Abstract
In 1959, Arthur Kornberg was awarded the Nobel Prize for his work on the principles by which DNA is duplicated by DNA polymerases. Since then, it has been confirmed in all branches of life that replicative DNA polymerases require a single-stranded template to build a complementary strand, but they cannot start a new DNA strand de novo. Thus, they also depend on a primase, which generally assembles a short RNA primer to provide a 3′-OH that can be extended by the replicative DNA polymerase. The general principles that (1) a helicase unwinds the double-stranded DNA, (2) single-stranded DNA-binding proteins stabilize the single-stranded DNA, (3) a primase builds a short RNA primer, and (4) a clamp loader loads a clamp to (5) facilitate the loading and processivity of the replicative polymerase, are well conserved among all species. Replication of the genome is remarkably robust and is performed with high fidelity even in extreme environments. Work over the last decade or so has confirmed (6) that a common two-metal ion-promoted mechanism exists for the nucleotidyltransferase reaction that builds DNA strands, and (7) that the replicative DNA polymerases always act as a key component of larger multiprotein assemblies, termed replisomes. Furthermore (8), the integrity of replisomes is maintained by multiple protein–protein and protein–DNA interactions, many of which are inherently weak. This enables large conformational changes to occur without dissociation of replisome components, and also means that in general replisomes cannot be isolated intact.
Many characteristics of replicative polymerases (e.g., the nucleotidyltransferase reaction) are conserved across all domains of life. But some features (e.g., quaternary structures) vary.
The genomes, from the smallest to the largest, provide an enormous challenge for the replicative DNA polymerases to faithfully copy to give the many generations that follow a comparable condition for life. In this article, we discuss the structural and functional bases by which replicative DNA polymerases are able to efficiently and faithfully build new copies of genomes in eubacteria, archaea, and eukaryotes.
BUILDING DNA
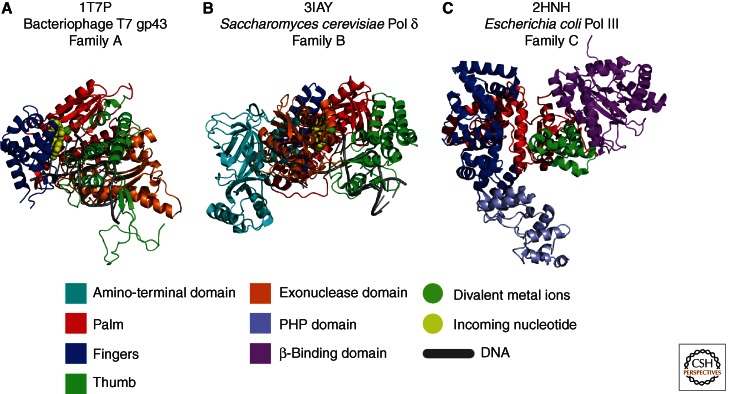
Considerable progress in the description of the chemical and structural basis for DNA synthesis has been made in recent years. A large variety of DNA polymerases have been isolated and studied since the initial discovery of Escherichia coli DNA Pol I. The DNA polymerases have been divided into families based on homology of their primary sequences. The replicative DNA polymerases from eukarya are found in family B, bacteria in families A and C, and archaea in families B and D. The structures of the catalytic subunits of these polymerases share a similar organization, and the nucleotidyl transferase reaction of adding nucleotides to the 3′-OH of the growing strand is conserved (Table 1). All known structures of DNA polymerases (examples in Fig. 1) appear to resemble a right hand, where the functional domains are depicted as fingers, palm, and thumb domains, and the replicative polymerases often contain additional domains for 3′–5′ exonucleolytic proofreading, interactions with other proteins, or other functions.
Table 1.
A current view of subunit compositions and roles of subunits of replicative DNA polymerases
| Human | Yeast (Sc) | Yeast (Sp) | (Eury)archaea | Eubacteria (Eco) | Firmicutes (Bsu) | |
|---|---|---|---|---|---|---|
| Chromosomal replicases | Pol δ (δ) and Pol ε (ε) | Pol δ (δ) and Pol ε (ε) | Pol δ (δ) and Pol ε (ε) | Pol B (B) Pol D (D) |
Pol III HE (DnaE + nine other subunits) | Pol C |
| Replicase core(s) | (δ): PolD1/PolD2/PolD3/PolD4 (ε): Pol E/PolE2/PolE3/PolE4 |
(δ): Pol3/Pol31/Pol32 (ε): Pol2/Dpb2/ Dpb3/Dpb4 |
(δ): Pol3/Cdc1/Cdc27/Cdm1 (ε): Pol2/Dpb2/ Dpb3/Dpb4 |
(B): Pol B (D): (DP1/DP2)2 |
αεθ | Pol C |
| Polymerase subunit | (δ): PolD1 (ε): Pol E |
(δ): Pol3 (ε): Pol2 |
(δ): Pol3 (ε): Pol2 |
(B): Pol B (D): DP2 |
α (DnaE) | Pol C |
| Proofreading subunit | (δ): PolD1 (ε): Pol E |
(δ) Pol3 (ε) Pol2 |
(δ): Pol3 (ε): Pol2 |
(B): Pol B (D): DP1 |
ε (DnaQ) | Pol C |
| Primase (subunits) | Pol α (Prim1/Prim2A) | Pol α (Pri1/Pri2) | Pol α (Pri1/Spp2) | PriSL | DnaG | DnaG |
| Primer extension (subunits) | Pol α (PolA1/Pol2A) | Pol α (Pol1/Pol12) | Pol α (Pol1/Pol12) | DnaE/DnaQ | ||
| Leading-strand Pol | Pol ε | Pol ε | Pol ε | Pol B | αεθ | Pol C |
| Lagging-strand Pol | Pol δ | Pol δ | Pol δ | Pol B or Pol D (?) | αεθ | Pol C |
| Clamp | PCNA | PCNA | PCNA | PCNA | β | β |
| Clamp loader | RFC (pentamer of five different subunits) | RFC (pentamer of five different subunits) | RFC (pentamer of five different subunits) | RFC (pentamer of two different subunits) | τ2γδδ′χψ | τ3δδ′′ |
| Okazaki fragment processing | Pol δ/FEN1/PCNA, Dna2 | Pol δ/FEN1/PCNA, Dna2 | Pol δ/FEN1/PCNA, Dna2 | FEN1 | Pol I | Pol I |
Isolable complexes are color coded. PCNA, proliferating cell nuclear antigen; RFC, replication factor C.
