Abstract
Caspases are the primary drivers of apoptotic cell death, cleaving cellular proteins that are critical for dismantling the dying cell. Initially translated as inactive zymogenic precursors, caspases are activated in response to a variety of cell death stimuli. In addition to factors required for their direct activation (e.g., dimerizing adaptor proteins in the case of initiator caspases that lie at the apex of apoptotic signaling cascades), caspases are regulated by a variety of cellular factors in a myriad of physiological and pathological settings. For example, caspases may be modified posttranslationally (e.g., by phosphorylation or ubiquitylation) or through interaction of modulatory factors with either the zymogenic or active form of a caspase, altering its activation and/or activity. These regulatory events may inhibit or enhance enzymatic activity or may affect activity toward particular cellular substrates. Finally, there is emerging literature to suggest that caspases can participate in a variety of cellular processes unrelated to apoptotic cell death. In these settings, it is particularly important that caspases are maintained under stringent control to avoid inadvertent cell death. It is likely that continued examination of these processes will reveal new mechanisms of caspase regulation with implications well beyond control of apoptotic cell death.
As the main drivers of apoptotic cell death, caspases are tightly regulated. They are synthesized as zymogens and regulated by various mechanisms (e.g., posttranslational modifications) both before and after activation.
Apoptosis is a form of programmed cell death that eliminates individual cells within an organism while preserving the overall structure of surrounding tissue. Many of the prominent morphological features of apoptosis were first described in 1972 by Kerr, Wyllie, and Currie (Kerr et al. 1972). However, it was not until the mid-1990s that apoptosis was linked to the activation of the cysteine-dependent aspartate-driven proteases (caspases), which cleave key intracellular substrates to promote cell death (Cerretti et al. 1992; Nicholson et al. 1995; Alnemri et al. 1996; Liu et al. 1996; Thornberry and Lazebnik 1998). Given the critical role that caspases play in dismantling the cell during apoptosis, their activation and subsequent activity are highly regulated. Failure of a cell to properly modulate caspase activity can cause aberrant or untimely apoptotic cell death, potentially leading to carcinogenesis, autoimmunity, neurodegeneration, and immunodeficiency (Thompson 1995; Hanahan and Weinberg 2000; Yuan and Yankner 2000; Li and Yuan 2008).
Caspases are synthesized within the cell as inactive zymogens that lack significant protease activity. Thus, caspases are, in essence, regulated from the moment of protein synthesis in that they are not activated until receipt of specific death stimuli (Earnshaw et al. 1999). The primary structure of a caspase is an amino-terminal prodomain and a carboxy-terminal protease domain, which contains the key catalytic cysteine residue. Caspases are categorized as initiator or effector caspases, based on their position in apoptotic signaling cascades. The initiator caspases (caspase-2, -8, -9, and -10) act apically in cell death pathways and all share long, structurally similar prodomains. This group of enzymes is activated through “induced proximity” when adaptor proteins interact with the prodomains and promote caspase dimerization (Boatright et al. 2003; Baliga et al. 2004; Pop et al. 2006; Riedl and Salvesen 2007; Wachmann et al. 2010). In contrast, the effector caspases (caspase-3, -6, and -7) have shorter prodomains and exist in the cell as preformed, but inactive, homodimers. Following cleavage mediated by an initiator caspase, effector caspases act directly on specific cellular substrates to dismantle the cell. Although many individual caspase substrates have been implicated in specific aspects of cellular destruction (e.g., lamin cleavage is required for the efficient packaging of nuclei into small membrane-bound vesicles), recent proteomic approaches have greatly expanded the known repertoire of proteolytic products generated during apoptosis (Van Damme et al. 2005; Dix et al. 2008; Mahrus et al. 2008). Further work will be needed to confirm these findings and to determine how (or if) all of these substrates participate in the apoptotic process (see Poreba et al. 2013), especially as new details emerge on the relationship between posttranslational modifications, like phosphorylation, and caspase cleavage (Dix et al. 2012).
CASPASE ACTIVATION
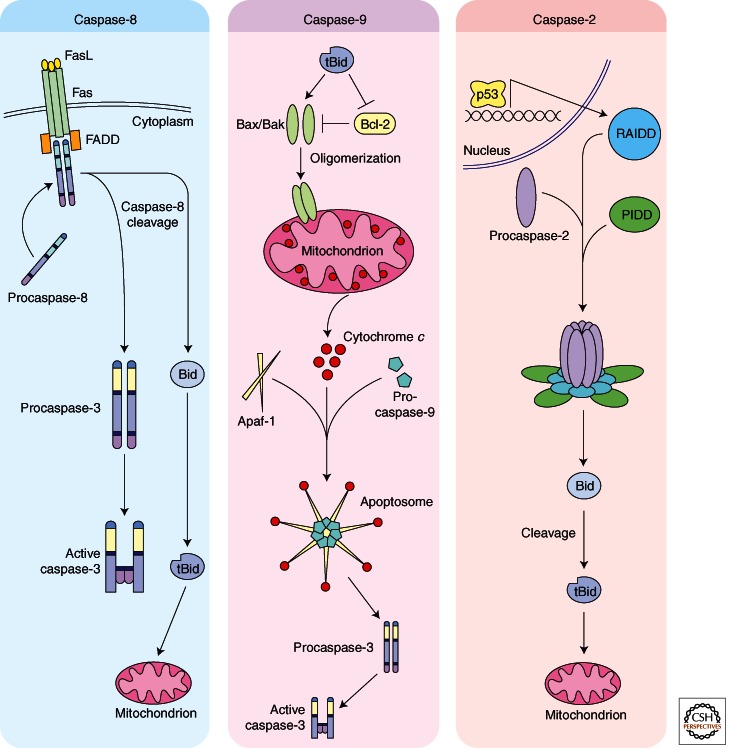
Initiation of apoptosis occurs through either a cell-intrinsic or cell-extrinsic pathway. Extrinsic pathway cell death signals originate at the plasma membrane where an extracellular ligand (e.g., FasL) binds to its cell surface transmembrane “death receptor” (e.g., Fas receptor), inducing oligomerization of the receptor (Trauth et al. 1989; Itoh and Nagata 1993; Danial and Korsmeyer 2004). This, in turn, promotes clustering of proteins that bind to the intracellular domain of the receptor (e.g., FADD, or Fas-associated death domain-containing protein), which then binds to the prodomain of initiator caspases (e.g., caspase-8 or -10) to promote their dimerization and activation; these complexes are referred to as DISCs, or death-induced signaling complexes (Kischkel et al. 1995). As initiator caspases, caspases-8 and -10 are activated within the DISC through induced proximity dimerization (Boatright et al. 2003; Wachmann et al. 2010). Active caspase-8/-10 can then directly cleave and activate effector caspases, such as caspase-3. In some cell types, this pathway is sufficient to cause cell death (Type I cells). However, in other cells, caspase-8 must also engage the mitochondria as described below for the intrinsic pathway (Type II cells; Fig. 1) (Li et al. 1998; Luo et al. 1998; Scaffidi et al. 1998). Other death receptors that, like FasL–Fas receptor, participate in extrinsic apoptotic pathways include the TNF ligand–TNF-R1 complex and the DR4/5–Apo2 L/TRAIL ligand complexes.
Figure 1.
Activation of initiator caspases. Caspase-8: The extrinsic pathway is initiated at the plasma membrane, where a receptor (FasL) interacts with its transmembrane receptor (Fas), causing oligomerization of the receptor. The Fas receptor has an intracellular DD (death domain), which allows for the recruitment of the adaptor protein FADD from the cytoplasm through its DD. Caspase-8 is then recruited to the complex through interactions between the DED (death effector domain) on FADD and similar DED sequences on caspase-8. Active caspase-8 can directly activate caspase-3, or it can cleave Bid, which facilitates mitochondrial cytochrome c release. Caspase-9: Caspase-9 is activated through the mitochondria-dependent intrinsic pathway. At the mitochondria, the antiapoptotic bcl-2 family members like Bcl-2 and Bcl-xL inhibit cytochrome c release from the mitochondrial intermembrane space. In contrast, proapoptotic bcl-2 family members, Bax and Bak, facilitate cytochrome c release. BH3-only proteins help regulate the balance between the pro- and antiapoptotic bcl-2 family members. After cytochrome c translocates into the cytosol, it interacts with Apaf-1, which undergoes a conformational change and oligomerization into a heptameric structure known as the apoptosome. The apoptosome recruits and activates the initiator caspase, caspase-9. Active caspase-9 then directly cleaves and activates effector caspases, such as caspase-3. Caspase-2: Caspase-2 appears to function as an initiator caspase upstream of the mitochondria. The most well-understood mechanism for caspase-2 activation involves the PIDDosome. p53-dependent transcription of the adaptor protein PIDD forms the backbone of the complex with RAIDD interaction through a DD–DD interaction. Caspase-2 is recruited to RAIDD through a CARD–CARD interaction. Active caspase-2 then cleaves and activates the BH3-only protein Bid, facilitating cytochrome c release from the mitochondria.
The intrinsic pathway proceeds through the mitochondria and involves release of the respiratory chain component, cytochrome c, from the intermembrane space of the mitochondria into the cytoplasm. Cytochrome c interacts with the adaptor protein, Apaf-1 (apoptotic protease activating factor-1), to form the heptameric backbone of the apoptosome complex, which recruits and activates caspase-9 through dimerization (Liu et al. 1996; Zou et al. 1997; Acehan et al. 2002; Boatright et al. 2003). The remarkably similar phenotypes of the Apaf-1–/– and caspase-9–/– mice suggest that caspase-9 is indeed dependent on this Apaf-1-based complex for its activation (Cecconi et al. 1998; Hakem et al. 1998; Kuida et al. 1998). Recent data suggest that each apoptosome backbone recruits and activates only two caspase-9 molecules, creating a 7:2 ratio between Apaf-1 and caspase-9 within the apoptosome (Malladi et al. 2009). Active caspase-9 cleaves and activates downstream effector caspases, such as caspase-3 (Slee et al. 1999) (Fig. 1). Although caspase-9 can autocleave and can also be directly cleaved by active caspases, this modification does not appear to be necessary for its activation (Renatus et al. 2001; Boatright et al. 2003). Cleavage of caspase-9 does, however, have a variety of implications on its regulation, including the generation of a new epitope that is required for subsequent caspase-9 inhibition by XIAP. Additionally, Bratton and colleagues have suggested a model whereby cleavage of caspase-9 lowers its affinity for the apoptosome (relative to procaspase-9), promoting its replacement by new incoming procaspase-9 molecules recruited to the Apaf-1 caspase recruitment domains (CARDs) for activation (Malladi et al. 2009).
