Abstract
Alternative excision repair (AER) is a category of excision repair initiated by a single nick, made by an endonuclease, near the site of DNA damage, and followed by excision of the damaged DNA, repair synthesis, and ligation. The ultraviolet (UV) damage endonuclease in fungi and bacteria introduces a nick immediately 5′ to various types of UV damage and initiates its excision repair that is independent of nucleotide excision repair (NER). Endo IV-type apurinic/apyrimidinic (AP) endonucleases from Escherichia coli and yeast and human Exo III-type AP endonuclease APEX1 introduce a nick directly and immediately 5′ to various types of oxidative base damage besides the AP site, initiating excision repair. Another endonuclease, endonuclease V from bacteria to humans, binds deaminated bases and cleaves the phosphodiester bond located 1 nucleotide 3′ of the base, leading to excision repair. A single-strand break in DNA is one of the most frequent types of DNA damage within cells and is repaired efficiently. AER makes use of such repair capability of single-strand breaks, removes DNA damage, and has an important role in complementing BER and NER.
DNA damage caused by UV light, for example, can be fixed by alternative excision repair (AER). AER involves an endonuclease that nicks the DNA near the damaged site, excision of the damaged DNA, repair synthesis, and ligation.
NER and base excision repair (BER) are the major excision repair pathways present in almost all organisms. In NER, dual incisions are introduced, the damaged DNA between the incised sites is then removed, and DNA synthesis fills the single-stranded gap, followed by ligation. In BER, an AP site, formed by depurination or created by a base damage-specific DNA glycosylase, is recognized by an AP endonuclease that introduces a nick immediately 5′ to the AP site, followed by repair synthesis, removal of the AP site, and final ligation. Besides these two fundamental excision repair systems, investigators have found another category of excision repair—AER—an example of which is the excision repair of UV damage, initiated by an endonuclease called UV damage endonuclease (UVDE). UVDE introduces a single nick immediately 5′ to various types of UV lesions as well as other types of base damage, and this nick leads to the removal of the lesions by an AER process designated as UVDE-mediated excision repair (UVER or UVDR). Genetic analysis in Schizosaccharomyces pombe indicates that UVER provides cells with an extremely rapid removal of UV lesions, which is important for cells exposed to UV in their growing phase.
Endo IV–type AP endonucleases from Escherichia coli and budding yeast and the Exo III–type human AP endonuclease APEX1 are able to introduce a nick at various types of oxidative base damage and initiate a form of excision repair that has been designated as nucleotide incision repair (NIR). Endonuclease V (ENDOV) from bacteria to humans recognizes deaminated bases, introduces a nick 1 nucleotide 3′ of the base, and leads to excision repair initiated by the nick. These endonucleases introduce a single nick near the DNA-damage site, leaving 3′-OH termini, and initiate repair of both the DNA damage and the nick. The mechanisms of AER may be similar to those of single-strand break (SSB) repair or BER except for the initial nicking process. However, how DNA damage is recognized determines the repair process within the cell. This article discusses the mechanisms and functional roles of AER. We begin with AER of UV damage, because genetic analysis has shown functional differences between this AER and NER in S. pombe.
