Abstract
Recognition of microbial nucleic acids is one strategy by which mammalian hosts respond to infectious agents. Intracellular DNA which is introduced into cells during infection elicits potent inflammatory responses by triggering the induction of antiviral type I interferons and the maturation and secretion of inflammatory cytokines such as tumor necrosis factor α (TNFα), interleukin (IL)-1β and IL-18. In addition, if nucleases such as DNase II or DNase III (Trex1) fail to clear self-DNA accumulated DNA gains access to intracellular compartments where it drives inflammatory responses leading to autoimmune disease. In this review, we discuss a rapidly evolving view of how cytosolic DNA sensing machineries coordinate antimicrobial immunity and if unchecked lead to autoimmune disease.
Introduction
The Innate immune system is the first line of defense against infectious agents. Germline-encoded pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs) and C-type lectins recognize a wide range of microbial products, often referred to as microbe-associated molecular patterns (MAMPs) (1). Recognition of MAMPs by these surveillance receptors turns on signaling pathways that coordinate transcription of hundreds of inflammatory genes, the products of which control infection directly and marshal the T- and B- cells of the adaptive immune system (2). In addition to classical microbial products such as bacterial lipopolysaccharide or lipoproteins, microbial nucleic acids have emerged as major triggers of innate immune defenses.
The best-characterized nucleic acid sensors are a subset of TLRs, type I transmembrane receptors localized to the endosomal compartment that sense double-strand (ds) RNA (TLR3) (3), ssRNA (TLR7 and TLR8) (4–6) and hypomethylated CpG DNA (TLR9) (7, 8). TLRs induce type I IFN and other inflammatory genes via Toll/Interleukin-1 receptor (TIR) domain containing adaptor molecules such as MyD88 (TLR7/8/9) or TRIF (TIR-domain-containing adaptor inducing IFN-β) for TLR3 (9). Cytosolic RNA sensors such as retinoic acid inducible-I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) have also been identified (10, 11), which signal via a unique adaptor mitochondrial antiviral signaling (MAVS; also known as IPS) to mediate NF-κB and IRF (IFN regulatory factor)-dependent transcription of inflammatory genes (12).
The cellular machinery that senses cytosolic DNA is still being elucidated. While significant progress has been made in understanding how DNA is recognized and how DNA signaling ensues and leads to inflammation, the molecular basis to cytosolic DNA recognition in innate immunity is still being worked out in detail. A number of new molecules (which will be described below) have been identified that contribute in various ways to recognition and signaling in response to DNA. TLR9 expressed in endosomal membranes was the first identified DNA receptor that recognizes hypomethylated CpG motifs (7). However, in humans, TLR9 expression is restricted to B cells (13) and plasmacytoid dendritic cells (DCs) (14) and therefore does not account for DNA-induced immune responses in other cell types such as macrophages. Two seminal studies demonstrated that the targeted delivery of synthetic dsDNA or a 45-nucleotide immunostimulatory DNA (ISD) into the cytosol of macrophages and DCs triggered Tank-binding kinase 1 (TBK1)/IRF3-dependent induction of type I IFN and other inflammatory genes in a TLR9-independent manner (15, 16). These findings led to a search in many labs for sensor(s) that could couple cytosolic DNA recognition to immune signaling. Two conceptually distinct signaling pathways have since emerged. The first of these leads to the proteolytic activation of the cysteine protease caspase-1 associated with maturation and secretion of the proinflammatory cytokines IL-1β and IL-18. A second pathway that is still being worked out leads to the transcriptional induction of type I interferon (IFN) and proinflammatory genes. These two pathways are depicted in their simplest form in Figure 1. Here, we briefly summarize and discuss the state of play of DNA sensing and signaling and the functional significance of these event in antimicrobial immunity and autoimmune diseases.