Although caspase-2 is structurally and functionally similar to the other initiator caspases, the role of caspase-2 in apoptotic processes is a bit less clear. It is activated similarly to other initiator caspases via induced proximity dimerization and autocatalytic processing within an adaptor protein complex, and it appears to function, at least under some circumstances, upstream of the mitochondria (Baliga et al. 2004). The current model for caspase-2 activation involves two adaptor proteins, PIDD (p53-induced protein with a death domain) and RAIDD (RIP-associated ICH-1/CED-3 homologous proteins with a death domain) (Duan and Dixit 1997; Read et al. 2002; Tinel and Tschopp 2004). Together, these proteins form the PIDDosome complex, which consists of five PIDDs, seven RAIDDs, and seven caspase-2 molecules (Park et al. 2007). Active caspase-2 is thought to facilitate apoptosis by cleaving the proapoptotic family member Bid to promote mitochondrial permeabilization and cytochrome c release, thereby propagating the apoptotic signal (Gao et al. 2005; Bonzon et al. 2006) (Fig. 1). Because one of its upstream activators, PIDD, is a p53 target gene, caspase-2 is thought to play a role in at least some p53-mediated cell deaths (Baptiste-Okoh et al. 2008; Sidi et al. 2008). However, caspase-2 also appears to be activated under conditions of heat shock in a p53-independent manner, which suggests that there may be PIDDosome-independent mechanisms for activating caspase-2 (Tu et al. 2006). Indeed, PIDD–/– mice were found to have no defects in caspase-2-initiated apoptosis following certain apoptotic stimuli, such as DNA damage and ER stress. Additionally, p53-induced caspase-2 activation following 5-FU treatment has been observed in the absence of PIDD (Vakifahmetoglu et al. 2006; Manzl et al. 2009). Conversely, cells from caspase-2 knockout mice are still susceptible to some actions of PIDD, suggesting that PIDD can use effectors other than caspase-2 to promote cell death (Berube et al. 2005).
CONSERVATION OF APOPTOTIC PATHWAYS
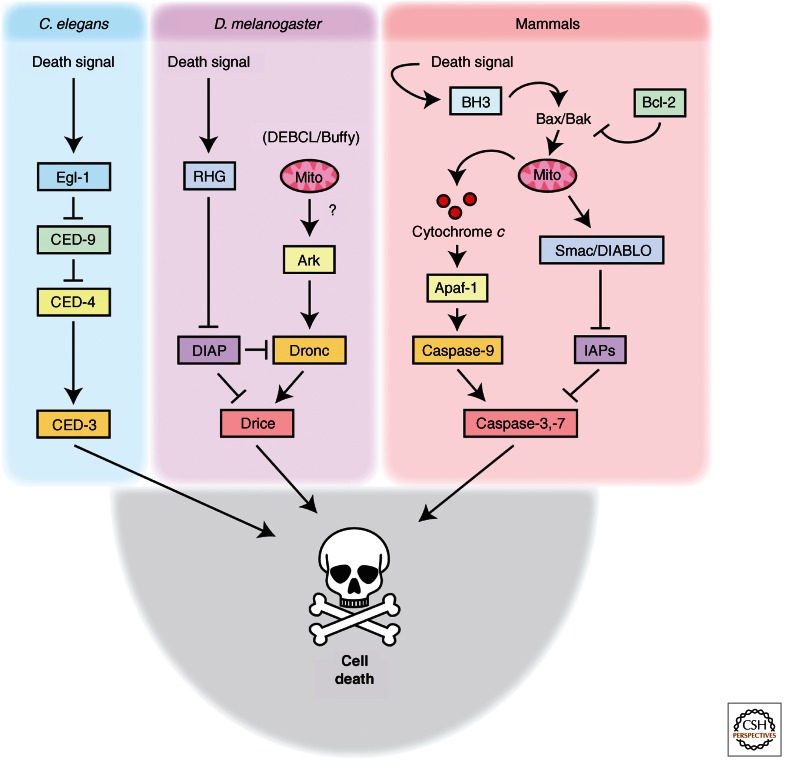
The caspase family of proteins is highly conserved between organisms, and, as seen in Figure 2, certain regulatory “modules” are conserved between nematodes, fruit flies, and mammals, although the precise ways in which these pathways are arranged and the individual factors involved vary in their importance/prominence in different organisms (Yan and Shi 2005). Genetic characterization of programmed cell death in the nematode identified ced-3, ced-4, ced-9, and egl-1 as central regulators of cell death in that organism (Yuan et al. 1993). EGL-1 is a Bcl-2 family member with a single block of Bcl-2 homology (a so-called BH3-only protein) that alleviates inhibition of the Apaf-1-like molecule CED-4, by CED-9, an antiapoptotic Bcl-2-like protein (Hengartner and Horvitz 1994; Conradt and Horvitz 1998). Oligomerization of the adaptor protein, CED-4 creates a high molecular weight complex that parallels those made by Apaf-1 in vertebrates (Yang et al. 1998; Qi et al. 2010; Yuan et al. 2010). This CED-4 complex activates the CED-3 caspase, which functions as a prominent executioner of cell death. Although alterations in mitochondrial dynamics have been reported to modulate apoptosis in C. elegans, there is no evidence that the apoptosis in this organism involves mitochondrial permeabilization, and the worm apoptosome is active once EGL-1 relieves apoptosome inhibition. Similar to the mammalian apoptosome, the C. elegans apoptosome appears to contain two CED-3 molecules, but with a ratio of eight CED-4 to two CED-3 (Qi et al. 2010).
Figure 2.
Similarities and differences in apoptotic machinery across evolution. Many of the core proteins of the apoptotic pathway are conserved in nematodes (C. elegans), fruit flies (D. melanogaster), and mammals, and these protein families share some functional similarities (which is noted by matching colors in the figure). As an example, Apaf-1, which forms the backbone of the caspase-9 activating apoptosome in response to cytochrome c, has a homolog, dApaf-1 (dArk), in the Drosophila pathway, which facilitates Dronc activation, although it does not appear to be activated by cytochrome c. In C. elegans, CED-4 also auto-oligomerizes to form a complex that enables activation of the CED-3 caspase. For a more complete description of each pathway, including highlights of similarities and differences between species, please see the text.
Drosophila cells share with their mammalian counterparts certain features of caspase activation, including activation through oligomerization of initiator caspases. As in worms, cytochrome c does not appear to be broadly involved in caspase activation. Rather, the fly apoptosome, consisting of an Apaf-1-like protein known as DARK and an initiator caspase called Dronc, has the capacity for constitutive activity, but is held in check by a caspase inhibitor, DIAP1. Following an apoptotic stimulus, Dronc is activated through removal of DIAP1 (it is both displaced and degraded), leading to cleavage of the executioner caspases, DCP-1 and Drice. Although there have been seven caspases identified thus far in Drosophila, the remaining four, Dredd, Decay, Damm, and Dream/Strica, appear to have functions outside of apoptosis (e.g., innate immunity) (Kornbluth and White 2005).
Given the crucial role that active caspases play in the demise of cells in all eukaryotic organisms, it is not surprising that both their activation and activity are highly regulated. Posttranslational modifications, including phosphorylation and ubiquitylation can block caspase activation and activity. Additionally, other proteins and molecules bind to either caspases or components of their activation platforms, modulating their activity. This work will focus on the mechanisms for direct regulation of the caspase proteins; additional details on the more indirect mechanisms, including effects on components of caspase-activating complexes, can be found in a variety of recent reviews (Schafer and Kornbluth 2006; Fadeel et al. 2008; Krumschnabel et al. 2009).
POSTTRANSLATIONAL MODIFICATIONS
Although synthesis of caspases in a zymogenic, inactive form keeps basal activity low until receipt of apoptotic stimuli, a variety of other regulatory mechanisms have been characterized for both initiator and effector caspases. These additional layers of control allow a finely tuned response to a death-inducing or survival signal. As described above, caspases are subject to posttranslational modification and also interact with modulatory proteins that can enhance or suppress their activities. Moreover, posttranslational modifications can indirectly affect caspase activity by altering the ability of caspases to interact with their binding partners (including the activation platforms described above). This chapter focuses on caspases and the direct mechanisms known to regulate their activation and/or activity.
Initiator Caspases
Caspase-9
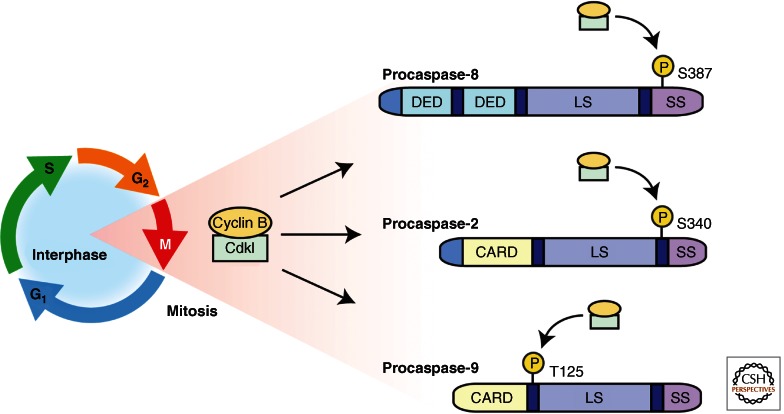
As the initiator caspase component of the apoptosome complex, caspase-9 is activated downstream of mitochondrial cytochrome c release and thus has a crucial role in activating effector caspases in response to a variety of death stimuli. Caspase-9 serves as an important locus of regulation at this point in the apoptotic pathway; indeed, it may be the most well-characterized caspase with regard to posttranslational modifications. A number of distinct phosphorylation sites have been reported for caspase-9, and they most frequently result in a reduction of caspase-9 activation and/or cleavage (Allan and Clarke 2009). One residue that appears to be the locus of caspase-9 regulation by multiple signaling pathways is Thr125, which is phosphorylated by several different kinases. Lying between the CARD domain and the large subunit of caspase-9, phosphorylated Thr125 decreases caspase-9 activation (and consequently decreases its proteolytic processing) through an undefined mechanism (Allan et al. 2003). Thr125 was first identified as a site of phosphorylation by the Erk kinase, but more recently, the cyclin- dependent kinase cdk1, DYRK1A, and p38α have all been reported to phosphorylate caspase-9 at this site, reducing its activation and the downstream activation of caspase-3 (Allan et al. 2003; Allan and Clarke 2007; Laguna et al. 2008; Seifert et al. 2008; Seifert and Clarke 2009) (Fig. 3). In all cases, this modification is thought to contribute to antiapoptotic signaling by the kinases involved, preventing full-blown caspase activation in the event of mitochondrial cytochrome c release. In some cases, the same kinases also act upstream of caspase-9 to inhibit cytochrome c release itself, suggesting that the caspase-9 phosphorylation may simply provide an extra layer of antiapoptotic protection in the event of inadvertent cytochrome c release. Phosphorylation of Thr125 can be reversed by protein phosphatase-1α (PP1α), a proapoptotic event (Dessauge et al. 2006). Thr125 is conserved among mice, rats, and some mammals such as primates. Although not all of the caspase phosphorylation sites discussed below are as well-conserved as Thr125, regulation of the Ser135 phosphorylation site on Xenopus capase-2 is conserved throughout mammals and there is similar regulation of the Drosophila caspase Dronc (Nutt et al. 2005, 2009; Yang et al. 2010). When taken together, examples of conserved modification sites help highlight the importance these posttranslational changes have in modulating caspase activity to affect the ultimate fate of a cell.
Figure 3.
Caspase regulation during mitosis. During the mitotic phase of the cell cycle, the apoptotic pathway appears to be inhibited at multiple steps in the pathway. The mitotic kinase, cdk1/cyclin B, has been shown to directly phosphorylate three of the initiator caspases (caspase-2, -8, and -9), leading to their inhibition. Caspase-8 is phosphorylated by cdk1 at Ser387, which is located in the small subunit of the protein. This modification diminishes caspase-8 processing in response to apoptotic stimuli. Cdk1 directly phosphorylates caspase-2 at Ser340 (in human, Ser308 in Xenopus). This site is located in the linker region between the large and small subunits, and phosphorylation at this site inhibits caspase-2 activation. Caspase-9 was the first caspase to be identified as a cdk1 substrate. Phosphorylation at Thr125, which is located between the CARD and the large subunit, blocks activation of the caspase through an unclear mechanism.
