
| Under 70 (71) | 17 | 54 | 1 | |
| Gender | ||||
| Male (78) | 16 | 62 | ||
| Female (22) | 8 | 14 | 1 | |
| Smoking status | ||||
| Smoker (69) | 17 | 52 | ||
| Nonsmoker (31) | 7 | 24 | 1 | |
| Alcohol exposure | ||||
| Ever (58) | 10 | 48 | ||
| Never (42) | 14 | 28 | .096 | |
| Tumor size | ||||
| T1–2 (49) | 10 | 39 | ||
| T3–4 (51) | 14 | 37 | .486 | |
| Lymph node status | ||||
| N0 (44) | 7 | 37 | ||
| N+ (56) | 17 | 39 | .105 | |
| Stage | ||||
| I, II, III (45) | 9 | 36 | ||
| IV (55) | 15 | 40 | .483 | |
| Recurrence events | ||||
| Positive (52) | 21 | 31 | ||
| Negative (48) | 3 | 45 | 6.52E-05† | |
| p16 methylation | ||||
| Yes (54) | 19 | 35 | ||
| No (46) | 5 | 41 | .005† | |
| RASSF1A methylation | ||||
| Yes (22) | 5 | 17 | ||
| No (78) | 19 | 59 | 1 | |
| MGMT methylation | ||||
| Yes (34) | 10 | 24 | ||
| No (66) | 14 | 52 | 1 | |
| DAPK methylation | ||||
| Yes (39) | 16 | 23 | ||
| No (61) | 8 | 53 | .002† | |
| E-cadherin methylation | ||||
| Yes (40) | 19 | 21 | ||
| No (60) | 5 | 55 | 1.03E-05† | |
| H-cadherin methylation | ||||
| Yes (37) | 17 | 20 | ||
| No (63) | 7 | 56 | .0002† | |
| DCC methylation | ||||
| Yes (58) | 16 | 42 | ||
| No (42) | 8 | 34 | .354 | |
| GALR1 methylation | ||||
| Yes (38) | 19 | 19 | ||
| No (62) | 5 | 57 | 1.88E-06† | |
Fisher exact probability test.
P < .05.
Prognostic Value of the Galanin and Promoter Methylation of Other Genes in Head and Neck Primary Tumors
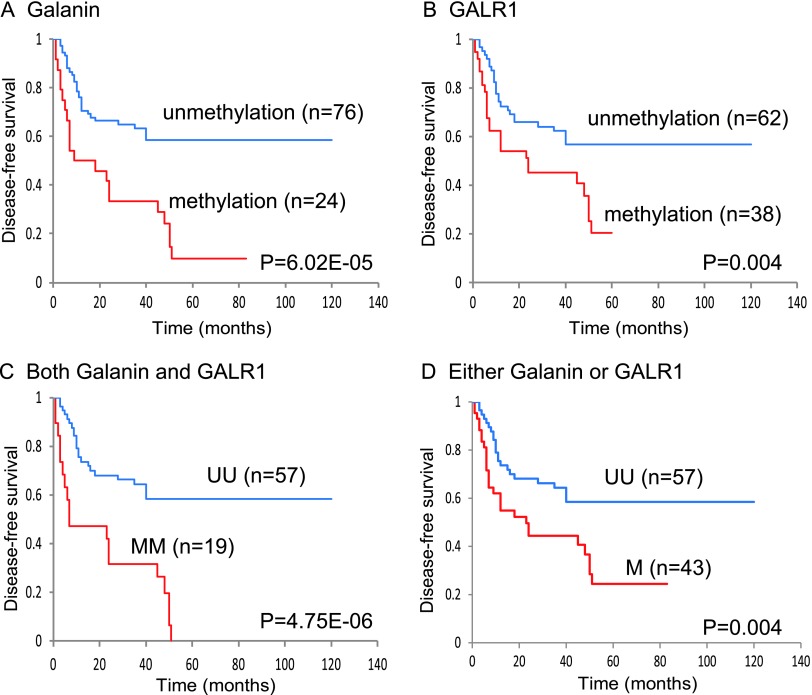
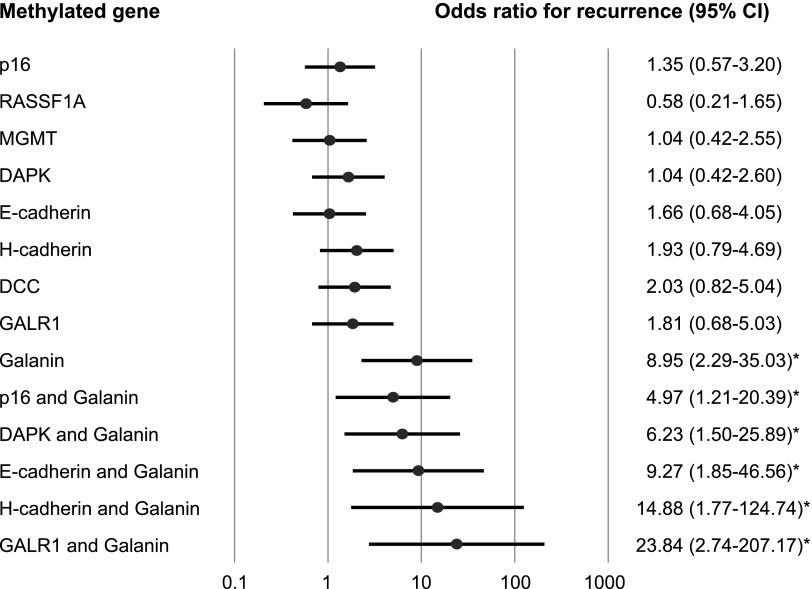
Kaplan-Meier plots indicated that the methylation of galanin in patients' tumors was related to the duration of DFS. The presence of galanin promoter methylation was associated with a statistically significant decrease in DFS (log-rank test, P = 6.02E-05; Figure 4A). Patients with a GALR1 methylation status had a significantly reduced DFS (log-rank test, P = .004; Figure 4B). Methylation of both galanin and GALR1 was associated with a DFS rate of 0%, in comparison to 58.5% in the absence of methylation of these genes (log-rank test, P = 4.75E-06; Figure 4C). Methylation of either galanin or GALR1 was associated with a DFS rate of 24.4%, in comparison to 58.5% in the absence of methylation of these genes (log-rank test, P = .004; Figure 4D). A multivariate logistic regression analysis showed the estimated odds of recurrence associated with methylation of galanin and eight other genes. The adjusted odds ratio for recurrence when galanin was methylated in the primary tumor was 8.95 (95% CI, 2.29 to 35.03; P = .002). Patients with methylation of both galanin and GALR1 had a significantly higher odds ratio for recurrence of 23.84 (95% CI, 2.74 to 207.17; P = .004) than those without any methylation of this pair of genes (Figure 5).