Figure 1.
Representative structures and domain architecture of DNA polymerases from the A, B, and C families. (A) Bacteriophage T7 gene 5 protein with primer-template DNA (family A; pdb 1T7P). (Data from Doublié et al. 1998.) (B) Saccharomyces cerevisiae Pol δ (family B; pdb 3IAY). (Data from Swan et al. 2009.) (C) E. coli Pol III α subunit, residues 1–917 (family C; pdb 2HNH). (Data from Lamers et al. 2006.) Figure drawn using PyMOL.
Both the growing DNA and template strands are integral components of their active sites, providing information about which nucleotide should be added, and the 3′-hydroxyl group acts as a nucleophile to attack the α phosphate of the correct incoming deoxyribonucleoside triphosphate (dNTP). This all occurs in the palm domain, which is the most highly conserved subdomain. Here, two magnesium ions are coordinated by two invariable aspartic acids. This is the basis for the two-metal catalytic mechanism (Steitz 1993), conserved among all replicative polymerases studied so far. The fingers domain is adjacent to the palm domain, contacts the incoming nucleotide, and goes through large conformational changes when the nucleotide is positioned near the metals, template, and the 3′ end of the growing strand in the active site (Yang et al. 1999; Franklin et al. 2001). The thumb domain, which interacts with the duplex DNA upstream of the polymerase active site, has been shown in A- and B-family polymerases to influence the partitioning between editing and polymerization modes (Doublié et al. 1998; Franklin et al. 2001). Mutations in the thumb domain can act both as antimutators and mutators, depending on how the balance has been shifted either toward the editing or the polymerization mode (Stocki et al. 1995; Wu et al. 1998). The thumb domain in C-family polymerases may have a similar function to that in A- and B-family polymerases, because it also contacts the duplex DNA (Bailey et al. 2006; Lamers et al. 2006; Wing et al. 2008).
Replicative DNA polymerases are highly accurate during the synthesis of DNA. In addition they have either a built-in 3′–5′ exonuclease site, located at a distance from the polymerase active site, or an associated subunit with 3′–5′ exonuclease activity (reviewed in Patel and Loeb 2001; Kunkel 2004; McHenry 2011). In either case, the distance from the polymerase site to the exonuclease site necessitates that at least three nucleotides of the double-stranded DNA are unwound to allow editing. This is a particularly important facet of the editing function because the exonuclease sites only accommodate single-stranded DNA.
At every replication fork, replicative polymerases cooperate in copying both DNA strands. Because the directions of the two template strands are opposite, and polymerases can only extend preexisting primers from their 3′-OH end, only one strand can be made continuously (the leading strand). The other (lagging) strand has to be synthesized in the direction opposite the fork movement as a series of short (Okazaki) fragments that are processed and subsequently joined together. In general, leading- and lagging-strand synthesis is coordinated, but how this is achieved varies somewhat among organisms.
Replicases from all domains of life function within larger molecular machines called replisomes that contain helicase(s), which separate the two DNA strands at the apex of the replication fork, a primase for repeated RNA priming in lagging-strand synthesis, sliding clamps that encircle the nascent double-stranded product to tether polymerases onto the DNA to ensure they dissociate rarely, clamp loaders that load the clamps, various single-stranded DNA-binding proteins, etc. The ring-shaped sliding clamps, called the β subunit (a homodimer) in eubacteria and PCNA (a homotrimer) in archaea and eukaryotes, have very similar structures (Kong et al. 1992; Krishna et al. 1994). They interact at the same site in each of their subunits with many other proteins including the replicative polymerases, via short peptide motifs called the clamp-binding motif in eubacteria (Dalrymple et al. 2001) and the PIP box in higher organisms (Warbrick 1998). The structures and functions of the sliding clamps and other accessory replisomal components are described in detail in the literature.
So far we have highlighted a few well-conserved features of the replicative polymerases. However, as we now proceed with a more detailed comparison, you will find that they have special features to function in their specific environments.
QUATERNARY STRUCTURE
The quaternary structures of replicative polymerases vary significantly (Table 1). In part, this variation rather arbitrarily depends on which subunits are isolated together to define the polymerase. From some species (e.g., Pol III in Escherichia coli [Eco]), a holoenzyme can be purified as an entity with 10 different subunits. In eukaryotes, the replicative polymerases appear to be more loosely associated within the replisome, and they are thus isolated in assemblies that contain fewer accessory subunits. We will discuss here the core catalytic subunit and accessory subunits that are considered to be parts of the DNA polymerase.
Eubacteria
Pol III holoenzyme is the E. coli chromosomal replicase that synthesizes both leading and lagging strands simultaneously. As isolated directly from cells, it has an average composition close to (αεθ)2–(τ2γδδ′ψχ)–(β2)2 (17 subunits), where αεθ is the polymerase core discussed in more detail below, β2 is the sliding clamp, and τ2γδδ′ψχ is the clamp loader complex that may contain two to three τ and one to zero γ subunits (McHenry 2011).
The αεθ core of Pol III is a tightly associated complex. The large α subunit is a family C polymerase, and ε is a separate 3′–5′ editing exonuclease subunit from the DnaQ family. The small θ subunit has a role in stabilizing ε, but it only occurs in a limited range of bacterial species. The α subunit is made up of a series of domains (Fig. 1C) (Bailey et al. 2006; Lamers et al. 2006): the amino-terminal PHP domain seems to be a vestigial exonuclease domain that may still be functional as a proofreader in some species (Stano et al. 2006). In E. coli, it has evolved to be the site of interaction of the ε subunit (Wieczorek and McHenry 2006). This domain is followed by the usual polymerase palm, thumb, and fingers domains, and a β-binding domain that contains a conserved clamp-binding motif that interacts with the β2 clamp to tether α to the product DNA to ensure its high processivity. This is followed by an OB-fold domain that is likely to interact with the single-stranded template DNA, and a carboxy-terminal domain that interacts tightly with the carboxy-terminal domain of the τ subunit of the clamp loader. Because the clamp loader contains two (or three) τ subunits, at least two αεθ cores are maintained in the replicase complex, one each for leading- and lagging-strand synthesis.