In addition to phosphorylation at Thr125, caspase-9 can be phosphorylated by PKCζ at Ser144 in response to hyperosmotic stress. This modification also suppresses caspase activity (Brady et al. 2005). In addition, three different PKA phosphorylation sites have been reported for caspase-9, Ser99, Ser183, and Ser195 (Martin et al. 2005). However, the importance of these sites in PKA-mediated suppression of caspase activity is unclear because mutating these sites failed to render the apoptosome resistant to inhibition by PKA signaling, suggesting that there might be other relevant sites or that the effect of PKA was indirect. Akt and CK2 kinases can phosphorylate caspase-9 at Ser196 and Ser348, respectively (Cardone et al. 1998; McDonnell et al. 2008). However, the overall relevance of these sites is unclear because they are not evolutionarily conserved (even from mouse to human). In contrast to the preponderance of caspase modifications, c-Abl-mediated caspase-9 phosphorylation at Tyr153 following DNA damage appears to enhance caspase activity (Raina et al. 2005). These caspase-9 phosphorylation sites, along with those affecting the other caspases (see further below), are summarized in Table 1.
Table 1.
Caspase regulation via phosphorylation
| Substrate | Kinase/phosphatase | Sites | +/−a | References |
|---|---|---|---|---|
| Initiator caspases | ||||
| Caspase-8 | Src, Fyn, Lyn | Tyr380 | − | Cursi et al. 2006; Senft et al. 2007; Jia et al. 2008 |
| Caspase-8 | Lyn | Tyr465 | − | Jia et al. 2008 |
| Caspase-8 | p38 MAPK | Ser364 | − | Alvarado-Kristensson et al. 2004 |
| Caspase-8 | CDK1 | Ser387 | − | Matthess et al. 2010 |
| Caspase-8 | RSK2 | Thr263 | + | Peng et al. 2011 |
| Caspase-9 | Erk1/2 | Thr125 | − | Allan et al. 2003 |
| Caspase-9 | CDK1 | Thr125 | − | Allan and Clarke 2007 |
| Caspase-9 | DYRK1A | Thr125 | − | Seifert et al. 2008 |
| Caspase-9 | p38α | Thr125 | − | Seifert and Clarke 2009 |
| Caspase-9 | PP1α | Thr125 | − | Dessauge et al. 2006 |
| Caspase-9 | PKCζ | Ser144 | − | Brady et al. 2005 |
| Caspase-9 | c-Abl | Tyr153 | + | Raina et al. 2005 |
| Caspase-9 | PKA | Ser99, Ser183, Ser195 | Unclear | Martin et al. 2005 |
| Caspase-9 | Akt | Ser196 (human) | − | Cardone et al. 1998 |
| Caspase-9 | CK2 | Ser348 (mouse) | − | McDonnell et al. 2008 |
| Caspase-2 | CDK1 | Ser340 | − | Andersen et al. 2009 |
| Caspase-2 | CK2 | Ser157 | − | Shin et al. 2005 |
| Caspase-2 | CaMKII | Ser135 (Xenopus) | − | Nutt et al. 2005 |
| Dronc | CaMKII | Ser130 | Yang et al. 2010 | |
| Effector caspases | ||||
| Caspase-3 | PKCδ | ND | + | Voss et al. 2005 |
| Caspase-3 | p38 | Ser150 | − | Alvarado-Kristensson et al. 2004 |
| Caspase-3 | PP2A | Ser150 | − | Alvarado-Kristensson and Andersson 2005 |
| Caspase-6 | ARK5 | Ser257 | − | Suzuki et al. 2004 |
| Caspase-7 | PAK2 | Ser30, Thr173, Ser239 | − | Li et al. 2011 |
aThe effect of phosphorylation at specific residues is listed as activating (+) or inactivating (−). ND, not determined.
In addition to phosphorylation, caspase-9 can be modified by nitrosylation, which occurs when intracellular nitric oxide (NO) levels are high (Torok et al. 2002). Treatment of cell lines with a pharmacological NO donor, S-Nitroso-N-acetyl-d,l-penicillamine (SNAP), does not block mitochondrial cytochrome c release but reduces caspase-9 activation in a cell-free system. In vitro, SNAP was also able to negatively regulate the activity of recombinant, human capsase-9 although the relevant site was not identified (Torok et al. 2002). Mannick et al. (2001) have also showed nitrosylation of caspase-9 in different subcellular compartments, finding that a mitochondrial fraction of caspase-9 was preferentially nitrosylated.
Besides modifications that alter its activity or activation, caspase-9 can be controlled at the level of protein stability through ubiquitylation. It has been reported that caspase-9 can be ubiquitylated by XIAP (X-linked inhibitor of apoptosis protein) E3 ubiquitin ligase, a member of the IAP family of proteins, which will be discussed further below for their role in blocking caspase activity through direct interaction (see Silke and Meier 2013). XIAP contains a carboxy-terminal RING domain critical for the protein’s ubiquitin ligase activity (Joazeiro and Weissman 2000). XIAP can polyubiquitylate the large subunit of active caspase-9 in vitro, but not the inactive procaspase-9 (Morizane et al. 2005). Consistent with these observations, treatment of cells with the proteasome inhibitor MG132, in conjunction with overexpression of XIAP, promotes accumulation of polyubiquitylated caspase-9 (Morizane et al. 2005). Additional studies are required to fully understand the means by which XIAP-mediated caspase-9 ubiquitylation can be controlled as well as the importance of this activity in controlling apoptotic progression.
Caspase-8
As the predominant initiator caspase in the extrinsic pathway, caspase-8 has a vital role in determining the fate of the cell following death receptor activation. Although rodents only contain caspase-8, other mammals also contain caspase-10, which appears to have at least a partially overlapping function with caspase-8 and is discussed in its own section below. Like caspase-9 in the intrinsic pathway, the activity of caspase-8 can also be altered by posttranslational modification, particularly phosphorylation.
A variety of serine/threonine kinases have been reported to directly phosphorylate caspase-8. For example, a recent report by Matthess et al. linked inhibition of the apoptotic machinery with the onset of mitosis, showing that active cdk1/cyclin B phosphorylates procaspase-8 at Ser387, which inhibits cleavage and activation of the caspase (Fig. 3). Accordingly, expression of a nonphosphorylatable caspase-8 mutant (Ser387 to Ala) rendered cells more sensitive to Fas-induced apoptosis during M phase (Matthess et al. 2010). Given the proximity of Ser387 to the caspase-8 cleavage sites, it was suggested that cdk1-mediated phosphorylation of caspase-8 blocks its autoprocessing, thereby protecting the cells from certain caspase-8-activating stimuli during mitosis.
Phosphorylation of caspase-8 by p38 MAPK at Ser364 inhibits the active caspase and protects neutrophils from Fas-induced death (Alvarado-Kristensson et al. 2004). Caspase-8 phosphorylation at Thr263 by the RSK2 kinase appears to both promote caspase-8 degradation and inhibit Fas-induced HeLa cell death (Peng et al. 2011). Interestingly, Peng and colleagues showed EGF-mediated caspase-8 ubiquitylation and degradation, which was reduced when caspase-8 phosphorylation of Thr263 was abrogated by mutation of Thr263 to Ala (Peng et al. 2011).
Caspase-8 also appears to be regulated directly by tyrosine kinases. Following EGF stimulation, the Src kinase phosphorylates caspase-8 at Tyr380, inhibiting Fas-induced caspase-8 activation and subsequent apoptosis (Cursi et al. 2006). Western blotting for Tyr380 phosphorylation revealed high levels of phosphorylated caspase-8 in colon cancer, where Src activity is often elevated (Cursi et al. 2006). This site is also phosphorylated by other kinases in this family, including Fyn and Lyn (Senft et al. 2007; Jia et al. 2008). In studies with the Lyn kinase, it was also determined that caspase-8 was phosphorylated at Tyr465, and both of these sites (Tyr380 and Tyr465) could be dephosphorylated by the Src-homology domain 2 (SH2)-containing tyrosine phosphatase 1 (SHP1) (Jia et al. 2008). In a model of neutrophil survival, this dephosphorylation rendered caspase-8 more responsive than the phosphorylated protein to apoptotic stimuli (Jia et al. 2008).
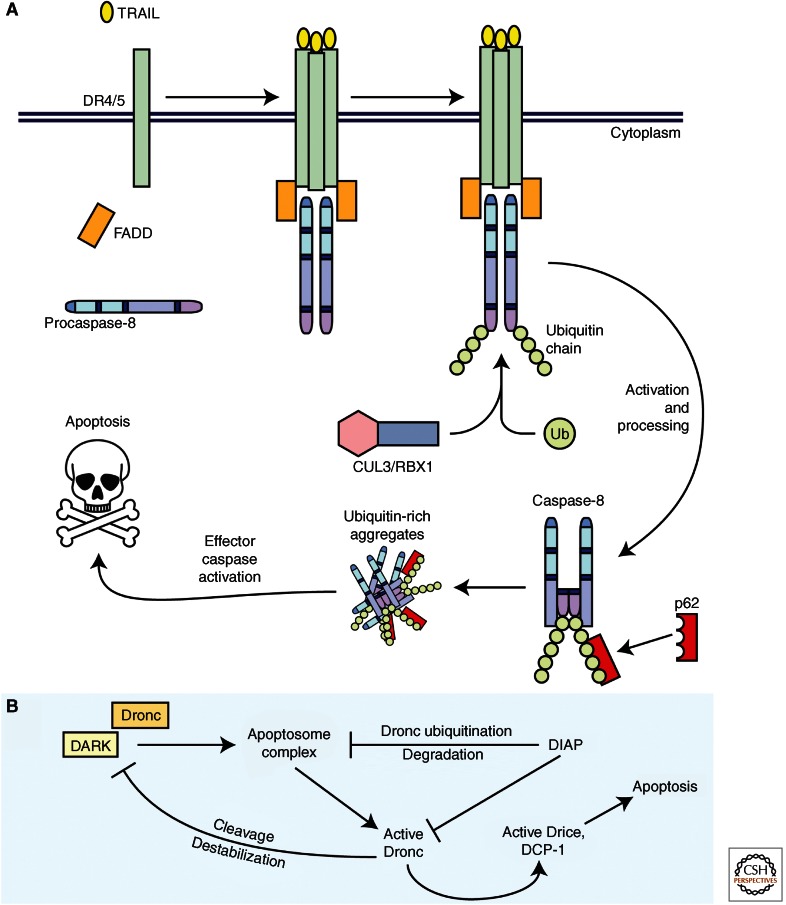
Ubiquitylation of caspase-8 also appears to have an interesting role in directly regulating the activity of the enzyme. Unlike ubiquitylation reported for other caspases, polyubiquitylation of caspase-8 by a cullin3-based E3 ligase enhances its enzymatic activity (Jin et al. 2009). This polyubiquitylation of caspase-8 occurs after recruitment of caspase-8 to the DISC, and the modification allows for the binding of active caspase-8 to the poly-Ub binding protein, p62 (Fig. 4A). Furthermore, p62 facilitates an association between this complex and other similar complexes in the cell, forming an aggregate of active caspase-8 and p62. Interestingly, caspase-8 activity appears to be enhanced within these aggregated foci, perhaps through increased stability of cleaved capase-8 (Jin et al. 2009). The deubiquitinating (DUB) enzyme A20 was reportedly involved in reversing this modification (Jin et al. 2009).
Figure 4.