Figure 4.
Kaplan-Meier estimates of DFS among 100 patients based on their galanin and GALR1 methylation status. (A) Galanin: 24 patients with HNSCC had methylated galanin status (red line), and 76 patients with HNSCC had unmethylated galanin status (blue line); this difference was statistically significant (P = 6.02E-05). (B) GALR1: 38 patients with HNSCC had methylated GALR1 status (red line), and 62 patients with HNSCC had unmethylated GALR1 status (blue line); this difference was statistically significant (P = .004). (C) Galanin and GALR1: 19 patients with HNSCC hadmethylation of both galanin and GALR1 (MM; red line), and 57 patients with HNSCC had no methylation of both galanin and GALR1 (UU; blue line); this difference was statistically significant (P = 4.75E-06). (D) Galanin and GALR1: 43 patients with HNSCC had methylation of either galanin or GALR1 (M; red line), and 57 patients with HNSCC had no methylation of both galanin and GALR1 (UU; blue line); this difference was statistically significant (P = .004).
Figure 5.
Odds ratios for recurrence are reported on the basis of the multivariate logistic regression model adjusted for age (70 and older vs <70), sex, smoking status, alcohol exposure, and stage (I, II, III vs IV). A multivariate logistic regression analysis showed the estimated odds of recurrence to be associated with the methylation of galanin and other eight genes. The methylation of the gene pair galanin and GALR1 in the primary tumor was associated with the most significant odds ratio of recurrence.
Discussion
Early reports from our group noted that the loss of chromosome 18q developed with tumor progression and that 18q is also associated with a significantly decreased survival in HNSCC patients [25,26]. Subsequently, it was shown that tumors with a loss of chromosome 18q23 have alterations in the GALR signaling pathway [27]. The expression of GALR1 (18q23) mRNA is lost in HNSCC as a consequence of chromosome loss and DNA methylation [15]. GALR1 induces cell cycle arrest and GALR2 induces both cell cycle arrest and apoptosis in HNSCC after galanin treatment [14,28]. Therefore, galanin, GALR1, and GALR2 might act as tumor suppressors, targets for therapy, and potential prognostic factors in HNSCC.