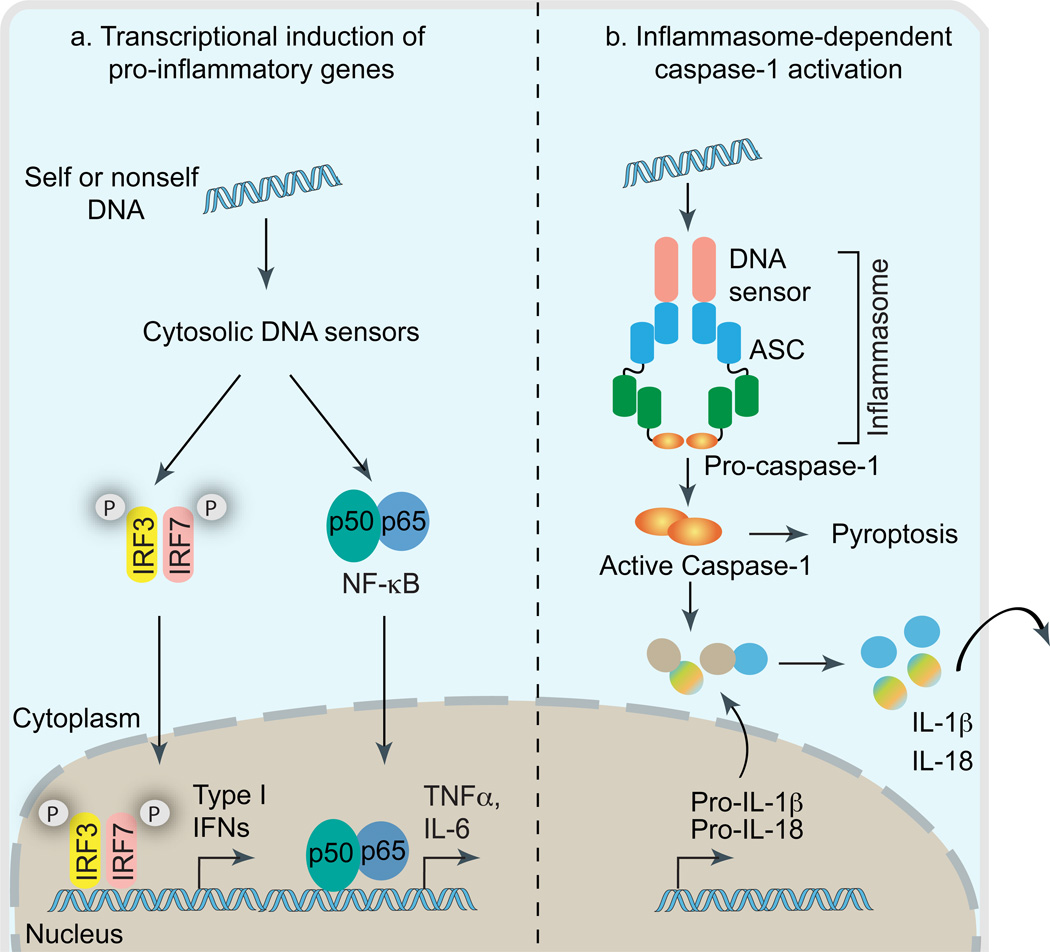
Figure 1. DNA in cytosol triggers transcription of inflammatory genes and inflammasome-dependent proteolytic activation of caspase-1.
Cytosolic DNA leads to the engagement of two conceptually distinct signaling pathways involved in host immune responses (a) Activation of IRF3, IRF7 and NF-κB leads to the transcriptional induction of type I interferon genes or proinflammatory genes such as IL-6 and TNFα. (b) Cytosolic DNA driven inflammasome assembly via homotypic PYD:PYD and CARD:CARD interactions leading to caspase-1 activation and subsequent proteolytic cleavage of pro-IL-1β and pro-IL-18 into biologically active, mature forms IL-1β and IL-18. Besides cytokine processing, caspase-1 also mediates cell-death under certain biological contexts.
Inflammasome activating cytosolic DNA sensors
Cytosolic DNA leads to the production of the pro-inflammatory cytokines IL-1β and IL-18. IL-1β is secreted by many cell-types and is essential for both the innate and adaptive arms of the immune response (17). IL-1β is important in activating neutrophils, macrophages, DCs and T cells while IL-18 is crucial for IFN-γ production by NK cells and T cells (18, 19). In contrast to the majority of inflammatory mediators that are regulated transcriptionally, IL-1β and IL-18 are regulated at both the transcriptional and post-translational levels. Upon transcriptional induction by TLRs and other sensor systems, IL-1β and IL-18 are synthesized as inactive precursor proteins, which are subsequently processed by the cysteine protease caspase-1 (IL-1β converting enzyme; ICE) (20). Conversion of procaspase-1 itself into an enzymatically active form, caspase-1 occurs upon formation of a multiprotein complex in the cytosol referred to as the inflammasome (21). The nucleotide binding and leucine-rich repeat containing receptor (NLR; aka NOD-like receptors) and PYHIN family proteins (discussed further below) are known to form inflammasome complexes by recruiting procaspase-1 via an adaptor protein ASC (22). ASC is a bipartite molecule containing an N-terminal pyrin domain (PYD) and the C-terminal caspase-1 activation/recruitment domain (CARD), and therefore acts as an adaptor to bring together NLRs (or PYHIN proteins) and procaspase-1 via homotypic PYD:PYD and CARD:CARD interactions, respectively (Figure 2). Several inflammasome complexes have been identified in recent years. Of all the known inflammasomes, Nlrp3, absent in melanoma 2 (AIM2) and most recently interferon inducible protein 16 (IFI16) inflammasomes have been linked to immune responses to intracellular DNA as well as bacterial or DNA virus infections.