Novel mechanisms of ubiquitylation in caspase regulation. (A) A novel mechanism in which caspase-8 activity is positively regulated via ubiquitylation. A proapoptotic ligand, such as TRAIL, initiates the extrinsic apoptotic pathway at the plasma membrane through formation of the DISC and recruitment of procaspase-8. Caspase-8 is then activated through induced-proximity dimerization. Once active, caspase-8 can be ubiquitylated by the neddylated form of CUL3/RBX1. This posttranslational modification on caspase-8 allows for its interaction with p62. Through autoprocessing, active caspase-8 releases the catalytic domains, which remain bound to p62. This active caspase-8 is moved into cytosolic aggregates rich in ubiquitin in which the caspase-8 remains active through stabilization of the dimer. (B) A unique mechanism for a negative feedback loop between DARK (dApaf-1) and Dronc. The Drosophila IAP, DIAP1, inhibits Dronc and Drice/DCP-1 by interacting with the caspases. DIAP1 also indirectly reduces downstream effector caspase activity by facilitating degradation of the apoptosome complex through ubiquitylation of DARK-bound active Dronc. Active Dronc can also negatively feedback on DARK by directly cleaving the protein, leading to destabilization of DARK and reduced protein levels. Thus, Dronc and DARK are involved in a DIAP1-dependent negative feedback loop.
Nitrosylation appears to also play a role in regulating caspase-8 activity. Nitric oxide (NO) induces S-nitroslyation of caspase-8, which has been reported to reduce the sensitivity of hepatocytes to TNF-α/ActD-induced apoptosis (Kim et al. 2000). Specifically, elevating levels of NO through an inducible NO synthase inhibited caspase-8 activity, caspase-3 activity, and cytochrome c release. In vivo studies in rats have showed that a liver-specific NO donor, V-PYRRO/NO, blocked caspase-8 activity, Bid cleavage, and mitochondrial cytochrome c release in the rat livers treated with TNF-α and d-galactosamine (Kim et al. 2000).
Caspase-2
Caspase-2 is a critical initiator caspase in apoptotic pathways activated by a number of cell stresses, including nutrient depletion, heat shock, DNA damage, and spindle disruption (Robertson et al. 2002; Nutt et al. 2005; Tu et al. 2006; Rudolf et al. 2009). In some of these situations, caspase-2 is controlled through posttranslational modification.
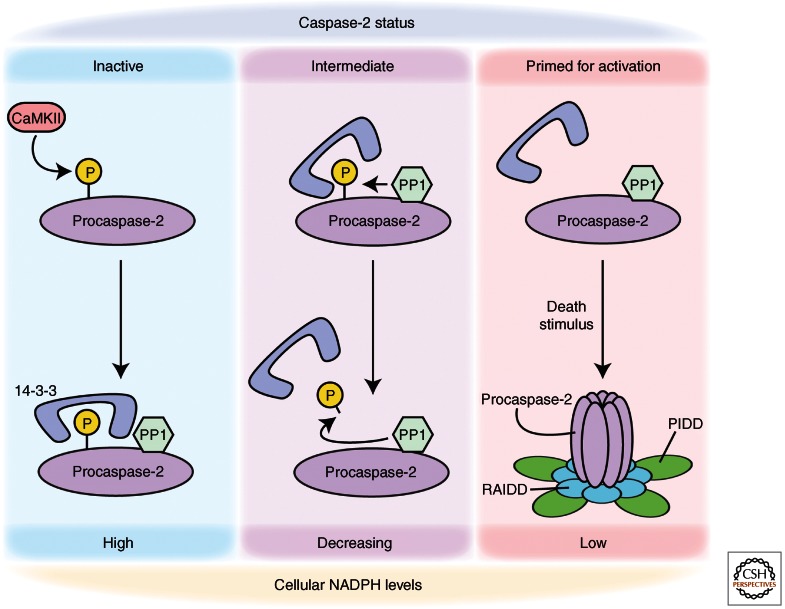
For example, it has been shown that caspase-2 is under metabolic control in the Xenopus oocyte; when there are sufficient nutrients available to drive NADPH production by the pentose phosphate pathway (PPP), a suppressive phosphorylation at Ser135 (Xenopus numbering) within the caspase-2 prodomain is catalyzed by calcium/calmodulin-dependent protein kinase II (CaMKII) (Nutt et al. 2005). This phosphorylation appears to block its binding to the activating adaptor protein, RAIDD (Nutt et al. 2005). Ser135 phosphorylation is also controlled by metabolism via binding of the phosphoserine/phosphothreonine-binding protein, 14-3-3. 14-3-3 binding protects Ser135 from being dephosphorylated by a constitutively bound PP1 (Fig. 5). As nutrients in the oocyte are depleted, 14-3-3 is removed from caspase-2, leaving it to be dephosphorylated and subsequently activated (Nutt et al. 2009). Interestingly, caspase-2–/– mice have excess oocytes resulting from a failure of apoptosis, confirming the importance of caspase-2 in mammalian oocyte cell death as well (Bergeron et al. 1998).
Figure 5.
Metabolic regulation of caspase-2. In conditions of high intracellular NAPDH, CaMKII has been shown to phosphorylate caspase-2 at Ser135 (Xenopus laevis site). This phosphorylation leads to the recruitment of the phosphoserine/phosphothreonine binding protein, 14-3-3. In this state, caspase-2 is held inactive. As NADPH levels begin to drop, 14-3-3 is released from caspase-2, allowing the constitutively bound phosphatase, PP1, to dephosphorylate Ser135. This dephosphorylation leaves caspase-2 primed for activation such that it can be recruited to its activation platform (as with the PIDDosome shown here) following a prodeath stimulus.
Phosphorylation of caspase-2 appears to also be involved in modulation of the extrinsic apoptotic pathway induced by engagement of TRAIL ligand. Specifically, CK2 was reported to suppress caspase-2 through phosphorylation at Ser157 in several cancer cell lines where low levels of casein kinase 2 (CK2) correlated with sensitization to TRAIL-induced death. Because CK2-mediated phosphorylation of caspase-2 at Ser157 blocked activation of the caspase, when CK2 activity was decreased, caspase-2 activity was enhanced (Shin et al. 2005). Although TRAIL-induced death in these cells requires caspase-8, active caspase-2 appeared to cleave procaspase-8, possibly priming these cancer cells for TRAIL-induced death.
Posttranslational modification of caspase-2 appears to also provide a means to tether caspase-2 activation to cell cycle status. Cdk1/cyclin B inhibits caspase-2 during mitosis because of direct phosphorylation at Ser340 (human numbering [Andersen et al. 2009]). This phosphorylation occurs in the linker region between the large and small subunits, suggesting that this phosphorylation may play a role in blocking full cleavage and activation of the caspase. Protein phosphatase 1, PP1, may play a role in dephosphorylating this site on caspase-2. This mitotic phosphorylation of caspase-2 provides a third example of direct caspase inhibition by cdk1 during mitosis (Fig. 3). It is interesting to note that while the cdk1 phosphorylation sites on caspase-2 and caspase-9 are located in “linker regions” that are not part of the mature, active caspase, the cdk1 phosphorylation site on caspase-8 site is part of the small subunit of the mature enzyme.
Caspase-10
Although caspase-8 and caspase-10 have both been placed in the extrinsic pathway downstream of death receptor ligands, it is still a bit unclear if these caspases are functionally equivalent with regard to activation and cleavage of downstream substrates. (Kischkel et al. 2001; Wang et al. 2001; Sprick et al. 2002; Fischer et al. 2006; Bae et al. 2008; Benkova et al. 2009; Chen et al. 2009). A recent paper by Wachmann and colleagues used in vitro dimerization assays to show that caspase-10 is also activated through induced proximity dimerization, as described for other initiator caspases (Wachmann et al. 2010). In this analysis of caspase-10 activation, active caspase-10 was able to cleave Bid, raising the possibility that capsase-10, like caspase-8, can stimulate mitochondrial cytochrome c release to aid in activation of effector caspases. Further studies are necessary to more carefully differentiate between caspase-10 and caspase-8 in response to activation of death receptor signaling and to identify specific modes of caspase-10 regulation that may or may not differ from caspase-8.
Dronc
The Drosophila initiator caspase Dronc has both caspase-9 and caspase-2-like properties. In its activation through induced proximity by an Apaf-1-homologous adaptor protein, Dronc appears to function in the fly apoptosome in a manner similar to caspase-9. However, in a paradigm similar to caspase-2, Dronc appears to be negatively regulated by CaMKII phosphorylation in response to nutrient flux through the pentose phosphate pathway (Yang et al. 2010). Specifically, high levels of NADPH support phosphorylation at a site within the prodomain of the caspase (in this case Ser130), which reduces Dronc activation by impeding the interaction between Dronc and DARK. Decreasing NADPH levels promotes Dronc’s dephosphorylation, resulting in its activation and subsequent cell death. In vivo experiments in which nonphosphorylatable Dronc was expressed in fly neurons support the observation that NADPH production attenuates Drosophila apoptosis and the importance of Dronc phosphorylation at Ser130 in maintaining metabolic suppression of Dronc (Yang et al. 2010).
Dronc is also regulated through ubiquitylation by the Drosophila IAP (DIAP1), and at least part of DIAP1’s antiapoptotic activity is attributable to this function (Muro et al. 2002; Wilson et al. 2002; Shapiro et al. 2008). Recently, work by Shapiro and colleagues showed an interesting feedback mechanism between Dronc and DARK whereby active Dronc cleaves and destabilizes its adaptor (Shapiro et al. 2008) (Fig. 4B). However, DARK also appears to play a feedback role in reducing Dronc protein levels. It was shown that Dronc could be ubiquitylated (presumably by DIAP1) only when present on the apoptosome (and thus bound to DARK). Interestingly, although details are still unclear, deubiquitylases have also been suggested to play a role in Drosophila apoptosis, likely helping to maintain a critical balance in the levels of certain apoptotic proteins (Ryoo et al. 2002; Wing et al. 2002; Ribaya et al. 2009).
Effector Caspases
Caspase-3
Caspase-3 is the most well-characterized effector caspase. Studies directed toward ordering of the caspase cascade and characterization of caspase substrates indicates that caspase-3 has both distinct and overlapping roles with caspase-7 and caspase-6 (Slee et al. 1999; Lakhani et al. 2006; Luthi and Martin 2007; Inoue et al. 2009). Although it might appear that by the time apoptotic signals reach the point of caspase-3 activation, there is no salvaging the cell, caspase-3 is still regulated through a variety of posttranslational modifications. First, both PKCδ and p38 have been shown to phosphorylate caspase-3. The PKCδ site has not been identified, but interestingly, this modification appears to enhance caspase-3 activity (Voss et al. 2005). This isoform of PKC is sometimes referred to as a proapoptotic kinase, and thus its role here may not be surprising. Caspase-mediated cleavage of PKCδ generates a shorter, active form of the kinase, which suggests the presence of a positive proapoptotic feedback loop between these two enzymes (Emoto et al. 1995; Ghayur et al. 1996; DeVries et al. 2002). Similarly to caspase-8, p38 phosphorylation of caspase-3 at Ser150, located in the large subunit of the protein, directly inhibits caspase-3 and has been shown to impede Fas-induced apoptosis in neutrophils (Alvarado-Kristensson et al. 2004). Protein phosphatase 2A (PP2A) dephosphorylates Ser150, restoring caspase-3 activity and rendering the neutrophils sensitive to Fas-induced apoptosis (Alvarado-Kristensson and Andersson 2005).