The ε subunit has a globular amino-terminal exonuclease domain (residues 7–180), the structure of which is reminiscent of many editing exonuclease domains from other polymerases (Hamdan et al. 2002). It contains a binuclear metal site for processing nucleotides misincorporated by α, followed by a poorly conserved 63-residue carboxy-terminal segment that by itself is unstructured. The last ∼40 residues are now known to be involved directly in binding to the PHP domain of α (Ozawa et al. 2008); a short segment that immediately follows the exonuclease domain contains a second clamp-binding motif (S Jergic and N Dixon, unpubl.), and the unstructured segment in between remains flexible even in the αεθ complex (Ozawa et al. 2008). This may suggest an unusual mechanism in which ε can occupy quite different positions in the complex when it is or is not being used for proofreading.
Most (or all) eubacterial species seem to have replicase subunits closely related to α (DnaE family) and ε, but some Gram-positive bacteria like Bacillus subtilis (Bsu) also have a second more distantly related polymerase/exonuclease called Pol C (Table 1). These latter enzymes have the proofreading and polymerase activities in a single polypeptide, and a different domain organization, with putative τ-binding and OB-fold domains preceding a discontinuous PHP domain that incorporates the proofreader, and the β-binding motif being right at the carboxyl terminus (Evans et al. 2008). Although it was for some time believed that Pol C was dedicated to leading-strand synthesis and DnaE to the lagging strand (Dervyn et al. 2001), recent work suggests that most chromosomal DNA synthesis in these bacteria is performed by Pol C. However, because only the DnaE polymerase can extend RNA primers like those made by the DnaG primase, it has a critical role in lagging-strand synthesis. The current model is that DnaE extends RNA primers for some length before passing them to Pol C (Sanders et al. 2010). This has some parallel with the eukaryotic Pol α (below). Note that in eubacteria, RNA priming is performed by a dedicated primase (DnaG) that does not associate directly with the polymerase; it is associated, sometimes firmly and often more transiently, with the replicative helicase to enable primer synthesis on both the leading and lagging strands.
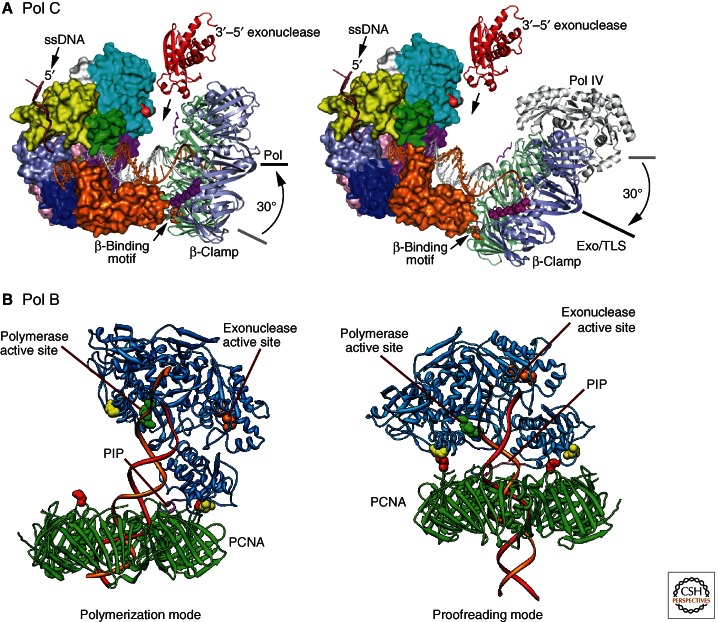
There is no crystal structure available of a complete Pol III core from any bacterium, but available structures of α subunits include E. coli α(1–917), which misses its internal clamp-binding motif and domains that follow it (Lamers et al. 2006), and full-length Thermus aquaticus (Taq) α (DnaE), both by itself and in complex with primer-template DNA (Bailey et al. 2006; Wing et al. 2008). The most recent structure is of a ternary complex containing primer-template DNA and incoming dNTP bound to Pol C from Geobacillus kaustophilus (Gka) (Evans et al. 2008). The Taq and Eco structures without DNA are very similar in the regions that can be compared (Fig. 1C), but DNA binding induces large conformational changes to a closed state, especially in the β-binding region of the Taq protein. The Gka Pol C ternary complex is also similar to the Taq α–DNA structure in this respect. Interestingly, the structure of a β2–DNA complex (Georgescu et al. 2008a) can be docked neatly into both the α and Pol C–DNA structures to give plausible models of the α or Pol C–β2–DNA replicases in the polymerization mode (Evans et al. 2008; Wing et al. 2008), and it has been suggested that the open structures might mimic the replicase structure in the editing mode (Fig. 2A). The position of the proofreading exonuclease has not been defined (it was deleted from the Pol C construct used for crystallization), but it is presumably between the PHP domains and β2 in the modeled structures. A second interesting aspect of the α/Pol C structures is that the palm domain (active site) architecture is distinct from that found in the A- and B-family polymerases, being closer to that of X family members like the eukaryotic repair polymerase, Pol β. This suggests that the replicative polymerases of eubacteria evolved independently of those in eukaryotes and archaea.
Figure 2.
Modeled structures of ternary complexes of polymerases, clamps, and DNA in the polymerization and editing modes. (A) Geobacillus kaustophilus Pol C with DNA and β sliding clamp. (Panel A is from Evans et al. 2008; reprinted, with express permission, from the authors.) (B) Pyrococcus furiosus Pol B with DNA and PCNA (Mayanagi et al. 2011). (Panel B is from Mayanagi et al. 2011; reprinted, with permission, from The National Academy of Science © 2011.)