Recent advances in molecular biology have made it possible to apply new strategies, such as gene and molecular targeted therapies for suitable cancer treatment. However, in comparison to other lesions such as breast, renal, and colorectal carcinomas, the application for HNSCC treatment has been lagging [29]. GPCRs belong to a superfamily of cell surface signaling proteins that have a pivotal role in many physiological functions and in multiple diseases, including the development of cancer and cancer metastasis [30]. Several studies have suggested that the neuropeptides function as tumor suppressor genes and possess potent antitumor activity in human cancers. Hypermethylation of the tachykinin-1 gene, which encodes the neuropeptides substance P, neurokinin A, and neuropeptide K and γ, is associated with a poor prognosis in esophageal squamous cell carcinoma, colon cancer, and HNSCC patients [5,31,32]. Somatostatin promoter hypermethylation is a common event in human esophageal adenocarcinoma and colon cancer [5,32]. The expression and promoter methylation of galanin is an area that still remains to be explored, despite the accumulated knowledge in tachykinin-1 and somatostatin. These findings provide a foundation for further studies on the role of neuropeptide genes in carcinogenesis and their potential as a biomarker for many types of tumors. Berger et al. reported that exogenous expression of GalR2 inhibited cell proliferation and induced apoptosis in neuroblastoma cells [33]. Tofighi et al. showed that galanin induced apoptosis in GalR2-transfected rat pheochromocytoma cells [34].
This study and previous reports from our group showed the restoration of galanin and GALR1 expression to be induced in HNSCC cell lines by 5-azacytidine and TSA. Selective drugs that modify specific epigenetic changes will thus become a crucial part of the therapeutic arsenal against cancer in the near future [35]. PI polyamide conjugates possessing a suberoylanilide hydroxamic acid moiety act as effective HDAC inhibitors with a broad spectrum of epigenetic activities [36]. These novel suberoylanilide hydroxamic acid-PI polyamide conjugates, which were designed to target the promoter region of p16 tumor suppressor gene, induce histone H3 Lys 9 acetylation [37]. Epigenetic therapy might be a novel therapeutic strategy for HNSCC treatment. The establishment of DNA methylation and histone modification profiles of the primary tumor specimen itself might be a valuable tool for determining the prognosis and predicting the patient's response to therapies.
Galanin expression is frequently absent in HNSCC. The galanin expression-negative UM-SCC cell lines (3 of 12, 25.0%) are hypermethylated in the CpG islands of the galanin promoter region. Furthermore, expression-negative squamous cancers and cell lines exhibit hypermethylation of CpG islands in the galanin promoter region. Moreover, galanin expression can be restored after treatment with the demethylating agent, 5-azacytidine, and the HDAC inhibitor, TSA. A survey of 100 tumor tissue samples using MSP shows that hypermethylation of the galanin promoter is as common as that of genes frequently thought of as being epigenetically regulated, such as p16 (54%), RASSF1A (22%), MGMT (34%), DAPK (39%), E-cadherin (40%), H-cadherin (37%), DCC (58%), and GALR1 (38%). Interestingly, there is a significant correlation between galanin promoter methylation and the GALR1 methylation status in primary tumor samples. GALR1 methylation was also found in 38 of 100 (38%) primary tumor specimens. A concurrent analysis showed that both galanin and GALR1 were completely methylated in 19 (19.0%), another 24 (24.0%) cases were methylated at either galanin or GALR1, and 43 (43.0%) cases were methylated at least one of the galanin or GALR1 promoters. Finally, galanin-GALR1 signaling are frequently inactivated in HNSCC and this process is associated with tumor development. Patients who had hypermethylation of both genes had a significantly shorter DFS than did patients without methylation of both genes. A multivariate logistic regression analysis showed that the methylation of both genes was associated with the most significant odds ratios of recurrence. Brock et al. reported the methylation of p16 and H-cadherin to be associated with early recurrence of stage I non-small cell lung cancer [38]. The significant association between galanin aberrant methylation and promoter hypermethylation of p16 and H-cadherin, respectively, observed in the current study supports the usefulness of galanin methylation as a potential molecular marker for the early recurrence of head and neck lesions. Moreover, exogenous galanin specifically suppresses colony-forming activity in the GALR1 expressing cell line. The current findings suggested that simultaneous methylation of the galanin and GALR1 genes occurs in a subset of HNSCC and may be used as a prognostic marker for patients.