Caspase-3 is also modified by ubiquitylation. Although the role of IAPs in regulating effector caspases such as caspase-3 is not entirely clear, at least one study has shown the ability of cIAP2 to monoubiquitylate caspase-3 in vitro (Huang et al. 2000). The physiological relevance of this modification has yet to be determined. More recent work has characterized cIAP1-dependent ubiquitylation of an intermediate processed form of caspase-3, leading to proteasome-dependent degradation of the effector caspase and increased resistance to TRAIL-induced apoptosis (Choi et al. 2009). Indeed, treatment of cells with the proteasome inhibitor lactacystin has been reported to stabilize cleaved caspase-3 (active subunits) enhancing apoptosis, while having only a small effect on caspase-8 and caspase-9 (Chen et al. 2003). Other work has shown that XIAP can polyubiquitylate active caspase-3 (not procaspase-3), leading to its proteasome-dependent degradation (Suzuki et al. 2001). Unfortunately, studies with mouse models have not provided clarity on this issue. Mice with a targeted deletion in the RING domain of XIAP have decreased levels of ubiquitylated caspase-3. They also exhibit elevated caspase-3 activity, consistent with lowered levels of caspase-3 ubiquitylation (Schile et al. 2008). However, compared with control mice, XIAP–/– mice do not express increased levels of caspase-3, although this may be attributable to compensation (Harlin et al. 2001).
It has been reported that caspase-3 can be nitrosylated on its active site cysteine (Mannick et al. 1999). In this same study, caspase-3 was shown to be denitrosylated in cells following treatment with Fas ligand, thus increasing caspase activity and cell death in response to Fas. Subsequent work by this group has suggested that only a subset of caspase-3 (and caspase-9, as discussed above), localized to mitochondria, is nitrosylated (Mannick et al. 2001). A recent study reported caspase-3 nitrosylation in primary adhesion fibroblasts isolated from patients, and those cells appeared to have diminished apoptotic responsiveness when caspase-3 was nitrosylated (Jiang et al. 2009). Other models have also been used to show a role for nitrosylation in regulating caspase-3, including treatment of cadriomyocytes with SNAP and doxorubicin (Dimmeler et al. 1997; Rossig et al. 1999; Maejima et al. 2005).
Caspase-7
As for caspase-8 and caspase-10, it is currently unclear the extent to which caspase-3 and caspase-7 share functions. Initial work using peptide libraries to characterize the substrate specificity of caspase-7 suggested that caspase-7 and caspase-3 have the same substrate preferences (Thornberry et al. 1997; Stennicke et al. 2000). Despite apparently overlapping substrate specificity, caspase-7 and caspase-3 have been shown to exhibit distinct subcellular distributions in mouse livers treated with anti-Fas antibody (Chandler et al. 1998). More recent studies have supported distinct roles of these caspases in cellular breakdown during apoptosis (Walsh et al. 2008). These findings are reinforced by a comparison of phenotypes from caspase-3–/–, caspase-7–/–, and double knockout mice (and MEFs), which suggests both separate and redundant roles for these caspases (Lakhani et al. 2006). As for caspase-3, it has been reported that cIAP1 can ubiquitylate caspase-7, although cIAP1 appears to act on the mature caspase-7 enzyme, rather than the partially processed form, as for caspase-3 (Choi et al. 2009). Although there do not appear to be parallel reports of caspases-3 and -7 phosphorylation, recent work has shown PAK2 (p21-activated kinase)-mediated phosphorylation of caspase-7 at Ser30, Thr173, and Ser239, which negatively regulate caspase-7 activity (Li et al. 2011).
Interestingly, although caspases-3 and -7 share both upstream activators and some overlapping substrate specificity, their divergent amino-terminal sequences appear to contribute to different modes of regulation. Amino-terminal sequences of caspase-7 present in the proenzyme but absent from the mature enzyme seem to negatively regulate capsase-7 function. Removal of this amino terminus potentiates caspase-7 activity, and Salvesen and colleagues have hypothesized that these residues play an as-yet undefined role in sequestering the enzyme from its upstream activators (Denault and Salvesen 2003).
Caspase-6
Although caspase-6 also functions as an effector caspase, it has been known for some time that caspase-6 has at least some unique cellular substrates. It has been reported that caspase-6 is activated only following activation of caspase-3 or caspase-7 and thus is not activated directly by an initiator caspase (Inoue et al. 2009). Once active, caspase-6 can cleave initiator caspases-2 and -8, although the functional consequences of this cleavage (away from the initiator caspase activation platform) are unclear (Slee et al. 1999; Inoue et al. 2009). In contrast, other studies have showed that caspase-6 can be activated in the absence of caspase-3 or -7 activity (LeBlanc et al. 1999; Allsopp et al. 2000; Doostzadeh-Cizeron et al. 2000). Subsequent work has provided support for the idea that caspase-6 can activate itself in vivo (Wang et al. 2010). Using the crystal structure of caspase-6, Wang and colleagues have revealed that caspase-6 can regulate its own activity through intramolecular caspase self-activation, although it is not clear how this process is engaged physiologically. Like caspase-7, the amino-terminal residues of caspase-6 appear to serve an inhibitory function (Klaiman et al. 2009; Wang et al. 2010).
Caspase-6 can be inhibited posttranslationally by kinase ARK5 (Suzuki et al. 2004). In the colon cancer cell line, SW480, which effectively evades Fas-induced apoptosis, it has been shown that caspase-6 is held inactive by the ARK5 kinase. Treatment of SW480 cells with ARK5 antisense RNA liberated caspase-6 to cleave c-FLIP to inhibit Fas signaling. Although there are two putative ARK5 sites on capase-6, the kinase appears to phosphorylate Ser257. Mutation of Ser257 to Ala overrides ARK5-mediated inhibition and allows c-FLIP cleavage and apoptosis to proceed in the SW480 cells (Suzuki et al. 2004).
DRICE AND DCP-1
The Drosophila effector caspases, Drice and DCP-1, are regulated through ubiquitylation, catalyzed by the IAP proteins, DIAP1 and DIAP2. Ribeiro and colleagues recently showed a role for DIAP2 in the ubiquitylation and inactivation of Drice (Ribeiro et al. 2007). Interestingly, DIAP2 acts as a pseudosubstrate of Drice; Drice’s attempt to cleave DIAP2 results in trapping of DIAP2 in a covalent association with Drice, which allows proximal ubiquitylation of the associated Drice by the DIAP2 RING domain.
In a distinct mechanism, the catalytic activity of Drice also appears to be required for its ubiquitylation by DIAP1. Drice cleaves DIAP1, resulting in a conformational change, which allows DIAP1 to interact directly with Drice/DCP-1 for caspase ubiquitylation by the DIAP1 RING (Ditzel et al. 2008). In S2 cells, Drice polyubiquitylation does not appear to facilitate its degradation. Rather, ubiquitylation catalytically inactivates the protein and inhibits its ability to cleave downstream substrates. Although it is currently unclear exactly how this polyubiquitylation inactivates Drice, several possibilities exist. The polyubiquitin chains may physically block substrate access to the caspase’s catalytic site, or the posttranslational modification may alter the confirmation of the catalytic domain to preclude productive enzyme–substrate interactions.
PROTEIN/PROTEIN INTERACTIONS
In addition to the changes that posttranslational modifications can have on caspase function, various proteins (by nature of their interaction with a caspase) can impact apoptotic pathways both positively and negatively. Some of the first proteins discovered as direct caspase inhibitors were of viral origin (such as p35 and CrmA) and do not appear to have direct cellular counterparts (Clem et al. 1991; Ray et al. 1992). In addition, there are many proteins that affect caspase activity indirectly by modulating, for example, the complex/platform on which the caspase is activated. For example, the backbone of the caspase-9-activating apoptosome, Apaf-1, is regulated by a complex of proteins, hsp70, CAS, and PHAPI (Kim et al. 2008). These proteins facilitate nucleotide exchange on Apaf-1, which is necessary for full apoptosome formation and subsequent caspase-9 activation. Interestingly, levels of other (nonprotein) cellular components, such nucleotides and ions, also appear to play critical roles in regulating certain steps in the activation of caspases and the apoptotic pathway (Cain et al. 2001; Chandra et al. 2006; Bao et al. 2007; Karki et al. 2007; Mei et al. 2010). Because there have been a variety of recent reviews on the proteins that regulate caspases indirectly, these circumstances are not covered here (Schafer and Kornbluth 2006; Fadeel et al. 2008; Krumschnabel et al. 2009). Instead, we focus on a few key examples of proteins that interact directly with caspases to alter either their activation or enzymatic activity.
FLIP
The FLIP (FLICE-like inhibitory protein, so-called because of the original caspase-8 name, FLICE) family of proteins was first discovered in a viral context, but homologs in other species, including mammals, were discovered nearly simultaneously (Thome et al. 1997). Interestingly, the FLIP proteins have been shown to function both as inhibitors and activators of the apoptotic process. Mammalian cellular FLIP (c-FLIP) is notably similar to procaspase-8 and procaspase-10 in primary sequence. Specifically, the long FLIP isoform contains the two death effector domains (DEDs) at the amino terminus of the protein, and at the carboxyl terminus, it has a pseudocaspase domain that lacks catalytic activity (Irmler et al. 1997). The short isoform (c-FLIPs) is similar to the viral FLIP, resembling a truncated caspase-8, with two DEDs and no region homologous with the catalytic domain. It has been shown that the long form of c-FLIP is recruited to the DISC and can be partially processed, with a portion of the FLIP protein retained at the DISC. This interaction with the DISC inhibits any subsequent recruitment of caspase-8 to that site (Goltsev et al. 1997; Han et al. 1997; Irmler et al. 1997; Scaffidi et al. 1999). In addition, it appears that the short isoform can completely prevent processing of DISC-bound caspase-8, whereas the long isoform appears to allow partial processing (Krueger et al. 2001).
In contrast to the studies above, which characterized FLIPL as an inhibitor of caspase activation, some studies have shown that FLIPL can, in fact, facilitate caspase activation (Chang et al. 2002; Micheau et al. 2002). It may be that the local concentration of c-FLIP at the DISC determines its role as an inhibitor or activator of caspase-8. As an activator of caspase-8, c-FLIPL appears to heterodimerize with casapse-8, activating the caspase portion of the dimer. Indeed, this hypothesis is supported by Boatright and colleagues who used kosmotropes to induce FLIP/caspase-8 dimers. These heterodimers appeared to have a lower kinetic barrier to activation than homodimers between two wild-type caspase-8 molecules (Boatright et al. 2004). Interestingly, in contrast to the proapoptotic caspase-8 homodimer, these heterodimers of caspase-8 and FLIPL have been shown to promote cell survival (Oberst et al. 2011; van Raam and Salvesen 2012).
IAPs
The inhibitor of apoptosis proteins (IAPs) are a conserved family of proteins that are defined by the presence of at least one baculovirus IAP repeat domain (BIR), and, as discussed above, some of the IAPs also contain a RING domain that confers E3 ubiquitin ligase function (Crook et al. 1993). In addition, XIAP and DIAP proteins have been shown to act as direct stoichiometric inhibitors of caspases. XIAP can inhibit caspase-3 and caspase-7 by binding to the active site, and caspase-9 by preventing or reversing dimerization (Deveraux et al. 1997; Chai et al. 2001; Riedl et al. 2001; Srinivasula et al. 2001; Shiozaki et al. 2003). This inhibitory action of XIAP can be antagonized by mitochondrial proteins Smac/DIABLO and Omi/HtrA2, which are released during an apoptotic stimulus along with cytochrome c (Chai et al. 2000; Du et al. 2000). These proteins are similar to the Reaper, Hid, Grim (RHG) family of Drosophila proteins, which can activate apoptosis in part by displacing the DIAP1 protein from Dronc and effector caspases (Fig. 2). As a direct inhibitor of these caspases, XIAP has been detected on the apoptosome (Bratton et al. 2001, 2002). In a recent study of melanoma cell lines, XIAP was also subject to caspase cleavage in a positive feedback loop that reduces caspase inhibition and XIAP levels through proteasomal degradation (Hornle et al. 2011). Although XIAP is the best-characterized vertebrate member of the IAP family with regard to direct binding and inhibition of caspases, one recent study showed that cIAP1 can specifically block apoptosis downstream of cytochrome c release by binding to and inhibiting active caspase-9 within the apoptosome, precluding downstream activation of procaspase-3 (Burke et al. 2010).