NICKING ACTIVITY AND GENES ENCODING UVDE
Genetic and biochemical evidence suggested that UV lesions were removed, even in the absence of NER, in the eukaryote, the fission yeast S. pombe. In several strains of S. pombe deleted in one of the NER genes, cyclobutane pyrimidine dimers (CPD) and 6-4 pyrimidine pyrimidone photoproducts (6-4 photoproducts) were efficiently removed from UV-damaged DNA (McCready et al. 1993). In addition, an enzymatic activity introducing a nick immediately 5′ to CPD and 6-4 photoproducts was reported in S. pombe cell extracts (Bowman et al. 1994). In the filamentous fungus Neurospora crassa, a mutant mus18 strain was isolated that was specifically sensitive to UV, suggesting the presence of a novel repair process for UV lesions in N. crassa. By introducing a wild-type N. crassa cDNA library into E. coli cells deficient in three UV-damage repair genes (uvrA, recA, phr), a novel gene was isolated that complemented the UV sensitivity of the host E. coli cells and was subsequently shown to be the mus18 gene (Yajima et al. 1995). A recombinant mus18 gene product introduced a nick 5′ at the three major UV-induced lesions—CPD (cyclobutane pyrimidine dimer), 6-4 photoproducts, and DEWAR-type photoproducts—leaving a 5′-P-UV lesion and 3′-OH. The enzyme was designated as UVDE to distinguish it from “T4 endo” or “UV endo,” names that were often used for CPD glycosylases from T4-phage–infected cells or from Micrococcus luteus cell extracts. The gene for the counterpart in S. pombe was isolated by the same means as mus18 and showed the expected nicking activity previously reported. The repair of UV damage initiated by UVDE was designated as UVDE-mediated excision repair (UVER or UVDR). This repair is present in various eukaryotic microorganisms, including S. pombe and in prokaryotes, such as the Gram-positive bacteria Bacillus subtilis and Deinococcus radiodurans. Figure 1 depicts a comparison of BER with AER initiated by UVDE or AP endonuclease (UVER or NIR, respectively; see below). Having the S. pombe uvde (or uve1) gene, the repair pathway of this AER could be genetically analyzed and compared with NER.
Figure 1.
CPD glycosylase, UVDE, and AP endonuclease in action. The first steps of base excision repair initiated by DNA glycosylase for base damage (A), and by CPD glycosylase for CPD (B), are compared with those of alternative excision repair initiated by UVDE (C), and AP endonuclease (D).
AER PATHWAY FOR UV DAMAGE IN S. Pombe
Genetic analysis of UVDE in S. pombe indicated that the rad2 gene, encoding a structure-specific flap endonuclease, is important for processing and repair of UV damage and UVDE-induced single-strand breaks (SSBs) (Yonemasu et al. 1997). Because a triple mutant lacking rad2, uvde, and rad13 (an S. pombe homolog of human XPG, an NER factor) is as sensitive as a rad13 uvde double mutant but is more sensitive than a rad13 rad2 double mutant, there may also be a rad2-independent repair pathway for repairing UVDE-nicked DNA, and this may be a form of recombination repair. Using purified UVDE, Rad2p, DNA polymerase δ, RFC (replication factor C), PCNA (proliferating cell nuclear antigen), and DNA ligase, the AER process was successfully reconstituted in vitro (Alleva et al. 2000).
BIOLOGICAL ROLES OF AER BY UVDE IN S. pombe
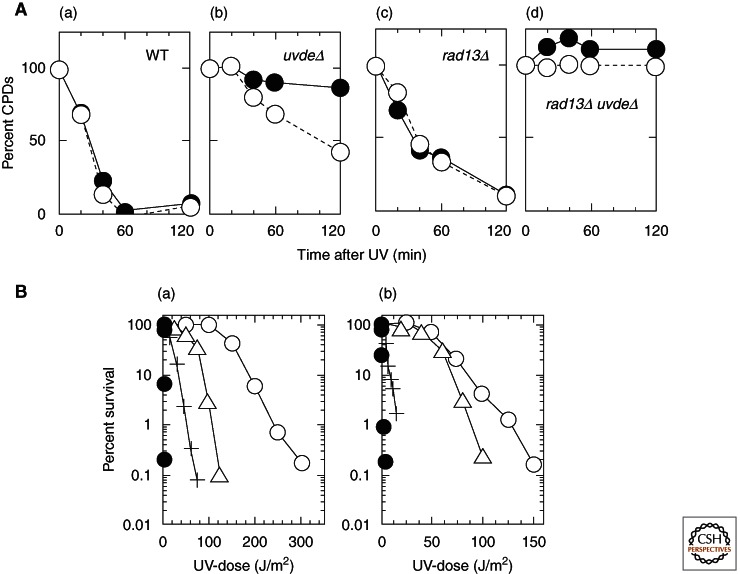
To understand the role of the AER pathway, the function of UVER was compared with NER for the repair of UV damage in S. pombe. Figure 2A shows the removal of CPD on the transcribed and nontranscribed strands after UV irradiation in NER (rad13, an XPG homolog) and UVER (uvde) mutants (Yasuhira et al. 1999). UVER recognizes and removes CPD equally on both strands, and it removes CPD much more rapidly than NER. Even on the transcribed strand, UVER removes CPD more rapidly than the sum of the transcription-coupled repair (TCR) and global genome repair (GGR) components of NER. In addition, it has been shown that UVDE recognizes and removes 6-4 photoproducts more rapidly than CPD. However, despite the rapid removal of UV damage by UVER, NER is more effective than UVER for UV survival in both stationary and growing cells (Fig. 2B), but UVER contributes more to cell survival in growing cells than in stationary cells. It is not known why the slow repair by NER contributes more to cell survival than rapid repair by UVER. A possible explanation is that, because UVDE produces a large number of nicks, unrepaired nicks might be disadvantageous to replication. However, despite whatever disadvantages there might be, the presence of both repair systems contributes to the extremely high UV resistance of the organisms harboring them.