In conclusion, the present study showed that the galanin promoter methylation profile appears to be an important marker predicting the clinical outcome of HNSCC. This information can be used to identify patients with high-risk HNSCC who may benefit from adjuvant therapy and cautious observation after the resection of primary tumors. Finally, galanin can be reactivated by altering chromatin modifications with methyltransferase and HDAC inhibitors, raising the promise of selective small molecule inhibitors of these enzymes as a potential therapeutic target for HNSCC.
Acknowledgments
The authors thank Yuko Mohri for her excellent technical support.
Footnotes
Grant support: Grant-in-aid for Scientific Research (No. 23592524) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Conflict of Interest: None declared.
References
- 1.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 2.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, Wolf GT, Urba SG, Chepeha DB, Teknos TN, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26:3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 4.Hill SJ. G-protein-coupled receptors: past, present and future. Br J Pharmacol. 2006;147(suppl 1):S27–S37. doi: 10.1038/sj.bjp.0706455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, Mori Y, Mantzur C, Paun B, Hamilton JP, et al. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin Cancer Res. 2007;13:6293–6300. doi: 10.1158/1078-0432.CCR-07-0818. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 7.Tatemoto K, Rokaeus A, Jornvall H, McDonald TJ, Mutt V. Galanin—a novel biologically active peptide from porcine intestine. FEBS Lett. 1983;164:124–128. doi: 10.1016/0014-5793(83)80033-7. [DOI] [PubMed] [Google Scholar]
- 8.Vrontakis ME, Torsello A, Friesen HG. Galanin. J Endocrinol Invest. 1991;14:785–794. doi: 10.1007/BF03347918. [DOI] [PubMed] [Google Scholar]
- 9.Ottlecz A, Snyder GD, McCann SM. Regulatory role of galanin in control of hypothalamic-anterior pituitary function. Proc Natl Acad Sci USA. 1988;85:9861–9865. doi: 10.1073/pnas.85.24.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartfai T, Langel U, Bedecs K, Andell S, Land T, Gregersen S, Ahren B, Girotti P, Consolo S, Corwin R, et al. Galanin-receptor ligand M40 peptide distinguishes between putative galanin-receptor subtypes. Proc Natl Acad Sci USA. 1993;90:11287–11291. doi: 10.1073/pnas.90.23.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutkind JS. Cell growth control by G protein-coupled receptors: from signal transduction to signal integration. Oncogene. 1998;17:1331–1342. doi: 10.1038/sj.onc.1202186. [DOI] [PubMed] [Google Scholar]
- 12.Gundlach AL. Galanin/GALP and galanin receptors: role in central control of feeding, body weight/obesity and reproduction? Eur J Pharmacol. 2002;440:255–268. doi: 10.1016/s0014-2999(02)01433-4. [DOI] [PubMed] [Google Scholar]
- 13.Henson BS, Neubig RR, Jang I, Ogawa T, Zhang Z, Carey TE, D'Silva NJ. Galanin receptor 1 has anti-proliferative effects in oral squamous cell carcinoma. J Biol Chem. 2005;280:22564–22571. doi: 10.1074/jbc.M414589200. [DOI] [PubMed] [Google Scholar]
- 14.Kanazawa T, Iwashita T, Kommareddi P, Nair T, Misawa K, Misawa Y, Ueda Y, Tono T, Carey TE. Galanin and galanin receptor type 1 suppress proliferation in squamous carcinoma cells: activation of the extracellular signal regulated kinase pathway and induction of cyclin-dependent kinase inhibitors. Oncogene. 2007;26:5762–5771. doi: 10.1038/sj.onc.1210384. [DOI] [PubMed] [Google Scholar]
- 15.Misawa K, Ueda Y, Kanazawa T, Misawa Y, Jang I, Brenner JC, Ogawa T, Takebayashi S, Grenman RA, Herman JG, et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res. 2008;14:7604–7613. doi: 10.1158/1078-0432.CCR-07-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, Bradford CR, Carey TE. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32:417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park NH, Min BM, Li SL, Huang MZ, Cherick HM, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–1631. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Mori M, Croce CM. Allele loss and promoter hypermethylation of VHL, RAR-β, RASSF1A, and FHIT tumor suppressor genes on chromosome 3p in esophageal squamous cell carcinoma. Cancer Res. 2003;63:3724–3728. [PubMed] [Google Scholar]