The other polymerase that plays a significant role in bacterial DNA replication is Pol I, the founding member of the family A polymerases (representative Pol A structure is shown in Fig. 1A). Its primary function in replication is in Okazaki fragment processing on the lagging strand. Pol III is capable of synthesizing Okazaki fragments right up to the 5′ end of a preceding RNA primer, whereupon it is recycled to a new primer terminus, leaving behind a nick or short gap (discussed in Dohrmann et al. 2011). Pol I has three separate activities in a single polypeptide chain. The amino-terminal domain is a 5′–3′ exonuclease capable of excising the RNA primers at the same time as the carboxy-terminal polymerase domain (with thumb, palm, and fingers subdomains) extends the DNA primer behind it. The central domain is a DnaQ family proofreading exonuclease used to ensure high fidelity. Thus, Pol I uses a process of “nick translation” to replace RNA primers with DNA, leaving a nick with a 3′-OH and 5′-phosphate that is a substrate for DNA ligase, which seals the nick to make a contiguous lagging strand.
Archaea
The replisome in archaebacteria is less complex than in eubacteria, and in most aspects is a simplified version of the eukaryotic system. All archaea contain single-subunit Pol B polymerases, with the proofreading activity included in the catalytic subunit as a domain near the amino terminus (Table 1). Some genera have multiple Pol Bs (family B), whose separate roles in chromosomal replication are yet to be clearly defined, whereas the euryarchaea uniquely also contain a more complex heterotetrameric Pol D polymerase, with subunit composition (DP1)2–(DP2)2. The smaller DP1 subunit contains a 3′–5′ exonuclease domain, whereas the larger DP2 is the (family D) polymerase. Although Pol D is a processive and efficient enzyme that, in common with Pol B, also interacts with the PCNA sliding clamp, the role of Pol D as a chromosomal replicase in euryarchaeal species is still uncertain (Tori et al. 2007). It has been suggested that in Pyrococcus abyssi, Pol B synthesizes the leading and Pol D the lagging strand (Henneke et al. 2005). High-resolution structural information on Pol D is limited to the crystal structure of the amino-terminal domain of Pyrococcus horikoshii DP2, which has a role in Pol D subunit oligomerization (Matsui et al. 2011).
The crenarchaea do not have Pol Ds, but often have multiple Pol Bs. For example, Sulfolobus solfataricus has three of them, and Pol B1 (Dpo1) is a high-fidelity polymerase thought to carry out both leading- and lagging-strand replication (Choi et al. 2011). The archaeal Pol Bs have been studied extensively, in part because some of them from thermophilic species are used extensively for high-fidelity PCR applications. Although crystal structures of Pol Bs from several archaeal species have been determined (the earliest are in Hopfner et al. 1999, Zhao et al. 1999, Rodriguez et al. 2000, and Hashimoto et al. 2001), there is none yet that shows primer-template DNA in the polymerization or proofreading modes. These structures can, however, be modeled using the structures of the T4-related phage RB69 Pol B, bound to DNA in both of these modes (Franklin et al. 2001).
The archaeal Pol Bs seem generally and uniquely among family B polymerases (Wardle et al. 2008) to contain an additional amino-terminal domain that precedes the proofreading exonuclease domain, whose function is to scan the template DNA ahead of the polymerase for the uracil base in dUMP produced by cytosine deamidation (and hypoxanthine, from adenosine) that has escaped repair by uracil-N-glycosylase (Greagg et al. 1999, Fogg et al. 2002). On recognition of dUMP in the template, the polymerase stalls four nucleotides before the lesion is encountered. Although how this might lead to subsequent repair of the lesion is not known, crystal structures have been reported of stalled complexes containing primer templates with appropriately incorporated dUMP or dIMP (Firbank et al. 2008, Killelea et al. 2010). The bases are buried within a specific pocket in the amino-terminal domain, and the primer-template DNA is thought to be in a position in the structure close to that in the editing mode, by comparison to the RB69 Pol B structure (Franklin et al. 2001).
In a recent study (Mayanagi et al. 2011), a series of available crystal structures was combined with low-resolution reconstructions from electron microscopy and single-particle analysis to generate plausible models of the structures of archaeal Pol B–DNA–clamp ternary complexes in both the polymerization and proofreading modes (Fig. 2B). As for the bacterial Pol III complex (Fig. 2A), these models generate new hypotheses that will be tested in the future, leading to further progressive improvement of our understanding of replisome structure and function.
A significant difference between the archaeal replisomes and those in eukaryotes (see below) is that archaea, like the bacteria, use primases that are not physically associated with the polymerases. Nevertheless, the archaeal primases contain two subunits (p41 and p46) that are homologs of the PriS and PriL subunits of the eukaryotic Pol α–primase complex (Table 1). Recent in vitro studies with purified proteins have shown quite clearly that the Thermococcus kodakarensis primase can carry out (DNA) primer synthesis in the total absence of ribonucleoside triphosphates (rNTPs), and in a rolling-circle assay with a synthetically primed minicircle DNA template, it can prime efficient lagging-strand Okazaki-fragment synthesis dependent on the hexameric MCM replicative helicase and Pol B (Chemnitz Galal et al. 2012). Thus, the stage is now set to use in vitro studies with the archaeal replisome to probe mechanistic questions in the way that has already been performed with the model bacterial systems.
Eukarya
Eukaryotes have evolved a more complex replisome that depends on three different B-family DNA polymerases: Pol α, Pol δ, and Pol ε (Kunkel and Burgers 2008; Burgers 2009). Each of these multisubunit polymerases (Table 1) has one catalytic core subunit and two to four accessory subunits. Pol α is built up by four different subunits: PriS, PriL, Pol1, and Pol12 (Muzi-Falconi et al. 2003); PriL has primase activity and Pol1 has DNA polymerase activity. Negative-stain EM reconstructions combined with high-resolution structures of domains of the Saccharomyces cerevisiae (Sc) Pol1–Pol12 complex have revealed two globular domains connected via a flexible linker (Klinge et al. 2009). It was suggested that an intramolecular hand-off occurs from the primase domain to the polymerase domain when the initial RNA primer has reached a specific length (Klinge et al. 2009; Nunez-Ramirez et al. 2011). The final result is a 30–35 nucleotide primer with RNA at the 5′ end and DNA at the 3′ end. Thus, Pol α synthesizes a substantial amount of DNA because the eukaryotic Okazaki fragments are on average only about 165 nucleotides long (Smith and Whitehouse 2012). Thus, the absence of a proofreading activity in Pol α requires that errors made by it are corrected by other mechanisms. For instance Pol δ, but not Pol ε, proofreads errors made by Pol α, in addition to error correction by the mismatch repair system (Pavlov et al. 2006; Nick McElhinny et al. 2010a).