TRANSCRIPTIONAL REGULATION OF CASPASES
Although we have focused thus far on the posttranslational regulation of caspases, some of the caspase proteins are also regulated at the level of expression through control of their mRNA transcription. Although much less is known about the mechanisms modulating mRNA expression levels, this type of regulation also appears to contribute to control of caspase activity before or during an apoptotic stimulus. Interestingly, caspase-2 transcriptional regulation appears to be coordinated with transcriptional induction of its activators, PIDD and RAIDD (Krumschnabel et al. 2009). One recent study showed a p53/p21-dependent pathway for down-regulating caspase-2 mRNA expression in resting cells and in response to DNA damage (Baptiste-Okoh et al. 2008). Although this finding seems a bit counterintuitive, the authors postulate that this mechanism reduces unwanted extraneous cell death. Following DNA damage, the caspase-2 activator, PIDD, is transcriptionally up-regulated, potentially overcoming the reduced expression of caspase-2 and facilitating cell death.
Alternative splicing of caspase-8 plays a role in modulating apoptotic induction via the extrinsic pathway. Similarly to FLIP, the caspsase-8 L splice variant has a functional DED but lacks key residues in the catalytic domain, which allows it to interact with the DISC at the plasma membrane without being able to induce downstream caspase activation (Curtin and Cotter 2003). Thus, caspase-8 L behaves like c-FLIP and acts like a caspase-8 inhibitor (Himeji et al. 2002). Methylation of the caspase-8 gene is thought to suppress caspase-8 expression in a variety of tumors (Harada et al. 2002; Curtin and Cotter 2003). Pediatric tumors, including rhabdomyosarcomas, medulloblastomas, retinoblastomas, and neuroblastomas, have all been reported to exhibit methylation of the caspase-8 gene, correlating with a decrease in caspase-8 expression. Interestingly, many of the cell lines used for this study also had down-regulated caspase-10 at a posttranscriptional level (Harada et al. 2002).
The caspase-10 gene has been characterized as a direct target of p53 following DNA damage (Rikhof et al. 2003). Treatment of both p53-wild type and p53-deficient cell lines with etopside or adriamycin-induced caspase-10 mRNA expression in a p53-dependent manner without an effect on caspase-8 expression. Caspase-10 mRNA levels are negatively regulated in Jurkat cells expressing the HIV tat gene when compared with control-treated Jurkat cells (Gibellini et al. 2005). Although the levels of caspase-8 were unchanged, the reduction of caspase-10 mRNA coupled with a concomitant increase in expression of cFLIP were sufficient to enhance the resistance of Tat-expressing T cells to TRAIL-induced apoptosis.
Two different caspase-9 transcripts can be derived from the caspase-9 gene, differing by four exons. Caspase-9a includes these four exons, whereas caspase-9b does not. Importantly, it appears that capsase-9a is proapoptotic, but the shorter isoform, caspase-9b, is antiapoptotic (Seol and Billiar 1999; Srinivasula et al. 1999). Recently, Shultz and colleagues have found that this alternative splicing of caspase-9 is defective in nonsmall cell lung carcinoma (NSCLC) (Shultz et al. 2010). Specifically, the authors showed that K-Ras12V overexpression increased the ratio of caspase-9a to capase-9b, while epidermal growth factor receptor (EGFR) overexpression lowered this ratio. Furthermore, they showed that the RNA splicing factor, SRp30a, is phosphorylated by Akt, which affects the inclusion of these four exons in the mature caspase-9 transcript. It is interesting to note that Apaf-1, the adapter protein for caspase-9, can also be transcriptionally regulated by E2F1 and p53 (Fortin et al. 2001; Moroni et al. 2001; Furukawa et al. 2002; Johnson et al. 2007).
CASPASE REGULATION IN NONAPOPTOTIC CELLULAR PROCESSES
In addition to the various mechanisms that regulate caspases during apoptosis, many studies have shown that active caspases play important roles in nonapoptotic cellular functions, including inflammation, protein secretion, and differentiation (reviewed in Kuranaga and Miura 2007; Li and Yuan 2008; Feinstein-Rotkopf and Arama 2009; Yi and Yuan 2009). Caspase activity in the absence of a cell death signal may have unintended deadly consequences in the absence of finely tuned regulation. Thus, caspase control is particularly critical in circumstances in which the proteases are activated for short, well-defined periods of time or at isolated subcellular locations. Phenotypes of the caspase knockout mice have provided additional evidence to support a role for caspases in nonapoptotic processes (reviewed in Li and Yuan 2008). An exhaustive discussion of nonapoptotic caspase activation/regulation is beyond the scope of this article, but a few recent studies are described below to illustrate the nature of such regulation and to highlight some outstanding questions.
In Drosophila melanogaster, tightly controlled spatial regulation of effector caspase activity is crucial during the terminal differentiation of spermatids, a process also known as individualization (Kaplan et al. 2010). Kaplan and colleagues have shown that a spermatid contains a distal-to-proximal gradient of the protein Soti, which results in a similar directional gradient of the IAP dBruce (because Soti inhibits an E3 ubiquitin ligase that ubiquitylates dBruce). During spermatid individualization, dBruce inhibits caspase activity, thus creating an opposing gradient of effector caspase activity (proximal-to-distal). This prevents unwanted death in the latest-individualizing region of the spermatid and helps to properly drive differentiation. As a result of the timing of individualization and the presence of dBruce, caspase activation is insufficient to initiate the full apoptotic program and subsequent cell death.
In hippocampal neurons, a role for transient activation of caspase-3 has been showed in synaptic long-term depression (LTD) and AMPA receptor internalization (Li et al. 2010). The authors provide evidence to support a model whereby caspase-3 is activated through a pathway similar to that which occurs during intrinsic apoptosis. Indeed, NMDA treatment facilitates mitochondrial release of cytochrome c and caspase-9 activation, which is necessary for the activation of caspase-3. Additional evidence suggests that cleavage and inactivation of Akt1 provides at least part of the link between caspase-3 activity and LTD; overexpression of a noncleavable Akt1 in hippocampal slice cultures inhibited LTD, confirming the importance of at least one caspase substrate during LTD and AMPA internalization. How this activity is limited to prevent neuronal death is not entirely clear.
Another recent study using olfactory sensory neurons (OSNs) has elucidated a nonapoptotic role for Apaf-1 and caspase-9 signaling in development (Ohsawa et al. 2010). This nonapoptotic caspase activity leads to cleavage of the membrane-bound protein, Semaphorin 7A, which is critical for appropriate formation of axonal projections (Pasterkamp et al. 2003). In mice lacking Apaf-1 or caspase-9 expression, a variety of problems exist, including OSN axons that are routed erroneously and OSNs that have not matured properly. However, there are no changes in the number of neurons, supporting the idea that Apaf-1 and caspase-9 are not impacting generation of OSNs, but rather specific formation of axonal projections.
CONCLUDING REMARKS
Because of the critical role that caspases play in executing the apoptotic program, to avoid unplanned cellular demise, their activation and activity must be tightly regulated. By their very structure, caspases are regulated as soon as they are expressed. They are synthesized as zymogens, which are only to be activated following the appropriate stimulus. Both before and after activation, caspases can be regulated through a variety of mechanisms including posttranslational modifications and protein/protein interactions. Although caspase activity was initially reported to occur only during apoptosis, more recent evidence suggests that caspases play critical roles in other, nonapoptosis cellular processes. Detailed analyses of these specific physiological/pathological circumstances may yield important insight into new and different modes of caspase regulation, some of which may also be important for understanding control of caspase activation in apoptosis.
ACKNOWLEDGMENTS
The authors thank Manabu Kurokawa and Reichen Yang for helpful feedback and discussion.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW 2002. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR 2007. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310 [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR 2009. Apoptosis and autophagy: Regulation of caspase-9 by phosphorylation. FEBS J 276: 6063–6073 [DOI] [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR 2003. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5: 647–654 [DOI] [PubMed] [Google Scholar]
- Allsopp TE, McLuckie J, Kerr LE, Macleod M, Sharkey J, Kelly JS 2000. Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ 7: 984–993 [DOI] [PubMed] [Google Scholar]
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J 1996. Human ICE/CED-3 protease nomenclature. Cell 87: 171. [DOI] [PubMed] [Google Scholar]
- Alvarado-Kristensson M, Andersson T 2005. Protein phosphatase 2A regulates apoptosis in neutrophils by dephosphorylating both p38 MAPK and its substrate caspase 3. J Biol Chem 280: 6238–6244 [DOI] [PubMed] [Google Scholar]
- Alvarado-Kristensson M, Melander F, Leandersson K, Ronnstrand L, Wernstedt C, Andersson T 2004. p38–MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J Exp Med 199: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S 2009. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J 28: 3216–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Ha TS, Yoon Y, Lee J, Cha HJ, Yoo H, Choe TB, Li S, Sohn I, Kim JY, et al. 2008. Genome-wide screening and identification of novel proteolytic cleavage targets of caspase-8 and -10 in vitro. Int J Mol Med 21: 381–386 [PubMed] [Google Scholar]
- Baliga BC, Read SH, Kumar S 2004. The biochemical mechanism of caspase-2 activation. Cell Death Differ 11: 1234–1241 [DOI] [PubMed] [Google Scholar]
- Bao Q, Lu W, Rabinowitz JD, Shi Y 2007. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol Cell 25: 181–192 [DOI] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C 2008. Caspase 2 is both required for p53-mediated apoptosis and downregulated by p53 in a p21-dependent manner. Cell Cycle 7: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Benkova B, Lozanov V, Ivanov IP, Mitev V 2009. Evaluation of recombinant caspase specificity by competitive substrates. Anal Biochem 394: 68–74 [DOI] [PubMed] [Google Scholar]
- Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham KE, Flaws JA, Salter JC, et al. 1998. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev 12: 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S 2005. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci 102: 14314–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. 2003. A unified model for apical caspase activation. Mol Cell 11: 529–541 [DOI] [PubMed] [Google Scholar]
- Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS 2004. Activation of caspases-8 and -10 by FLIPL. Biochem J 382: 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzon C, Bouchier-Hayes L, Pagliari LJ, Green DR, Newmeyer DD 2006. Caspase-2-induced apoptosis requires bid cleavage: A physiological role for bid in heat shock-induced death. Mol Biol Cell 17: 2150–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SC, Allan LA, Clarke PR 2005. Regulation of caspase 9 through phosphorylation by protein kinase C ζ in response to hyperosmotic stress. Mol Cell Biol 25: 10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM 2001. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20: 998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Lewis J, Butterworth M, Duckett CS, Cohen GM 2002. XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ 9: 881–892 [DOI] [PubMed] [Google Scholar]
- Burke SP, Smith L, Smith JB 2010. cIAP1 cooperatively inhibits procaspase-3 activation by the caspase-9 apoptosome. J Biol Chem 285: 30061–30068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain K, Langlais C, Sun XM, Brown DG, Cohen GM 2001. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J Biol Chem 276: 41985–41990 [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC 1998. Regulation of cell death protease caspase-9 by phosphorylation. Science 282: 1318–1321 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P 1998. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94: 727–737 [DOI] [PubMed] [Google Scholar]
- Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Van Ness K, Greenstreet TA, March CJ, Kronheim SR, Druck T, Cannizzaro LA, et al. 1992. Molecular cloning of the interleukin-1 β converting enzyme. Science 256: 97–100 [DOI] [PubMed] [Google Scholar]
- Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y 2000. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406: 855–862 [DOI] [PubMed] [Google Scholar]
- Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y 2001. Structural basis of caspase-7 inhibition by XIAP. Cell 104: 769–780 [DOI] [PubMed] [Google Scholar]
- Chandler JM, Cohen GM, MacFarlane M 1998. Different subcellular distribution of caspase-3 and caspase-7 following Fas-induced apoptosis in mouse liver. J Biol Chem 273: 10815–10818 [DOI] [PubMed] [Google Scholar]
- Chandra D, Bratton SB, Person MD, Tian Y, Martin AG, Ayres M, Fearnhead HO, Gandhi V, Tang DG 2006. Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome c and inhibiting apoptosome. Cell 125: 1333–1346 [DOI] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X 2002. c-FLIPL is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J 21: 3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Smith L, Wang Z, Smith JB 2003. Preservation of caspase-3 subunits from degradation contributes to apoptosis evoked by lactacystin: Any single lysine or lysine pair of the small subunit is sufficient for ubiquitination. Mol Pharmacol 64: 334–345 [DOI] [PubMed] [Google Scholar]
- Chen H, Xia Y, Fang D, Hawke D, Lu Z 2009. Caspase-10-mediated heat shock protein 90 β cleavage promotes UVB irradiation-induced cell apoptosis. Mol Cell Biol 29: 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Jin CY, Han MH, Kim GY, Kim ND, Lee WH, Kim SK, Choi YH 2009. Sanguinarine sensitizes human gastric adenocarcinoma AGS cells to TRAIL-mediated apoptosis via down-regulation of AKT and activation of caspase-3. Anticancer Res 29: 4457–4465 [PubMed] [Google Scholar]
- Clem RJ, Fechheimer M, Miller LK 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254: 1388–1390 [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529 [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol 67: 2168–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursi S, Rufini A, Stagni V, Condo I, Matafora V, Bachi A, Bonifazi AP, Coppola L, Superti-Furga G, Testi R, et al. 2006. Src kinase phosphorylates Caspase-8 on Tyr380: A novel mechanism of apoptosis suppression. EMBO J 25: 1895–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JF, Cotter TG 2003. Live and let die: Regulatory mechanisms in Fas-mediated apoptosis. Cell Signal 15: 983–992 [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ 2004. Cell death: Critical control points. Cell 116: 205–219 [DOI] [PubMed] [Google Scholar]
- Denault JB, Salvesen GS 2003. Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem 278: 34042–34050 [DOI] [PubMed] [Google Scholar]
- Dessauge F, Cayla X, Albar JP, Fleischer A, Ghadiri A, Duhamel M, Rebollo A 2006. Identification of PP1α as a caspase-9 regulator in IL-2 deprivation-induced apoptosis. J Immunol 177: 2441–2451 [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388: 300–304 [DOI] [PubMed] [Google Scholar]
- DeVries TA, Neville MC, Reyland ME 2002. Nuclear import of PKCδ is required for apoptosis: Identification of a novel nuclear import sequence. EMBO J 21: 6050–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmeler S, Haendeler J, Nehls M, Zeiher AM 1997. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1β-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J Exp Med 185: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Elliott R, Zvelebil M, Blagoev B, Bergmann A, et al. 2008. Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell 32: 540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF 2008. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Cravatt BF 2012. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 150: 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doostzadeh-Cizeron J, Yin S, Goodrich DW 2000. Apoptosis induced by the nuclear death domain protein p84N5 is associated with caspase-6 and NF-κB activation. J Biol Chem 275: 25336–25341 [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102: 33–42 [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM 1997. RAIDD is a new “death” adaptor molecule. Nature 385: 86–89 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH 1999. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68: 383–424 [DOI] [PubMed] [Google Scholar]
- Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong WW, Kamen R, Weichselbaum R, et al. 1995. Proteolytic activation of protein kinase C δ by an ICE-like protease in apoptotic cells. EMBO J 14: 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B, Ottosson A, Pervaiz S 2008. Big wheel keeps on turning: Apoptosome regulation and its role in chemoresistance. Cell Death Differ 15: 443–452 [DOI] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y, Arama E 2009. Can’t live without them, can live with them: Roles of caspases during vital cellular processes. Apoptosis 14: 980–995 [DOI] [PubMed] [Google Scholar]
- Fischer U, Stroh C, Schulze-Osthoff K 2006. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene 25: 152–159 [DOI] [PubMed] [Google Scholar]
- Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, Hakim A, Albert PR, Cecconi F, Helin K, et al. 2001. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J Cell Biol 155: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Nishimura N, Satoh M, Endo H, Iwase S, Yamada H, Matsuda M, Kano Y, Nakamura M 2002. Apaf-1 is a mediator of E2F-1-induced apoptosis. J Biol Chem 277: 39760–39768 [DOI] [PubMed] [Google Scholar]
- Gao Z, Shao Y, Jiang X 2005. Essential roles of the Bcl-2 family of proteins in caspase-2-induced apoptosis. J Biol Chem 280: 38271–38275 [DOI] [PubMed] [Google Scholar]
- Ghayur T, Hugunin M, Talanian RV, Ratnofsky S, Quinlan C, Emoto Y, Pandey P, Datta R, Huang Y, Kharbanda S, et al. 1996. Proteolytic activation of protein kinase C δ by an ICE/CED 3-like protease induces characteristics of apoptosis. J Exp Med 184: 2399–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini D, Re MC, Ponti C, Vitone F, Bon I, Fabbri G, Grazia Di Iasio M, Zauli G 2005. HIV-1 Tat protein concomitantly down-regulates apical caspase-10 and up-regulates c-FLIP in lymphoid T cells: A potential molecular mechanism to escape TRAIL cytotoxicity. J Cell Physiol 203: 547–556 [DOI] [PubMed] [Google Scholar]
- Goltsev YV, Kovalenko AV, Arnold E, Varfolomeev EE, Brodianskii VM, Wallach D 1997. CASH, a novel caspase homologue with death effector domains. J Biol Chem 272: 19641–19644 [DOI] [PubMed] [Google Scholar]
- Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, Elia A, de la Pompa JL, Kagi D, Khoo W, et al. 1998. Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell 94: 339–352 [DOI] [PubMed] [Google Scholar]
- Han DK, Chaudhary PM, Wright ME, Friedman C, Trask BJ, Riedel RT, Baskin DG, Schwartz SM, Hood L 1997. MRIT, a novel death-effector domain-containing protein, interacts with caspases and BclXL and initiates cell death. Proc Natl Acad Sci 94: 11333–11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Harada K, Toyooka S, Shivapurkar N, Maitra A, Reddy JL, Matta H, Miyajima K, Timmons CF, Tomlinson GE, Mastrangelo D, et al. 2002. Deregulation of caspase 8 and 10 expression in pediatric tumors and cell lines. Cancer Res 62: 5897–5901 [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB 2001. Characterization of XIAP-deficient mice. Mol Cell Biol 21: 3604–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO, Horvitz HR 1994. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76: 665–676 [DOI] [PubMed] [Google Scholar]
- Himeji D, Horiuchi T, Tsukamoto H, Hayashi K, Watanabe T, Harada M 2002. Characterization of caspase-8 L: A novel isoform of caspase-8 that behaves as an inhibitor of the caspase cascade. Blood 99: 4070–4078 [DOI] [PubMed] [Google Scholar]
- Hornle M, Peters N, Thayaparasingham B, Vorsmann H, Kashkar H, Kulms D 2011. Caspase-3 cleaves XIAP in a positive feedback loop to sensitize melanoma cells to TRAIL-induced apoptosis. Oncogene 30: 575–587 [DOI] [PubMed] [Google Scholar]
- Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem 275: 26661–26664 [DOI] [PubMed] [Google Scholar]
- Inoue S, Browne G, Melino G, Cohen GM 2009. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ 16: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, et al. 1997. Inhibition of death receptor signals by cellular FLIP. Nature 388: 190–195 [DOI] [PubMed] [Google Scholar]
- Itoh N, Nagata S 1993. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J Biol Chem 268: 10932–10937 [PubMed] [Google Scholar]
- Jia SH, Parodo J, Kapus A, Rotstein OD, Marshall JC 2008. Dynamic regulation of neutrophil survival through tyrosine phosphorylation or dephosphorylation of caspase-8. J Biol Chem 283: 5402–5413 [DOI] [PubMed] [Google Scholar]
- Jiang ZL, Fletcher NM, Diamond MP, Abu-Soud HM, Saed GM 2009. S-nitrosylation of caspase-3 is the mechanism by which adhesion fibroblasts manifest lower apoptosis. Wound Repair Regen 17: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A 2009. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137: 721–735 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM 2000. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Johnson CE, Huang YY, Parrish AB, Smith MI, Vaughn AE, Zhang Q, Wright KM, Van Dyke T, Wechsler-Reya RJ, Kornbluth S, et al. 2007. Differential Apaf-1 levels allow cytochrome c to induce apoptosis in brain tumors but not in normal neural tissues. Proc Natl Acad Sci 104: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E 2010. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev Cell 19: 160–173 [DOI] [PubMed] [Google Scholar]
- Karki P, Seong C, Kim JE, Hur K, Shin SY, Lee JS, Cho B, Park IS 2007. Intracellular K+ inhibits apoptosis by suppressing the Apaf-1 apoptosome formation and subsequent downstream pathways but not cytochrome c release. Cell Death Differ 14: 2068–2075 [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR 1972. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26: 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Kim TH, Chung HT, Talanian RV, Yin XM, Billiar TR 2000. Nitric oxide prevents tumor necrosis factor α-induced rat hepatocyte apoptosis by the interruption of mitochondrial apoptotic signaling through S-nitrosylation of caspase-8. Hepatology 32: 770–778 [DOI] [PubMed] [Google Scholar]
- Kim HE, Jiang X, Du F, Wang X 2008. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell 30: 239–247 [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME 1995. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 14: 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, Gazdar A, Blenis J, Arnott D, Ashkenazi A 2001. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 276: 46639–46646 [DOI] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, LeBlanc AC 2009. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta 1793: 592–601 [DOI] [PubMed] [Google Scholar]
- Kornbluth S, White K 2005. Apoptosis in Drosophila: Neither fish nor fowl (nor man, nor worm). J Cell Sci 118: 1779–1787 [DOI] [PubMed] [Google Scholar]
- Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S 2001. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem 276: 20633–20640 [DOI] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A 2009. The enigma of caspase-2: The laymen’s view. Cell Death Differ 16: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA 1998. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94: 325–337 [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Miura M 2007. Nonapoptotic functions of caspases: Caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol 17: 135–144 [DOI] [PubMed] [Google Scholar]
- Laguna A, Aranda S, Barallobre MJ, Barhoum R, Fernandez E, Fotaki V, Delabar JM, de la Luna S, de la Villa P, Arbones ML 2008. The protein kinase DYRK1A regulates caspase-9-mediated apoptosis during retina development. Dev Cell 15: 841–853 [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA 2006. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science 311: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc A, Liu H, Goodyer C, Bergeron C, Hammond J 1999. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer’s disease. J Biol Chem 274: 23426–23436 [DOI] [PubMed] [Google Scholar]
- Li J, Yuan J 2008. Caspases in apoptosis and beyond. Oncogene 27: 6194–6206 [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94: 491–501 [DOI] [PubMed] [Google Scholar]
- Li Z, Jo J, Jia JM, Lo SC, Whitcomb DJ, Jiao S, Cho K, Sheng M 2010. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 141: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wen W, Liu K, Zhu F, Malakhova M, Peng C, Li T, Kim HG, Ma W, Cho YY, et al. 2011. Phosphorylation of caspase-7 by P21-activated protein kinase (PAK) 2 inhibits chemotherapeutic drugs-induced apoptosis of breast cancer cell lines. J Biol Chem 286: 22291–22299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X 1996. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 86: 147–157 [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94: 481–490 [DOI] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ 2007. The CASBAH: A searchable database of caspase substrates. Cell Death Differ 14: 641–650 [DOI] [PubMed] [Google Scholar]