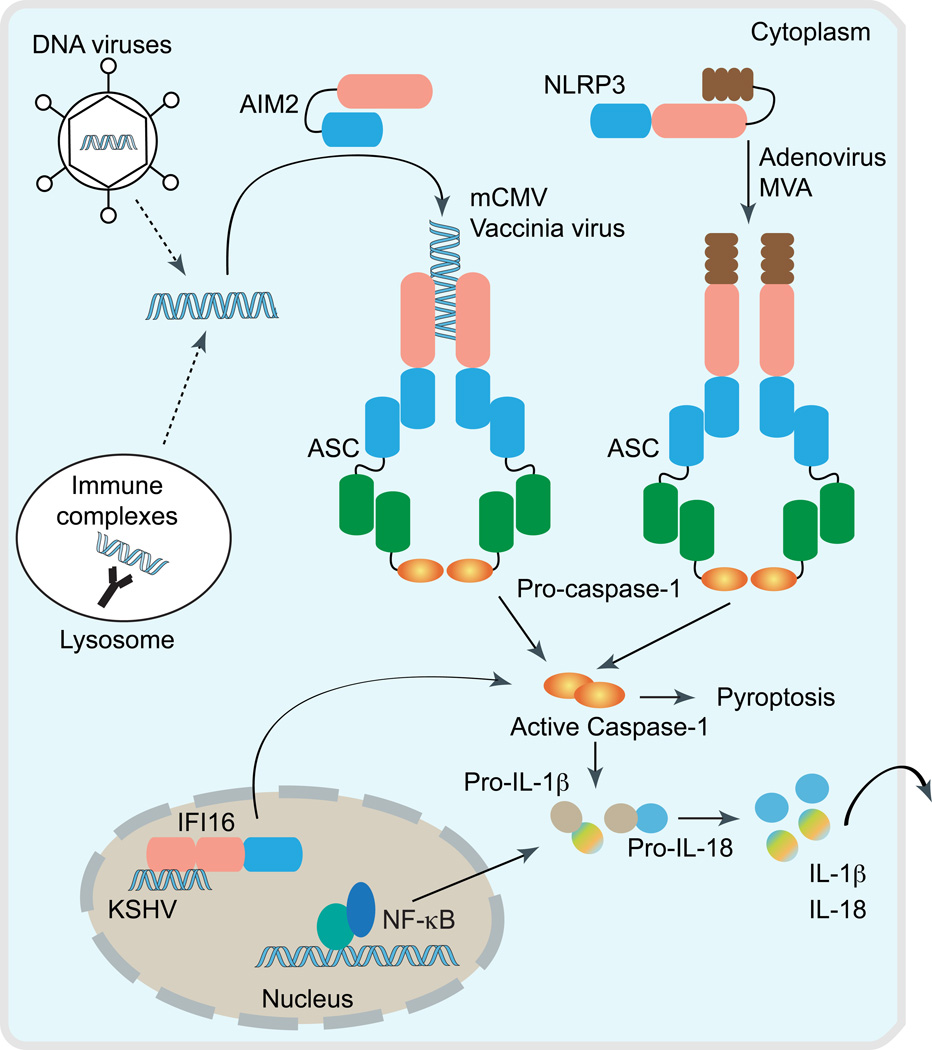
Figure 2. Cytosolic DNA triggers inflammasome activation.