Pol ε and Pol δ are responsible for the bulk synthesis of DNA on the leading and lagging strands (Kunkel and Burgers 2008; Burgers 2009). They are both highly accurate DNA polymerases with built-in 3′–5′ exonuclease activities for proofreading (Morrison et al. 1991; Morrison and Sugino 1994). They are also highly processive enzymes when interacting with the clamp, PCNA (Burgers 1991; Lee et al. 1991; Chilkova et al. 2007). The exact roles of Pols ε and δ at the replication fork have been debated (Kunkel and Burgers 2008; Burgers 2009). Over the past few years, evidence has accumulated that in S. cerevisiae and Schizosaccharomyces pombe (Sp), Pol ε is primarily responsible for leading-strand and Pol δ for lagging-strand synthesis (Pursell et al. 2007; Nick McElhinny et al. 2008; Miyabe et al. 2011). This suggests that the roles of Pol ε and Pol δ are evolutionarily conserved among eukaryotes.
Pol δ is constituted of three subunits (Pol3, Pol31, and Pol32) in S. cerevisiae, and four in S. pombe and H. sapiens (Table 1) (Gerik et al. 1998; Reynolds et al. 1998; Liu et al. 2000). This may be species variations but could also reflect how tightly specific subunits are associated with the catalytic core subunit. The hydrodynamic properties of ScPol δ and SpPol δ suggested that Pol δ has an elongated structure, heavily influenced by the Pol32 subunit (Johansson et al. 2001; Bermudez et al. 2002). This was later confirmed by small-angle X-ray scattering (SAXS) analysis of ScPol3 in complex with Pol31 and the amino terminus of Pol32 and a high-resolution structure of the human accessory subunits Pold2 together with the amino terminus of Pold3 (Baranovskiy et al. 2008; Jain et al. 2009). The catalytic subunit ScPol3 is located at one end of the complex; Pol31 acts as a bridge to the very elongated Pol32 that carries an important interaction motif with PCNA at the far end of the elongated structure extending away from the catalytic core subunit Pol3 (Johansson et al. 2004; Baranovskiy et al. 2008; Jain et al. 2009). A high-resolution structure of ScPol3 (amino acid residues 68–985) was solved which included both the exonuclease and polymerase domains (Fig. 1B), but excluded the carboxy-terminal domain with the CysA and CysB motifs discussed below. Overall, the structure of ScPol3 is highly reminiscent of a typical family B polymerase, with a fold very similar to that of RB69 despite the limited sequence identity (Swan et al. 2009).
Pol ε consists of four subunits. In S. cerevisiae, these are called Pol2, Dpb2, Dpb3, and Dpb4 (Hamatake et al. 1990; Chilkova et al. 2003). Structural information on Pol ε is limited to a low-resolution cryo-electron microscopy (cryo-EM) structure. ScPol ε has two domains, a globular domain with the catalytic subunit and an extended tail domain that has been suggested to harbor the three accessory subunits Dpb2, Dpb3, and Dpb4 (Asturias et al. 2006). Dpb2 is essential in S. cerevisiae and S. pombe (Araki et al. 1991; Feng et al. 2003). However, the Dpb2 subunit does not appear to influence the polymerase activity in human Pol ε (Bermudez et al. 2011). It is possible that the essential function of Dpb2 is during the initiation of DNA replication, as described in Tanaka and Araki (2013) (also reviewed in Araki 2010). Dpb3 and Dpb4 form a heterodimer that has a high affinity for double-stranded DNA (Tsubota et al. 2006). In contrast to Dpb2, ScPol ε depends on Dpb3 and Dpb4 for full processivity in the absence of PCNA (Aksenova et al. 2010).
The eukaryotic replicative polymerases all have two conserved motifs with cysteines (CysA and CysB) located at the carboxyl terminus of the catalytic subunit (Netz et al. 2012). For a long time they have been considered to be Zn-finger motifs and Zn was found in both Pol ε and Pol α (Dua et al. 2002; Klinge et al. 2009). Their primary role was suggested to form an interaction surface with the B subunit of the DNA polymerase. Recently it was shown that the two motifs CysA and CysB have differential functions and coordinate different metals in ScPol δ (Netz et al. 2012). CysA coordinates Zn and is required to support processive synthesis by Pol δ via a direct interaction with PCNA, whereas CysB coordinates a Fe–S center and is important for the interaction with the Pol31 subunit. Loss of interaction with Pol31 led to loss of Pol32, and as a result an important interaction with the PCNA clamp was lost. Thus, Pol δ has at least two separate interactions with PCNA, CysA in Pol3 and the PIP box in Pol32, to support efficient loading and synthesis of DNA (Netz et al. 2012). Experiments with carboxy-terminal domains of Pol α and Pol ε implied that these replicative polymerases may also have an Fe–S cluster. However, conflicting results with expression of the carboxy-terminal domains of the catalytic subunits from human Pols α and ε in E. coli were recently published (Baranovskiy et al. 2012) and it remains to be shown whether the ScPols α and ε holoenzymes carry Fe–S clusters that mediate the interactions between the catalytic subunit and Pol12 and Dpb2, respectively. The study of the carboxy-terminal domain of the catalytic subunit in human Pol δ confirmed that there is an Fe–S center coordinated by CysB (Baranovskiy et al. 2012).
As discussed above, ScPol δ has at least two well-defined motifs that mediate the contact with PCNA. Two additional motifs, one in Pol3 and one in Pol31, were recently reported, emphasizing that ScPol δ has multiple surfaces to stabilize the interaction with PCNA when synthesizing DNA (Acharya et al. 2011). Less is known about how Pol ε interacts with PCNA. In S. cerevisiae, the catalytic subunit and Dpb2 both have a PIP box, but they are not located at the amino or carboxyl terminus as found typically in PCNA-interacting proteins (Dua et al. 2002). Biochemical characterizations and genetic experiments in yeast suggested that these are not functional motifs during DNA replication (Dua et al. 2002; Chilkova et al. 2007). Nevertheless, Pol ε is stimulated by PCNA in vitro, suggesting that either the exact motif(s) remains to be found or there are multiple weak interactions that together stabilize the interaction with PCNA (Maga et al. 1999). Pol ε also has an unusually high intrinsic processivity in part mediated by the Dpb3 and Dpb4 subunits (Aksenova et al. 2010). In addition, the catalytic subunit has by itself an unusually high processivity and it was speculated that a 66 amino acid insertion in the vicinity of the palm domain might give Pol ε this unusual property (Shcherbakova et al. 2003).