Figure 2.
Comparison of AER with NER for repair of UV damage in S. pombe. (A) Strand bias of CPD removal on the myo2 locus in the nuclear genome in WT (wild-type) (a), uvdeΔ (b), rad13Δ (c), and rad13Δ uvdeΔ (d). Cells with or without functional NER (rad13Δ) or AER (uvdeΔ) were irradiated with 100 J/m2 of 254-nm UV and allowed to repair damage for 0–120 min at 30°C. Each point represents a mean value calculated from four hybridizations with two independent UV irradiations and DNA isolations. (Open circles) Transcribed strand, (closed circles) nontranscribed strand. (B) Contribution of NER and UVER to UV resistance of S. pombe in growth phase (a), or in stationary phase (b). uvde disruption causes a larger increase of UV sensitivity in growth phase. In addition, note that wild-type cells in exponential phase are more UV sensitive than those in stationary phase. (Open circles) Wild-type, (triangles) uvdeΔ, (plus signs), rad13Δ, (closed circles) rad13Δ uvdeΔ. (From Yasuhira et al. 1999 and Yasuhira and Yasui 2000; adapted, with permission, © American Society for Biochemistry and Molecular Biology.)
EXPRESSION OF UVDE IN HUMAN NER-DEFICIENT XPA CELLS FOR ANALYSIS OF AER AND SSB REPAIR IN HUMAN CELLS
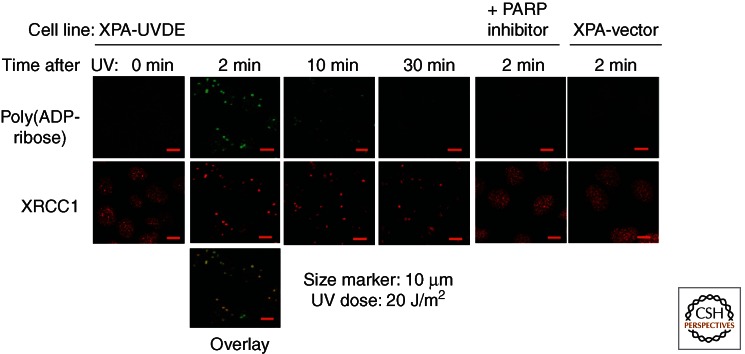
AER is an excision repair process initiated by a nick, that is, a single-strand break (SSB). The SSB is one of the most frequent types of DNA damage caused by reactive oxygen species or during enzymatic reactions of DNA metabolism, including repair. However, repair mechanisms for the SSB have not been well characterized because of the difficulty in producing only SSBs in cells. To characterize AER and SSB repair in human cells, a human fully NER-deficient XPA cell line (XP12ROSV) stably expressing N. crassa UVDE (XPA-UVDE cell line) was established, and UVER was characterized by UV irradiation of the cells (Okano et al. 2000). XPA-UVDE cells showed UV resistance at almost the wild-type level, suggesting that increased GGR of UV lesions by UVER complemented UV sensitivity of the XPA cell line. The repair patch size was shown to be ∼7 nucleotides, which is much shorter than that of NER, and repair synthesis was decreased to 30% of normal by aphidicolin, suggesting that DNA polymerase δ/ε-dependent long-patch repair is a major repair process. To further characterize AER initiated by UVDE, XPA-UVDE cells were UV irradiated through 3-μm micropores on a filter; several spots of this diameter within a nuclei were UV irradiated, and SSBs were produced by UVDE only at the irradiated sites (Fig. 3) (Okano et al. 2003). Proteins accumulating at SSBs created by UV irradiation and UVDE were identified by immunostaining or by real-time observation of expressed green fluorescent protein (GFP)-tagged repair proteins under the microscope. Thus, poly(ADP-ribose) immediately accumulated at the UV-irradiated sites in a UVDE-dependent manner and disappeared rapidly because of degradation of poly(ADP-ribose) by poly(ADP-ribose) glycohydrolase (PARG). XRCC1 accumulated at the UV-irradiated sites in a poly(ADP-ribose)-dependent manner and remained at the sites even after poly(ADP-ribose) disappeared. LigaseIIIα, Pol