Intracellular DNA following microbial infection or phagocytosis of immune complexes can potentially trigger the assembly of either NLR (e.g. NLRP3) or PYHIN inflammasomes (AIM2 and IFI16). Upon activation, these cytosolic DNA sensors recruit the inflammasome adaptor ASC to activate caspase-1, which leads to the processing of pro-IL-1β and pro-IL-18 into their biologically active forms. Notably, AIM2 and IFI16 bind directly to DNA via its C-terminal HIN200 domain, and therefore act as the true receptors for cytosolic DNA. However, the precise mechanism triggering Nlrp3 inflammasome assembly, and whether microbial DNA is the ligand for Nlrp3 is still unclear. Interestingly, IFI16 inflammasome is thought to assemble inside the nucleus in response to KSHV infection in endothelial cells.
The Nlrp3 inflammasome
The Nlrp3 inflammasome is the best-studied inflammasome and is associated with bacterial, viral, parasitic and fungal infections (23). NLRP3 is also activated in response to endogenous danger signals (24). The Nlrp3 inflammasome in macrophages responds to modified vaccinia virus Ankara strain (MVA) (25) and adenovirus (26) to facilitate caspase-1 activation and IL-1β secretion. For adenovirus, intracellular viral DNA, but not viral capsids activate the Nlrp3 inflammasome. Caspase-1 activation and IL-1β secretion in response to transfected non-viral cytosolic DNA (synthetic, bacterial or mammalian origin) however was Nlrp3-independent, but ASC-dependent indicating that cytosolic DNA-driven inflammasome activation requires an ASC-dependent sensor (26). Whether adenoviral DNA is the ligand required to trigger assembly of the Nlrp3 inflammasome activation is unclear, but an indirect mode is more likely as suggested by a recent study that perturbations in cell membrane associated with adenoviral entry is the trigger for Nlrp3 activation (27). The Nlrp3 inflammasome also recognizes influenza A infection, a negative stranded RNA virus (28). Therefore, the Nlrp3 inflammasome is involved in mediating the inflammatory responses to both DNA and RNA viruses. It is likely that the Nlrp3 inflammasome senses cytosolic nucleic acids indirectly.
The AIM2 inflammasome
Several groups independently identified AIM2 as the receptor for cytosolic DNA that leads to caspase-1 activation and IL-1β secretion (29–31). Contrary to other cytosolic DNA sensors, which are primarily involved in the induction of type I IFN, AIM2 triggers the activation of the inflammasome. Notably, the AIM2 inflammasome is the first among all inflammasome-activating proteins identified so far wherein a direct receptor: ligand interaction has been demonstrated. AIM2 binds cytosolic DNA of self and nonself origin including bacterial, viral and mammalian DNA in a sequence-independent manner. Upon DNA binding via its HIN200 domain, AIM2 undergoes oligomerization and thereby recruits caspase-1 via ASC. In vivo studies in Aim2-deficient mice clearly indicate that the Aim2 inflammasome is essential and functions in a non-redundant manner for innate defense responses to DNA viruses and intracellular bacterial infections (32, 33). Caspase-1 activation and IL-1β/IL-18 secretion in response to mCMV and vaccinia virus infection in murine macrophages is dependent upon Aim2 inflammasome (32). Consistently, Aim2 dependent IL-18 secretion and NK cell activation is essential for an early control of mCMV infection in vivo. Similarly, Aim2 plays a crucial role in controlling F. tularensis infection in vivo because Aim2 deficient mice are more susceptible to infection and show higher bacterial counts in infected organs (33, 34). Activation of the Aim2 inflammasome is also reported in other bacterial infections including L. monocytogenes (35, 36) and M. tuberculosis (37). How Aim2 is exposed to bacterial DNA during infection is not well understood. There is some evidence that DNA released into the cytosol following lysosomal bacteriolysis is the trigger for Aim2 activation (38). Recent evidence indicates that the AIM2-related protein IFI16 also forms an inflammasome complex following Kaposi-sarcoma associated herpes virus (KSHV) infection of endothelial cells (39).
Type I interferon producing cytosolic DNA sensors
Type I interferon production is another major consequence of cytosolic DNA sensing, and is essential for anti-viral immunity and immunity to many different classes of infectious agents. While we understand in detail how DNA via AIM2 leads to IL-1β production, the molecular bases to DNA induced type I IFN gene transcription is less clear. A core signaling module consisting of STING (Stimulator of type I IFN gene)-TBK1-IRF3 is engaged and is absolutely essential for type I IFN responses to DNA. TBK1, an IKK-related kinase originally characterized as the kinase responsible for the phosphorylation-induced activation of IRF3 in TLR and then RLR signaling is also central for DNA signaling pathways (40, 41). TBK1 activation therefore acts as a point of convergence of multiple PRR-driven pathways that result in IRF3 phosphorylation and transcription of type I IFN genes and related ISGs (interferon-stimulated genes).