FIDELITY OF THE REPLICATIVE POLYMERASES
The high fidelity of replicative polymerases is determined by the tolerance for the incoming nucleotide when performing chemistry and the 3′–5′ exonucleolytic proofreading activity. In addition, the concentration of each dNTP in the cell and the quality of the template influence the rate at which errors are made (Kunkel 2011). Until recently, not much attention was paid to how the relatively large pools of rNTPs in eukaryotic cells influence the fidelity of the replicative DNA polymerases. The polymerases have a built-in steric gate that should select for dNTPs and against rNTPs (Joyce 1997). However, the concentration of dNTPs is very low in comparison to the rNTPs and it appears that Pols α, δ, and ε misincorporate ribonucleotides at a surprisingly high rate when synthesizing DNA (Nick McElhinny et al. 2010c). All three Pols also incorporated rNTPs in vivo in S. cerevisiae and S. pombe and gave a significantly increased elevation of 2–5 nucleotide deletions (Nick McElhinny et al. 2010b; Miyabe et al. 2011). To avoid mutagenic events, ribonucleotides must be removed by a repair mechanism that involves RNase H2 before the next S phase (Nick McElhinny et al. 2010b; Miyabe et al. 2011). The incorporation of ribonucleotides and the dependence on RNase H2 for the removal of ribonucleotides was also found in mice, suggesting that this repair mechanism is conserved among all eukaryotes (Reijns et al. 2012). The exact mechanism behind the error signature in S. cerevisiae is unknown, but Pol α, Pol δ, and Pol ε stall when encountering a ribonucleotide in the template and this may lead to deletions in repetitive sequences (Watt et al. 2011). In case ribonucleotides are not removed, postreplication repair will be activated during the next S phase (Lazzaro et al. 2012). Pol ζ is capable of replicating across 1–4 ribonucleotides in the template and also MMS2-dependent template switching contributes to the tolerance for unrepaired ribonucleotide lesions (Lazzaro et al. 2012).
It is yet unclear if archaeal DNA polymerases also frequently misinsert ribonucleotides in the genome. However, as described earlier, the archaeal replicative polymerases (in, e.g., P. furiosus and S. solfataricus) have a unique capacity to proofread the template for uracil and other deamidated bases (Greagg et al. 1999; Choi et al. 2011). A binding pocket for uracil is found in their unique amino-terminal domains where the single-stranded template is scanned before entering the polymerase catalytic site (Fogg et al. 2002; Shuttleworth et al. 2004; Firbank et al. 2008).
ANTIBIOTICS/INHIBITORS
DNA replication is an essential process in all organisms. Although there are sufficient variations in structures among replisomal proteins from bacteria and humans to make replisomes a very good target for discovery of new antibacterial therapeutics, it is a target that is surprisingly underexploited both by pharmaceutical companies and in nature by other organisms (reviewed in Robinson et al. 2010, 2012). Although there are no known inhibitors of DnaE-type Pol IIIs, there have been substantial efforts to target Pol C from Gram-positive bacteria, including Staphylococcus aureus, using a range of 6-anilinouracils (6-AUs) and quinazolin-2-ylamino-quinazolin-4-ols (BisQuinols). The 6-AUs act as Pol C inhibitors by competing with dGTP for base pairing with cytosines in the template DNA, whereas the BisQuinols appear to interfere with template DNA binding. Another promising target is the β clamp, which makes essential interactions with the clamp-binding motifs in a large number of other proteins (including all five DNA polymerases, the clamp loader, and DNA ligase in E. coli, for example) at a single highly conserved binding site that is structurally distinct from the PIP-binding sites in PCNA (Dalrymple et al. 2001). Development of resistance to antibacterials that inhibit interactions at this site by target mutagenesis would be expected to be slow because it would require simultaneous mutations to arise in genes encoding several different proteins. There have been recent reports of identification through screening campaigns of new inhibitors of interactions with β, including two crystal structures (Georgescu et al. 2008b; Wijffels et al. 2011), but there is much to be done to develop these compounds into useful leads.
CONCLUDING REMARKS
Almost 60 years have passed since the first isolation of a replicative DNA polymerase by Arthur Kornberg in the mid-1950s. In this long history of research, many modern tools in structural and molecular biology have provided fragments of information for progressive synthesis of coherent pictures of how these complex multifunctional enzymes work in organisms from all three domains of life. There are common aspects that are well established, like the common catalytic mechanism of the nucleotidyltransferase reaction, but our pictures are still far from complete. The main reason for this is that these enzymes never work alone. They are key parts of larger, very dynamic nucleoprotein machines (replisomes) that undergo large structural changes as they function, with proteins coming and going and often changing binding partners. Sometimes this occurs at common binding sites (as on the sliding clamps) to ensure ordered sequences of events, and often interactions involve intrinsically unstructured regions of proteins that only become ordered as they interact with their binding partners. This latter fact alone has made structural studies slow and difficult, and it also means that multiple structures need to be solved to really understand function. A second basic operating principle for replisomes is that their integrity is maintained by a very large number of protein–protein and protein–nucleic acid interactions, many of which are inherently weak and can easily be transiently broken. These interactions are often difficult to detect individually, but sum to be strong enough to hold the replisomes together as they function.
ACKNOWLEDGMENTS
We apologize to those colleagues whose work is not cited because of formatting and space restrictions. We thank Matthew Hogg for help with Figure 1, and Rick Lewis for comments on the manuscript. Research in our laboratories is funded by the Swedish Cancer Society, the Swedish Research Council, the Knut and Alice Wallenberg foundation (E.J.), the Australian Research Council, including an Australian Professorial Fellowship, and the Australian National Health and Medical Research Council (N.D.).