β, Pol δ, PCNA, FEN1, and CAF1 complex followed XRCC1 and accumulated at the sites in a time- and process-dependent manner. This process was visualized in real time by applying laser microirradiation, because at a low-laser-light dose, SSBs are predominantly produced (Lan et al. 2004). Although AER initiated by UVDE at UV lesions in human cells is an artificial repair of SSB with 5′-attached UV damage, the results suggest the presence of a common repair process between AER and SSB repair. This type of repair occurs frequently at SSBs within human cells, and, therefore, the PARP inhibitor has a potent killing effect on cells with defective homologous recombination activity, because unrepaired SSBs are processed by homologous recombination. These data suggest that PARP-dependent repair of SSBs may be a major pathway for AER in human cells and other higher eukaryotes.
Figure 3.
Accumulation of poly(ADP-ribose) and XRCC1 at UVDE-induced SSB in the nucleus of human NER-deficient XPA cell line expressing N. crassa UVDE. The cell was irradiated with UVC light of 20 J/m2 through pores of 3 µm in diameter in a filter covered over the cell. (Upper panels, green) Poly(ADP-ribose) accumulates immediately at the irradiated site and dissociates rapidly, whereas XRCC1 (red) accumulates and retains at the irradiated site. In cells treated with PARP inhibitor (DIQ) or cells transfected with vector plasmid, neither poly(ADP-ribose) nor XRCC1 accumulation was observed. (Adapted from Lan et al. 2004.)
STRUCTURAL BIOLOGY OF UVDE
How does UVDE recognize various types of UV lesions? Except for proteins involved in NER, there has been no report of a protein other than UVDE recognizing both CPD and 6-4 photoproducts. It should be noted that UVDE also recognizes AP sites, platinum G–G diadducts, uracil, dihydrouracil, and other oxidative DNA-damage products and introduces a nick immediately 5′ to the lesions (Avery et al. 1999; Kanno et al. 1999). A crystal structure of UVDE derived from the thermophilic bacterium Thermus thermophilus was solved (Paspaleva et al. 2007) and showed essential structural features of a TIM-barrel fold shared with that of Endo IV (Hosfield et al. 1999; Garcin et al. 2008). Although both enzymes share some characteristic features of a DNA-binding groove, where both the damaged bases and their opposite bases are thought to flip out of the double helix into the binding groove, the predicted residues in the structure of UVDE make a larger space for the flipped-out UV lesions than in Endo IV. Furthermore, the structural flexibility of UVDE after DNA binding is thought to be essential for its broader substrate specificity. UVDE, but not Endo IV, rotates 2 nucleotides into its protein pocket near the active site and possesses a carboxylated lysine residue that is required for proper catalysis and prevention of increased incision of undamaged DNA (Paspaleva et al. 2007, 2009). Considering the fact that Endo IV–type AP endonucleases are able to recognize base damage in addition to AP sites (see below), UVDE has probably evolved from an Endo IV–type AP endonuclease in Gram-positive bacteria to adjust to a UV-rich environment.