Several groups independently identified STING (also known as MPYS, TMEM173, ERIS and MITA) as a key component of the DNA sensing pathway (42–44). STING was shown to localize to the endoplasmic reticulum (ER) and outer mitochondrial membranes via five transmembrane spanning regions. In response to DNA, STING translocates to perinuclear regions where it interacts with TBK1 to relay downstream signals to IRF3. STING-deficiency in macrophages or dendritic cells leads to a markedly impaired type I IFN response to B-DNA and ISD or to infection with DNA viruses including herpes simplex virus type I (HSV-1), human cytomegalovirus (HCMV) and vaccinia virus (VACV) (42, 45). STING is also essential for type I IFN induction in response to intracellular bacteria including Francisella Tularensis (46, 47), Listeria monocytogenes (46, 48) and Brucella abortus (49) and in response to extracellular bacteria such as Streptococcus pneumonia (50) and Streptococcus pyogenes (51). Initial studies showed that STING also interacted with components of the RNA recognition machinery such as RIG-I, where it was linked to type I IFN induction in response to vesicular stomatitis virus (VSV), a negative-strand RNA virus (42). Subsequent studies in STING-deficient cells however suggested that viruses or ligands that engaged RIG-I have normal IFN responses in STING-deficient murine macrophages (48, 52). Additional studies however have linked STING to DNA and RNA recognition pathways by uncovering a role for STING in the activation of STAT6 and induction of STAT6-dependent chemokines (52).
The assumption from these earlier studies is that analogous to MyD88 or MAVS, STING is functioning as an adapter molecule. This assumption implies that DNA binding proteins that recognize microbial DNA engage STING and activate TBK1 and downstream signaling. Several candidate sensors have been discovered and implicated to varying degrees in this DNA dependent TBK1-IRF3 and type I IFN production pathway (Figure 3).
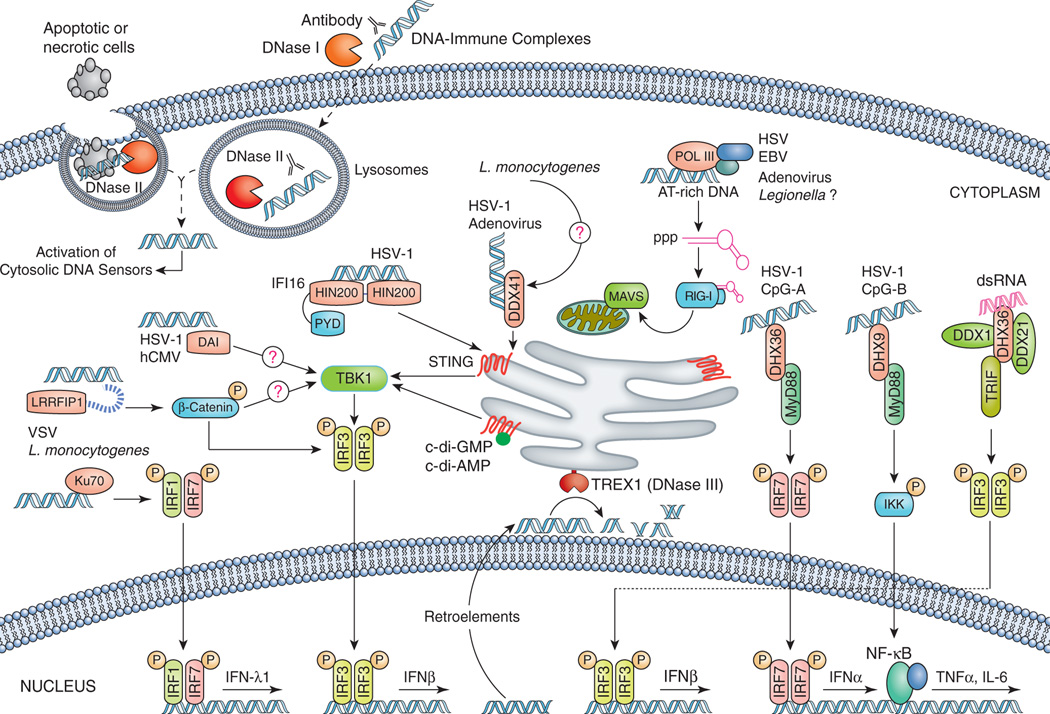
Figure 3. Cytosolic DNA sensors activate the transcription of type I IFN and other inflammatory genes.