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Acharya N, Klassen R, Johnson RE, Prakash L, Prakash S 2011. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc Natl Acad Sci 108: 17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova A, Volkov K, Maceluch J, Pursell ZF, Rogozin IB, Kunkel TA, Pavlov YI, Johansson E 2010. Mismatch repair—Independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase ε. PLoS Genet 6: e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol 22: 766–771 [DOI] [PubMed] [Google Scholar]
- Araki H, Hamatake RK, Johnston LH, Sugino A 1991. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci 88: 4601–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E 2006. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol 13: 35–43 [DOI] [PubMed] [Google Scholar]
- Bailey S, Wing RA, Steitz TA 2006. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126: 893–904 [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH 2008. X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle 7: 3026–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH 2012. DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J Biol Chem 287: 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, MacNeill SA, Tappin I, Hurwitz J 2002. The influence of the Cdc27 subunit on the properties of the Schizosaccharomyces pombe DNA polymerase δ. J Biol Chem 277: 36853–36862 [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J 2011. Studies on human DNA polymerase ε and GINS complex and their role in DNA replication. J Biol Chem 286: 28963–28977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM 1991. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ε. J Biol Chem 266: 22698–22706 [PubMed] [Google Scholar]
- Burgers PM 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284: 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz Galal W, Pan M, Kelman Z, Hurwitz J 2012. Characterization of DNA primase complex isolated from the archaeon, Thermococcus kodakearaensis, J Biol Chem 287: 16209–16219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilkova O, Jonsson BH, Johansson E 2003. The quaternary structure of DNA polymerase ε from Saccharomyces cerevisiae. J Biol Chem 278: 14082–14086 [DOI] [PubMed] [Google Scholar]
- Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E 2007. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res 35: 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP 2011. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J Biol Chem 286: 31180–31193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci 98: 11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervyn E, Suski C, Daniel R, Bruand C, Chapuis J, Errington J, Jannière L, Ehrlich SD 2001. Two essential DNA polymerases at the bacterial replication fork. Science 294: 1716–1719 [DOI] [PubMed] [Google Scholar]
- Dohrmann PR, Manhart CM, Downey CD, McHenry CS 2011. The rate of polymerase release upon filling the gap between Okazaki fragments is inadequate to support cycling during lagging strand synthesis. J Mol Biol 414: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391: 251–258 [DOI] [PubMed] [Google Scholar]
- Dua R, Levy DL, Li CM, Snow PM, Campbell JL 2002. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase ε in insect cells. Purification and characterization. J Biol Chem 277: 7889–7896 [DOI] [PubMed] [Google Scholar]
- Evans RJ, Davies DR, Bullard JM, Christensen J, Green LS, Guiles JW, Pata JD, Ribble WK, Janjic N, Jarvis TC 2008. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci 105: 20695–20700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Rodriguez-Menocal L, Tolun G, D’Urso G 2003. Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication. Mol Biol Cell 14: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA 2008. Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J Mol Biol 381: 529–539 [DOI] [PubMed] [Google Scholar]
- Fogg MJ, Pearl LH, Connolly BA 2002. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat Struct Biol 9: 922–927 [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA 2001. Structure of the replicating complex of a pol α family DNA polymerase. Cell 105: 657–667 [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Kim S-S, Yurieva O, Kuriyan J, Kong X-P, O’Donnell M 2008a. Structure of a sliding clamp on DNA. Cell 132: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Yurieva O, Kim S-S, Kuriyan J, Kong X-P, O’Donnell M 2008b. Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc Natl Acad Sci 105: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PM 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem 273: 19747–19755 [DOI] [PubMed] [Google Scholar]
- Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH 1999. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc Natl Acad Sci 96: 9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatake RK, Hasegawa H, Clark AB, Bebenek K, Kunkel TA, Sugino A 1990. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J Biol Chem 265: 4072–4083 [PubMed] [Google Scholar]
- Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE 2002. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure 10: 535–546 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Inoue T, Kai Y 2001. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J Mol Biol 306: 469–477 [DOI] [PubMed] [Google Scholar]
- Henneke G, Flament D, Hübscher U, Querellou J, Raffin JP 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J Mol Biol 350: 53–64 [DOI] [PubMed] [Google Scholar]
- Hopfner K-P, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B 1999. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci 96: 3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Hammel M, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2009. Structural insights into yeast DNA polymerase δ by small angle X-ray scattering. J Mol Biol 394: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Majka J, Burgers PM 2001. Structure of DNA polymerase δ from Saccharomyces cerevisiae. J Biol Chem 276: 43824–43828 [DOI] [PubMed] [Google Scholar]
- Johansson E, Garg P, Burgers PM 2004. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem 279: 1907–1915 [DOI] [PubMed] [Google Scholar]
- Joyce CM 1997. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc Natl Acad Sci 94: 1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA 2010. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry 49: 5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L 2009. 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. Embo J 28: 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X-P, Onrust R, O’Donnell M, Kuriyan J 1992. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69: 425–437 [DOI] [PubMed] [Google Scholar]
- Krishna TSR, Kong X-P, Gary S, Burgers PM, Kuriyan J 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Kunkel TA 2004. DNA replication fidelity. J Biol Chem 279: 16895–16898 [DOI] [PubMed] [Google Scholar]
- Kunkel TA 2011. Balancing eukaryotic replication asymmetry with replication fidelity. Curr Opin Chem Biol 15: 620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM 2008. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J 2006. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell 126: 881–892 [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell 45: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Pan ZQ, Kwong AD, Burgers PM, Hurwitz J 1991. Synthesis of DNA by DNA polymerase ε in vitro. J Biol Chem 266: 22707–22717 [PubMed] [Google Scholar]
- Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY 2000. Identification of a fourth subunit of mammalian DNA polymerase delta. J Biol Chem 275: 18739–18744 [DOI] [PubMed] [Google Scholar]
- Maga G, Jonsson ZO, Stucki M, Spadari S, Hubscher U 1999. Dual mode of interaction of DNA polymerase ε with proliferating cell nuclear antigen in primer binding and DNA synthesis. J Mol Biol 285: 259–267 [DOI] [PubMed] [Google Scholar]
- Matsui I, Urushibata Y, Shen Y, Matsui E, Yokoyama H 2011. Novel structure of an N-terminal domain that is crucial for the dimeric assembly and DNA-binding of an archaeal DNA polymerase D large subunit from Pyrococcus horikoshii. FEBS Lett 585: 452–458 [DOI] [PubMed] [Google Scholar]
- Mayanagi K, Kiyonari S, Nishida H, Saito M, Kohda D, Ishino Y, Shirai T, Morikawa K 2011. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc Natl Acad Sci 108: 1845–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS 2011. DNA replicases from a bacterial perspective. Annu Rev Biochem 80: 403–436 [DOI] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM 2011. The major roles of DNA polymerases ε and δ at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet 7: e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Sugino A 1994. The 3′ → 5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet 242: 289–296 [DOI] [PubMed] [Google Scholar]
- Morrison A, Bell JB, Kunkel TA, Sugino A 1991. Eukaryotic DNA polymerase amino acid sequence required for 3′–5′ exonuclease activity. Proc Natl Acad Sci 88: 9473–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi-Falconi M, Giannattasio M, Foiani M, Plevani P 2003. The DNA polymerase α-primase complex: Multiple functions and interactions. ScientificWorldJournal 3: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ 2012. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol 8: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA 2008. Division of labor at the eukaryotic replication fork. Mol Cell 30: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kissling GE, Kunkel TA 2010a. Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc Natl Acad Sci 107: 21070–21075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA 2010b. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6: 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA 2010c. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci 107: 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez-Ramirez R, Klinge S, Sauguet L, Melero R, Recuero-Checa MA, Kilkenny M, Perera RL, Garcia-Alvarez B, Hall RJ, Nogales E, et al. 2011. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res 39: 8187–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Jergic S, Park AY, Dixon NE, Otting G 2008. The proofreading exonuclease subunit ε of Escherichia coli DNA polymerase III is tethered to the polymerase subunit alpha via a flexible linker. Nucleic Acids Res 36: 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Loeb LA 2001. Getting a grip on how DNA polymerases function. Nat Struct Biol 8: 656–659 [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA 2006. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr Biol 16: 202–207 [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA 2007. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149: 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Watt A, Fantes PA, MacNeill SA 1998. Cdm1, the smallest subunit of DNA polymerase d in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr Genet 34: 250–258 [DOI] [PubMed] [Google Scholar]
- Robinson A, Brzoska AJ, Turner KM, Withers R, Harry EJ, Lewis PJ, Dixon NE 2010. Essential biological processes of an emerging pathogen: DNA replication, transcription and cell division in Acinetobacter spp. Microbiol Mol Biol Rev 74: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Causer RJ, Dixon NE 2012. Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr Drug Targets 13: 352–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AC, Park HW, Mao C, Beese LS 2000. Crystal structure of a pol α family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9° N-7. J Mol Biol 299: 447–462 [DOI] [PubMed] [Google Scholar]
- Sanders GM, Dallmann HG, McHenry CS 2010. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell 37: 273–281 [DOI] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA 2003. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem 278: 43770–43780 [DOI] [PubMed] [Google Scholar]
- Shuttleworth G, Fogg MJ, Kurpiewski MR, Jen-Jacobson L, Connolly BA 2004. Recognition of the pro-mutagenic base uracil by family B DNA polymerases from archaea. J Mol Biol 337: 621–634 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I 2012. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483: 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano NM, Chen J, McHenry CS 2006. A coproofreading Zn2+-dependent exonuclease within a bacterial replicase. Nat Struct Mol Biol 13: 458–459 [DOI] [PubMed] [Google Scholar]
- Steitz TA 1993. DNA- and RNA-dependent DNA polymerases. Curr Opin Struct Biol 3: 31–38 [Google Scholar]
- Stocki SA, Nonay RL, Reha-Krantz LJ 1995. Dynamics of bacteriophage T4 DNA polymerase function: Identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J Mol Biol 254: 15–28 [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK 2009. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Tanaka S, Araki H 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tori K, Kimizu M, Ishino S, Ishino Y 2007. DNA polymerase B1 and D from the hyperthermophilic archaeon Pyrococcus furiosus both bind proliferating cell nuclear antigen with their C-terminal PIP-box motifs. J Bacteriol 189: 5652–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota T, Tajima R, Ode K, Kubota H, Fukuhara N, Kawabata T, Maki S, Maki H 2006. Double-stranded DNA binding, an unusual property of DNA polymerase ε, promotes epigenetic silencing in Saccharomyces cerevisiae. J Biol Chem 281: 32898–32908 [DOI] [PubMed] [Google Scholar]
- Warbrick E 1998. PCNA binding through a conserved motif. BioEssays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Wardle J, Burgers PM, Cann IK, Darley K, Heslop P, Johansson E, Lin LJ, McGlynn P, Sanvoisin J, Stith CM, et al. 2008. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res 36: 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DL, Johansson E, Burgers PM, Kunkel TA 2011. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 10: 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek A, McHenry CS 2006. The NH2-terminal php domain of the α subunit of the Escherichia coli replicase binds the ε proofreading subunit. J Biol Chem 281: 12561–12567 [DOI] [PubMed] [Google Scholar]
- Wijffels G, Johnson WM, Oakley AJ, Turner K, Epa VC, Briscoe SJ, Polley M, Liepa A, Hofmann A, Buchardt J, et al. 2011. Binding inhibitors of the bacterial sliding clamp by design. J Med Chem 54: 4831–4838 [DOI] [PubMed] [Google Scholar]
- Wing RA, Bailey S, Steitz TA 2008. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α-subunit. J Mol Biol 382: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Nossal N, Benkovic SJ 1998. Kinetic characterization of a bacteriophage T4 antimutator DNA polymerase. Biochemistry 37: 14748–14755 [DOI] [PubMed] [Google Scholar]
- Yang G, Lin T, Karam J, Konigsberg WH 1999. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry 38: 8094–8101 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jeruzalmi D, Moarefi I, Leighton L, Lasken R, Kuriyan J 1999. Crystal structure of an archaebacterial DNA polymerase. Structure 7: 1189–1199 [DOI] [PubMed] [Google Scholar]