NIR, AER, AND SSB REPAIR
The substrate of AP endonuclease has been thought to be only AP sites. However, a direct incision activity immediately 5′ to oxidized bases was identified in the Endo IV–type AP endonuclease (encoded by the nfo gene) from E. coli as well as a similar activity in budding yeast and human cell extracts (Ischenko and Saparbaev 2002). This incision leaves 3′-hydroxyl termini for further DNA synthesis and repair, which was designated as nucleotide incision repair (NIR) by the authors. The human APEX1 is able to incise DNA containing 5,6-dihydro-2′-deoxyuridine, 5,6-dihydrothymidine, 5-hydroxy-2′-deoxyuridine, α-2′-deoxyadenosine, and α-thymidine adducts, generating 3′-hydroxyl and 5′-phosphate termini (Gros et al. 2003). Thus, human APEX1, an Exo III–type AP endonuclease that is evolutionarily distinct from Endo IV–type enzymes, incises base damage directly. Several questions were addressed as to the new functions of AP endonuclease: whether endonuclease activity for base damage is different from AP endonuclease activity, how active NIR is within human cells as compared with BER, and how AP endonucleases are able to recognize various types of base damage. For the first question, mutagenesis of the E. coli nfo gene showed that three mutants—H69A, H109A, and G149D—were proficient in the AP endonuclease and 3′ phosphodiesterase activities but deficient in NIR activity, indicating that AP endonuclease and base damage nicking activities are separate mechanisms (Ishchenko et al. 2006). These nfo mutants complemented the hypersensitivity of an E. coli nfo-deletion mutant to alkylation but not to oxidative DNA damage, suggesting that NIR is responsible for repair of oxidative base damage in E. coli cells. The repair process after the incision step of NIR is believed to be the same as that of BER after the glycosylase step, but binding of AP endonuclease at base damage may influence the following steps of repair synthesis.
ANOTHER HUMAN AP ENDONUCLEASE AND AER
A nuclear AP endonuclease other than APEX1 was recently reported in human cells and higher eukaryotes and was designated as PALF (APLF, Xip1). This enzyme possesses a direct incision activity at various types of base damage in addition to the AP site (Kanno et al. 2007). PALF interacts with many proteins for DNA single- and double-strand break repair in human cells and is involved in both SSB repair and NHEJ. PALF interacts strongly with PARP1 and additionally possesses 3′–5′ endonuclease and exonuclease activities that produce a gap at the nicked site. This gap dimerizes and activates PARP1, leading to SSB repair. This is another example of an AER in human cells.
ENDONUCLEASE V, AN ENDONUCLEASE THAT DIRECTLY NICKS THE PHOSPHODIESTER BOND 1 NUCLEOTIDE 3′ TO A DEAMINATED BASE
Nitrosative deamination produces various deaminated bases—hypoxanthine from adenine, xanthine or oxanine from guanine, and uracil from cytosine—and this can cause transition mutations. Endonuclease V (Endo V), encoded by the nfi gene in E. coli, cleaves the phosphodiester bond located 1 nucleotide 3′ of a deaminated base. The deaminated base is removed and repaired by subsequent excision repair. How this AER process proceeds, after Endo V introduced a nick, is not well understood. However, in vitro experiments have recently shown that Endo V, DNA polymerase I, and DNA ligase from E. coli are sufficient to reconstitute the repair process, in which DNA polymerase I has a 3′-exonuclease activity and is able to remove the damaged site (Lee et al. 2010). Substrate specificity differs among species. Although Endo V from E. coli has a broad substrate specificity, human Endo V is limited to deoxyinosine and deoxyxanthosine (Mi et al. 2012). Human Endo V suppresses mutation frequency to one-third in E. coli cells, but its biological significance in human cells remains unknown. The crystal structure of Thermotoga maritima Endo V was determined and shown to consist of an αβα protein with a central eight-stranded β sheet of parallel and antiparallel strands flanked on either side by α helices (Dalhus et al. 2009). Interestingly, Endo V contains a wedge structure consisting of a PYIP motif that may act as a damage sensor for small helical distortions and base mismatches and separate DNA strands at the lesion. Especially interesting for the AER process is that the insertion of the wedge into DNA extrudes the base damage into a binding pocket, where DNA ends interact with metal ions and possibly with downstream proteins in the repair pathway. The repair pathway of Endo V–initiated AER in human cells requires a 3′–5′ exonuclease activity that remains to be identified. Figure 4 depicts the relationship between BER and AER including NIR. These pathways are still to be fully characterized in cells.
Figure 4.
Repair pathways of BER, AER, and SSBR.