Cytosolic DNA of microbial or self-origin is a potent trigger of type I IFN production via STING-TBK1-IRF3 axis, as well as other proinflammatory genes (e.g. TNFα and IL-6) by engaging NF-κB signaling. Distinct cytosolic DNA sensors along with their select activating conditions are shown, which are discussed in detail in text. The DNA-induced signaling pathway converges on the adaptor STING and the kinase TBK1, which phosphorylates IRF3 to mediate downstream signaling events leading to transcriptional induction of inflammatory genes. Beyond cytosolic DNA, bacterial small molecules, c-di-AMP and c-di-GMP acts as a potent stimulator of type I IFN a response by engaging STING either as a direct sensor or co-activator (discussed in text). Host cells employ distinct nucleases to eliminate both self and non-self DNA from extracellular space (DNase I), phago-lysosomes (DNase II) and cytosol (DNase III; Trex1) to avoid deleterious effects of excess DNA-induced immune responses.
DAI (DNA-dependent activator of IRFs)
DAI also known as Z-DNA binding protein, ZBP, or DLM1 is an IFN-stimulated gene, and the first cytosolic DNA sensor identified (53, 54). DAI binds Z-form DNA via two N-terminal Z-DNA-binding domains, and an adjacent protein region carrying B-DNA binding potential. DAI was shown to mediate DNA-induced type I IFN production in cell-based studies. However, subsequent studies in DAI−/− mice revealed that DAI-deficient cells and mice retain normal type I IFN responses to DNA viruses and various types of synthetic DNA (55), indicating that DAI is either dispensable or a redundant sensor of cytosolic DNA. Whether DAI impacts other aspects of DNA-induced signaling is still unclear.
RNA polymerase III
Following the discovery of DAI, RNA polymerase III was linked to DNA recognition, adding another layer of complexity to the cytosolic DNA recognition pathway (56, 57). RNA polymerase III present in the cytosol transcribes AT-rich DNA into immunostimulatory dsRNA transcripts characterized by uncapped 5’-triphosphate moieties, which acts as a ligand for RIG-I (58). Subsequently, RIG-I signals via MAVS to induce the expression of type I IFN and other cytokines. Therefore, RNA polymerase III does not sense cytosolic DNA directly, but generates a ligand to engage the RIG-pathway. Involvement of RNA polymerase III/RIG-I pathway in response to AT-rich DNA explains the partial dependence upon MAVS for DNA-induced induction of type I IFN in certain cell-types. The RNA polymerase III pathway is functional in both human and mouse cells, and is engaged in response to the intracellular dsDNA mimetic poly(dA:dT), and with certain DNA viruses like adenovirus and Epstein-Barr virus. Additional studies have also linked RNA polymerase III and type I IFN responses during Legionella pneumophila and HSV-1 infections, however there is some controversy over these latter data (59, 60).