- Maejima Y, Adachi S, Morikawa K, Ito H, Isobe M 2005. Nitric oxide inhibits myocardial apoptosis by preventing caspase-3 activity via S-nitrosylation. J Mol Cell Cardiol 38: 163–174 [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA 2008. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134: 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB 2009. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J 28: 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS 1999. Fas-induced caspase denitrosylation. Science 284: 651–654 [DOI] [PubMed] [Google Scholar]
- Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B 2001. S-Nitrosylation of mitochondrial caspases. J Cell Biol 154: 1111–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A 2009. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol 185: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MC, Allan LA, Lickrish M, Sampson C, Morrice N, Clarke PR 2005. Protein kinase A regulates caspase-9 activation by Apaf-1 downstream of cytochrome c. J Biol Chem 280: 15449–15455 [DOI] [PubMed] [Google Scholar]
- Matthess Y, Raab M, Sanhaji M, Lavrik IN, Strebhardt K 2010. Cdk1/cyclin B1 controls Fas-mediated apoptosis by regulating caspase-8 activity. Mol Cell Biol 30: 5726–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell MA, Abedin MJ, Melendez M, Platikanova TN, Ecklund JR, Ahmed K, Kelekar A 2008. Phosphorylation of murine caspase-9 by the protein kinase casein kinase 2 regulates its cleavage by caspase-8. J Biol Chem 283: 20149–20158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Stonestrom A, Hou YM, Yang X 2010. Apoptotic regulation and tRNA. Protein Cell 1: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grutter MG 2002. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 277: 45162–45171 [DOI] [PubMed] [Google Scholar]
- Morizane Y, Honda R, Fukami K, Yasuda H 2005. X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J Biochem 137: 125–132 [DOI] [PubMed] [Google Scholar]
- Moroni MC, Hickman ES, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol 3: 552–558 [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ 2002. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 277: 49644–49650 [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. 1995. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376: 37–43 [DOI] [PubMed] [Google Scholar]
- Nutt LK, Margolis SS, Jensen M, Herman CE, Dunphy WG, Rathmell JC, Kornbluth S 2005. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell 123: 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt LK, Buchakjian MR, Gan E, Darbandi R, Yoon SY, Wu JQ, Miyamoto YJ, Gibbons JA, Andersen JL, Freel CD, et al. 2009. Metabolic control of oocyte apoptosis mediated by 14–3-3ζ-regulated dephosphorylation of caspase-2. Dev Cell 16: 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR 2011. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S, Hamada S, Kuida K, Yoshida H, Igaki T, Miura M 2010. Maturation of the olfactory sensory neurons by Apaf-1/caspase-9-mediated caspase activity. Proc Natl Acad Sci 107: 13366–13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H 2007. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL 2003. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature 424: 398–405 [DOI] [PubMed] [Google Scholar]
- Peng C, Cho YY, Zhu F, Zhang J, Wen W, Xu Y, Yao K, Ma WY, Bode AM, Dong Z 2011. Phosphorylation of caspase-8 (Thr-263) by ribosomal S6 kinase 2 (RSK2) mediates caspase-8 ubiquitination and stability. J Biol Chem 286: 6946–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS 2006. The apoptosome activates caspase-9 by dimerization. Mol Cell 22: 269–275 [DOI] [PubMed] [Google Scholar]
- *.Poreba M, Strozyk A, Salvesen GS, Drag M 2013. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. 2010. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141: 446–457 [DOI] [PubMed] [Google Scholar]
- Raina D, Pandey P, Ahmad R, Bharti A, Ren J, Kharbanda S, Weichselbaum R, Kufe D 2005. c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J Biol Chem 280: 11147–11151 [DOI] [PubMed] [Google Scholar]
- Ray CA, Black RA, Kronheim SR, Greenstreet TA, Sleath PR, Salvesen GS, Pickup DJ 1992. Viral inhibition of inflammation: Cowpox virus encodes an inhibitor of the interleukin-1 β converting enzyme. Cell 69: 597–604 [DOI] [PubMed] [Google Scholar]
- Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S 2002. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol 159: 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS 2001. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci 98: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribaya JP, Ranmuthu M, Copeland J, Boyarskiy S, Blair AP, Hay B, Laski FA 2009. The deubiquitinase emperor’s thumb is a regulator of apoptosis in Drosophila. Dev Biol 329: 25–35 [DOI] [PubMed] [Google Scholar]
- Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P 2007. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol 179: 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS 2007. The apoptosome: Signalling platform of cell death. Nat Rev Mol Cell Biol 8: 405–413 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS 2001. Structural basis for the inhibition of caspase-3 by XIAP. Cell 104: 791–800 [DOI] [PubMed] [Google Scholar]
- Rikhof B, Corn PG, El-Deiry WS 2003. Caspase 10 levels are increased following DNA damage in a p53-dependent manner. Cancer Biol Ther 2: 707–712 [PubMed] [Google Scholar]
- Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem 277: 29803–29809 [DOI] [PubMed] [Google Scholar]
- Rossig L, Fichtlscherer B, Breitschopf K, Haendeler J, Zeiher AM, Mulsch A, Dimmeler S 1999. Nitric oxide inhibits caspase-3 by S-nitrosation in vivo. J Biol Chem 274: 6823–6826 [DOI] [PubMed] [Google Scholar]
- Rudolf K, Cervinka M, Rudolf E 2009. Cytotoxicity and mitochondrial apoptosis induced by etoposide in melanoma cells. Cancer Invest 27: 704–717 [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H 2002. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 4: 432–438 [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J 17: 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME 1999. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 274: 1541–1548 [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Kornbluth S 2006. The apoptosome: Physiological, developmental, and pathological modes of regulation. Dev Cell 10: 549–561 [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H 2008. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev 22: 2256–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A, Clarke PR 2009. p38α- and DYRK1A-dependent phosphorylation of caspase-9 at an inhibitory site in response to hyperosmotic stress. Cell Signal 21: 1626–1633 [DOI] [PubMed] [Google Scholar]
- Seifert A, Allan LA, Clarke PR 2008. DYRK1A phosphorylates caspase 9 at an inhibitory site and is potently inhibited in human cells by harmine. FEBS J 275: 6268–6280 [DOI] [PubMed] [Google Scholar]
- Senft J, Helfer B, Frisch SM 2007. Caspase-8 interacts with the p85 subunit of phosphatidylinositol 3-kinase to regulate cell adhesion and motility. Cancer Res 67: 11505–11509 [DOI] [PubMed] [Google Scholar]
- Seol DW, Billiar TR 1999. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem 274: 2072–2076 [DOI] [PubMed] [Google Scholar]
- Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD 2008. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol 10: 1440–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Lee Y, Kim W, Ko H, Choi H, Kim K 2005. Caspase-2 primes cancer cells for TRAIL-mediated apoptosis by processing procaspase-8. EMBO J 24: 3532–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y 2003. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11: 519–527 [DOI] [PubMed] [Google Scholar]
- Shultz JC, Goehe RW, Wijesinghe DS, Murudkar C, Hawkins AJ, Shay JW, Minna JD, Chalfant CE 2010. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res 70: 9185–9196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. 2008. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 133: 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Silke J, Meier P 2013. Inhibitor of apoptosis (IAP) proteins—modulators of cell death and inflammation. Cold Spring Harb Perspect Biol 5: a008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ 1999. Serial killers: Ordering caspase activation events in apoptosis. Cell Death Differ 6: 1067–1074 [DOI] [PubMed] [Google Scholar]
- Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H 2002. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 21: 4520–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Guo Y, Zhan Y, Lazebnik Y, Fernandes-Alnemri T, Alnemri ES 1999. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res 59: 999–1002 [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, et al. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410: 112–116 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Renatus M, Meldal M, Salvesen GS 2000. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J 350 (Pt 2): 563–568 [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R 2001. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci 98: 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Kusakai G, Kishimoto A, Shimojo Y, Miyamoto S, Ogura T, Ochiai A, Esumi H 2004. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene 23: 7067–7075 [DOI] [PubMed] [Google Scholar]
- Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, et al. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386: 517–521 [DOI] [PubMed] [Google Scholar]
- Thompson CB 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267: 1456–1462 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y 1998. Caspases: Enemies within. Science 281: 1312–1316 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272: 17907–17911 [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843–846 [DOI] [PubMed] [Google Scholar]
- Torok NJ, Higuchi H, Bronk S, Gores GJ 2002. Nitric oxide inhibits apoptosis downstream of cytochrome c release by nitrosylating caspase 9. Cancer Res 62: 1648–1653 [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, Debatin KM, Krammer PH 1989. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245: 301–305 [DOI] [PubMed] [Google Scholar]
- Tu S, McStay GP, Boucher LM, Mak T, Beere HM, Green DR 2006. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol 8: 72–77 [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B 2006. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25: 5683–5692 [DOI] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K 2005. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods 2: 771–777 [DOI] [PubMed] [Google Scholar]
- van Raam BJ, Salvesen GS 2012. Proliferative versus apoptotic functions of caspase-8 Hetero or homo: The caspase-8 dimer controls cell fate. Biochim Biophys Acta 1824: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss OH, Kim S, Wewers MD, Doseff AI 2005. Regulation of monocyte apoptosis by the protein kinase Cδ-dependent phosphorylation of caspase-3. J Biol Chem 280: 17371–17379 [DOI] [PubMed] [Google Scholar]
- Wachmann K, Pop C, van Raam BJ, Drag M, Mace PD, Snipas SJ, Zmasek C, Schwarzenbacher R, Salvesen GS, Riedl SJ 2010. Activation and specificity of human caspase-10. Biochemistry 49: 8307–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JG, Cullen SP, Sheridan C, Luthi AU, Gerner C, Martin SJ 2008. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci 105: 12815–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ 2001. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci 98: 13884–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, LeBlanc AC, Su XD 2010. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep 11: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P 2002. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 4: 445–450 [DOI] [PubMed] [Google Scholar]
- Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N, Dong CK, Ogdahl JL, Schwartz LM, White K, et al. 2002. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol 4: 451–456 [DOI] [PubMed] [Google Scholar]
- Yan N, Shi Y 2005. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21: 35–56 [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D 1998. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281: 1355–1357 [DOI] [PubMed] [Google Scholar]
- Yang CS, Thomenius MJ, Gan EC, Tang W, Freel CD, Merritt TJ, Nutt LK, Kornbluth S 2010. Metabolic regulation of Drosophila apoptosis through inhibitory phosphorylation of Dronc. EMBO J 29: 3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CH, Yuan J 2009. The Jekyll and Hyde functions of caspases. Dev Cell 16: 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Yankner BA 2000. Apoptosis in the nervous system. Nature 407: 802–809 [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR 1993. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 β-converting enzyme. Cell 75: 641–652 [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW 2010. Structure of an apoptosome-procaspase-9 CARD complex. Structure 18: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X 1997. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90: 405–413 [DOI] [PubMed] [Google Scholar]