CONCLUDING REMARKS
AER is an excision repair pathway for DNA damage initiated by an endonuclease-induced nick near a DNA-damage site. AER, BER, and SSB repair differ in the first step of the repair process, which determines the further repair process. SSBs are a very common DNA damage within cells and at least partly repaired by poly(ADP-ribose) polymerase (PARP)-dependent SSB repair. It is important to know whether and how PARP is involved in AER, BER, and SSB repair processes within cells.
ACKNOWLEDGMENTS
I thank Dr. Shirley J. McCready for editing the text. This work is supported by Grants-in-Aid for Scientific Research and from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, to A.Y. (grants 24310037 and 22131005).
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Alleva JL, Zuo S, Hurwitz J, Doetsch PW 2000. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry 39: 2659–2666 [DOI] [PubMed] [Google Scholar]
- Avery AM, Kaur B, Taylor JS, Mello JA, Essigmann JM, Doetsch PW 1999. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res 27: 2256–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KK, Sidik K, Smith CA, Taylor JS, Doetsch PW, Freyer GA 1994. A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Res 22: 3026–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B, Arvai AS, Rosnes I, Olsen OE, Backe PH, Alseth I, Gao H, Cao W, Tainer JA, Bjoras M 2009. Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nat Struct Mol Biol 16: 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Bjoras M, Cunningham RP, Tainer JA 2008. DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol 15: 515–522 [DOI] [PubMed] [Google Scholar]
- Gros L, Ishchenko AA, Saparbaev M 2003. Enzymology of repair of etheno-adducts. Mutat Res 531: 219–229 [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA 1999. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98: 397–408 [DOI] [PubMed] [Google Scholar]
- Ischenko AA, Saparbaev MK 2002. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415: 183–187 [DOI] [PubMed] [Google Scholar]
- Ishchenko AA, Deprez E, Maksimenko A, Brochon JC, Tauc P, Saparbaev MK 2006. Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc Natl Acad Sci 103: 2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Iwai S, Takao M, Yasui A 1999. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res 27: 3096–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, Nakajima S, Yasui A 2007. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 26: 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A 2004. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci 101: 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Yang YC, Goodman SD, Yu YH, Lin SB, Kao JT, Tsai KS, Fang WH 2010. Endonuclease V–mediated deoxyinosine excision repair in vitro. DNA Repair 9: 1073–1079 [DOI] [PubMed] [Google Scholar]
- McCready S, Carr AM, Lehmann AR 1993. Repair of cyclobutane pyrimidine dimers and 6-4 photoproducts in the fission yeast Schizosaccharomyces pombe. Mol Microbiol 10: 885–890 [DOI] [PubMed] [Google Scholar]
- Mi R, Alford-Zappala M, Kow YW, Cunningham RP, Cao W 2012. Human endonuclease V as a repair enzyme for DNA deamination. Mutat Res 735: 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Kanno S, Nakajima S, Yasui A 2000. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J Biol Chem 275: 32635–32641 [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A 2003. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23: 3974–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspaleva K, Thomassen E, Pannu NS, Iwai S, Moolenaar GF, Goosen N, Abrahams JP 2007. Crystal structure of the DNA repair enzyme ultraviolet damage endonuclease. Structure 15: 1316–1324 [DOI] [PubMed] [Google Scholar]
- Paspaleva K, Moolenaar GF, Goosen N 2009. Damage recognition by UV damage endonuclease from Schizosaccharomyces pombe. DNA Repair 8: 600–611 [DOI] [PubMed] [Google Scholar]
- Yajima H, Takao M, Yasuhira S, Zhao JH, Ishii C, Inoue H, Yasui A 1995. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J 14: 2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhira S, Yasui A 2000. Alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe operates both in nucleus and in mitochondria. J Biol Chem 275: 11824–11828 [DOI] [PubMed] [Google Scholar]
- Yasuhira S, Morimyo M, Yasui A 1999. Transcription dependence and the roles of two excision repair pathways for UV damage in fission yeast Schizosaccharomyces pombe. J Biol Chem 274: 26822–26827 [DOI] [PubMed] [Google Scholar]
- Yonemasu R, McCready SJ, Murray JM, Osman F, Takao M, Yamamoto K, Lehmann AR, Yasui A 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res 25: 1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]