IFI16 and p204
IFN-inducible protein 16 (IFI16) was identified as a DNA-binding protein in human monocytes in affinity-purification studies (61). Like AIM2, IFI16 is a member of the PYHIN (pyrin and HIN200 domain-containing; also known as p200 protein family). IFI16 is primarily localized in the nucleus. In some cell types such as macrophages, a small pool of cytoplasmic IFI16 colocalizes with transfected DNA or viral DNA that gains access to the cytosolic compartment during HSV-1 infection. In other cell types such as fibroblasts that are permissive to HSV1 infection, IFI16 engages viral DNA that accumulates during productive infection in the nucleus (62). IFI16 does not leave the nucleus in HSV-1 infected human foreskin fibroblast (HFF) cells, however this nuclear sensing of HSV-1 requires cytoplasmic STING signaling. HSV-1 DNA recognition therefore may differ in permissive versus non-permissive cells, but in both cases IFI16 dependent recognition of HSV-1 DNA is coupled to STING signaling. It is important to understand the role of nuclear and cytoplasmic IFI16 function during HSV infection in these contexts and to better understand how signaling from these compartments is initiated and regulated. IFI16 has two C-terminal DNA-binding HIN200 domains and like AIM2, IFI16 binds DNA via non-sequence-specific DNA recognition accomplished through electrostatic attractions between the positively charged HIN200 domain residues and the negatively charged dsDNA sugar-phosphate backbone (63). IFI16 then interacts with STING to activate TBK-1 to trigger IFN-β induction. Whether IFI16 directly or indirectly binds STING is unknown. Humans have 4 PYHIN proteins (IFI16, AIM2, MNDA and IFIX) while mice have 13 members (64). The murine PYHIN protein Ifi204 (p204) is proposed to function in an analogous manner to IFI16. Knockdown of IFI16 (or Ifi204) compromise DNA as well as HSV-1 induced IRF3 activation and IFN induction (61). Evidence is also accumulating for a functional role of IFI16/Ifi204 in controlling HSV infection since p204 plays a role in resistance to HSV-1 infection in the corneal epithelium (65). Interestingly, the HSV viral nuclear ICP0 protein can target IFI16 for degradation thereby documenting a mechanism by which HSV evades IFI16 mediated detection (62).
DExD/H box helicases
Aspartate-glutamate-any amino acid-aspartate/histidine (DExD/H)-box containing helicases are also beginning to emerge as important players in the recognition of cytosolic nucleic acids. DHX36 and DHX9 have been identified as TLR9-independent, but MyD88-dependent sensors of CpG-A and CpG-B DNA, respectively in human pDCs (66). These two distinct oligodeoxynucleotides (ODNs) are known inducers of either type I IFN (CpG-A) or pro-inflammatory cytokines such as TNFα and IL-6 (CpG-B) in pDCs (67). Consistently, DHX36 is involved in the production of IFNα whereas DHX9 mediates TNFα/IL-6 production in HSV-1 infected (or CpG treated) human pDCs. DHX36 and DHX9 interact directly with the TIR domain of MyD88 to trigger downstream signaling to activate IRF7 and NF-κB p50 respectively. Subsequent studies have also ascribed a role for DHX36 and DHX9 in the recognition of cytosolic RNA. DHX36 together with DDX1 and DDX21 forms a complex with the adaptor TRIF to mediate type I IFN induction in response to synthetic dsRNA mimic poly(I:C) (68). Similarly, DHX9 interacts with MAVS to induce type I IFN in response to dsRNA in myeloid DCs (69). Whether DHX36 and DHX9 are involved in the recognition of RNA viruses is unclear at present. In a comprehensive shRNA screen for all 59 members of the DExD/H family, Zhang and colleagues identified DDX41 as a cytosolic DNA receptor in both mouse and human DCs (70). DDX41 binds to dsDNA through its helicase domain, and triggers the activation of IRF3, NF-κB and mitogen-activated protein kinase (MAPK) signaling. Knockdown of DDX41 in DCs led to impaired type IFN and pro-inflammatory cytokine production in response to transfected dsDNA or DNA viruses including HSV-1 and adenovirus, but not against poly(I:C) or influenza virus infection. Moreover, DDX41 binds STING in DCs in response to transfected poly(dA:dT) or HSV-1 DNA suggesting that STING is involved in signaling downstream of DDX41 recognition of the cytosolic DNA. Unlike IFI16, which is a type I IFN inducible gene, DDX41 is expressed at relatively high levels in immune cells. Therefore, DDX41 has been proposed to recognize DNA in the early stages of infection, while IFI16, a type I IFN inducible gene is induced at later stages (as a result of DDX41-dependent IFN production) where these two sensors together coordinate DNA induced IFN responses.
Ku70
Several additional DNA binding proteins have been linked to DNA recognition and innate immunity. A recent study by Zhang et al. identified Ku70, a component of the DNA-repair and telomere maintenance pathway, as the newest member of the cytosolic DNA sensing machinery (71). Ku70 was identified as a DNA-binding protein in HEK293 cells by DNA-affinity purification followed by mass spectrometry. Notably, Ku70 is involved in the production of type III IFN (IFN-λ1; also known as IL29) and not type I IFN (IFNα or IFNβ) in response to a variety of transfected DNA (>500 bp) in HEK293, murine macrophages and DCs. Subsequent studies indicated that the DNA-induced IFN-λ1 induction required IRF1 and IRF7 binding to the IFN-λ1 promoter, which implicates Ku70 in a signaling pathway involving IRF1/IRF7. Although Ku70 was shown to mediate HSV-induced IFN-λ1 induction in HEK293 cells, whether Ku70 recognizes DNA viruses and bacterial infections in primary immune cells remain to be fully elucidated. As the biological activity and the signaling pathway employed by both type I - and III IFN are very similar, it is also unclear how the engagement of Ku70-mediated IFN-λ1 production contributes to the overall DNA-induced innate immune responses.