- 20.Martone T, Gillio-Tos A, De Marco L, Fiano V, Maule M, Cavalot A, Garzaro M, Merletti F, Cortesina G. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res. 2007;13:5089–5094. doi: 10.1158/1078-0432.CCR-07-0119. [DOI] [PubMed] [Google Scholar]
- 21.Park HL, Kim MS, Yamashita K, Westra W, Carvalho AL, Lee J, Jiang WW, Baek JH, Liu J, Osada M, et al. DCC promoter hypermethylation in esophageal squamous cell carcinoma. Int J Cancer. 2008;122:2498–2502. doi: 10.1002/ijc.23434. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–4236. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 23.Katz MH. Multivariable Analysis: A Practical Guide for Clinicians and Public Health Researchers. Cambridge, United Kingdom: Cambridge University Press; 2011. [Google Scholar]
- 24.Kofler B, Berger A, Santic R, Moritz K, Almer D, Tuechler C, Lang R, Emberger M, Klausegger A, Sperl W, et al. Expression of neuropeptide galanin and galanin receptors in human skin. J Invest Dermatol. 2004;122:1050–1053. doi: 10.1111/j.0022-202X.2004.22418.x. [DOI] [PubMed] [Google Scholar]
- 25.Frank CJ, McClatchey KD, Devaney KO, Carey TE. Evidence that loss of chromosome 18q is associated with tumor progression. Cancer Res. 1997;57:824–827. [PubMed] [Google Scholar]
- 26.Pearlstein RP, Benninger MS, Carey TE, Zarbo RJ, Torres FX, Rybicki BA, Dyke DL. Loss of 18q predicts poor survival of patients with squamous cell carcinoma of the head and neck. Genes Chromosomes Cancer. 1998;21:333–339. doi: 10.1002/(sici)1098-2264(199804)21:4<333::aid-gcc7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Takebayashi S, Ogawa T, Jung KY, Muallem A, Mineta H, Fisher SG, Grenman R, Carey TE. Identification of new minimally lost regions on 18q in head and neck squamous cell carcinoma. Cancer Res. 2000;60:3397–3403. [PubMed] [Google Scholar]
- 28.Kanazawa T, Kommareddi PK, Iwashita T, Kumar B, Misawa K, Misawa Y, Jang I, Nair TS, Iino Y, Carey TE. Galanin receptor subtype 2 suppresses cell proliferation and induces apoptosis in p53 mutant head and neck cancer cells. Clin Cancer Res. 2009;15:2222–2230. doi: 10.1158/1078-0432.CCR-08-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanazawa T, Misawa K, Carey TE. Galanin receptor subtypes 1 and 2 as therapeutic targets in head and neck squamous cell carcinoma. Expert Opin Ther Targets. 2010;14:289–302. doi: 10.1517/14728221003598922. [DOI] [PubMed] [Google Scholar]
- 30.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 31.Misawa K, Kanazawa T, Misawa Y, Imai A, Uehara T, Mochizuki D, Endo S, Takahashi G, Mineta H. Frequent promoter hypermethylation of tachykinin-1 and tachykinin receptor type 1 is a potential biomarker for head and neck cancer. J Cancer Res Clin Oncol. 2013 doi: 10.1007/s00432-013-1393-5. [DOI] [PubMed] [Google Scholar]
- 32.Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP, Jin Z, Sato F, Berki AT, Kan T, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131:797–808. doi: 10.1053/j.gastro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Berger A, Lang R, Moritz K, Santic R, Hermann A, Sperl W, Kofler B. Galanin receptor subtype GalR2 mediates apoptosis in SH-SY5Y neuroblastoma cells. Endocrinology. 2004;145:500–507. doi: 10.1210/en.2003-0649. [DOI] [PubMed] [Google Scholar]
- 34.Tofighi R, Joseph B, Xia S, Xu ZQ, Hamberger B, Hokfelt T, Ceccatelli S. Galanin decreases proliferation of PC12 cells and induces apoptosis via its subtype 2 receptor (GalR2) Proc Natl Acad Sci USA. 2008;105:2717–2722. doi: 10.1073/pnas.0712300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 36.Pandian GN, Shinohara K, Ohtsuki A, Nakano Y, Masafumi M, Bando T, Nagase H, Yamada Y, Watanabe A, Terada N, et al. Synthetic small molecules for epigenetic activation of pluripotency genes in mouse embryonic fibroblasts. Chembiochem. 2011;12:2822–2828. doi: 10.1002/cbic.201100597. [DOI] [PubMed] [Google Scholar]
- 37.Ohtsuki A, Kimura M, Minoshima M, Suzuki T, Ikeda M, Bando T, Nagase H, Shinohara K, Sugiyama H. Synthesis and properties of PI polyamide-SAHA conjugate. Tetrahedron Lett. 2009;50:7288–7292. [Google Scholar]
- 38.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glockner S, Piantadosi S, Gabrielson E, Pridham G, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]