LRRFIP1
LRRFIP1 (leucine-rich repeat in Flightless-I interacting protein 1) was originally identified by Wilson and colleagues using a cDNA library to screen for cellular interacting proteins of HIV-1 RNA, and was shown to bind RNA and with lower affinity to DNA (72). Subsequent studies indicated that the LRRFIP1 recognizes both RNA and DNA in the cytosol, and enhances the transcription of the Ifnβ gene through a novel β-catenin-dependent signaling pathway (73). In a subsequent RNAi screen against LRR-containing, and LRR-interacting proteins in primary murine macrophages, Yang et al. identified LRRFIP1 as a cytosolic sensor involved in the production of Ifnβ in response to Listeria monocytogenes or VSV infection (73). LRRFIP1 is similarly involved in the Ifnβ production in response to transfected dsRNA or dsDNA. Interestingly, LRRFIP1 utilizes β-catenin, an integral component of the Wnt signaling pathway, to turn on Ifnβ gene transcription. Upon recruitment by LRRFIP1 by yet to be identified mechanisms, β-catenin is phosphorylated at serine 552, which then translocates to the nucleus, and binds to IRF3 leading to enhanced recruitment of the histone acetyltransferase p300, thereby enhancing the transcription of the Ifnβ gene.
Role of STING and DDX41 in sensing cyclic di-nucleotides
In addition to its role as a signaling intermediate in DDX41 and IFI16 signaling pathways, STING also acts as a receptor for the bacterial second messenger molecules, cyclic diadenosine monophosphate (c-di-AMP) and cyclic diguanosine monophosphate (c-di-GMP) (74). These secondary messenger molecules are potent inducers of type I IFNs and in the case of Listeria monocytogenes are proposed to represent the major trigger of IFN production in macrophages (75). Forward genetic studies in N-etyhyl-N-nitrosourea (ENU) mutagenized mice revealed that STING was essential for type I IFN response to c-di-AMP/GMP and following Listeria infection (48). STING binds these small molecules directly through its C-terminal domain (CTD), and leads to the activation of TBK1-IRF3 axis for the induction of Ifn-β genes. Recently, several groups have resolved the crystal structures of human STING-CTD bound to c-di-AMP/GMP (76–78). Collectively, these studies impart dual functions to STING where it either serves as an adaptor for DNA sensing, or as a direct sensor of bacterial second messenger molecules like c-di-AMP. Whether, c-di-AMP or DNA is the major microbial trigger involved in IFNβ response during Listeria infection remains to be determined. Since genes encoding c-di-AMP are also predicted to exist in several other pathogenic bacteria including Staphylococci, Streptococci, Mycobacterium and Chlamydia (79), it will be interesting to define the importance of STING/c-di-AMP interactions in host type I IFN responses to these pathogens. Very recently, Chen and colleagues showed that STING directly binds cyclic GMP-AMP (cGAMP), a novel endogenous second messenger generated in response to DNA to trigger IRF3 activation and the induction of IFNβ in response to transfected DNA or DNA viruses (80). They further identify cGAMP synthase (cGAS), a mammalian nucleotidyltransferase family member, which binds to cytosolic DNA and catalyzes the generation of cGAMP to trigger STING-dependent induction of type I IFN response (81). Therefore, STING recognition of cGAMP represents a new avenue for mounting host immune responses to cytosolic DNA.
Surprising recent evidence indicates that DDX41 also serves as the PRR involved in detecting c-di-GMP and c-di-AMP to trigger TBK1-IRF3 signaling via STING (82). Surprisingly, the interaction of c-di-GMP with DDX41 was shown to be greater than that observed for STING binding. These data indicate that DDX41 is required to facilitate c-di-GMP signaling via STING. The solved crystal structure of the STING-CTD in complex with c-di-GMP has shown that one molecule of c-di-GMP binds one dimer of STING (76–78). Cheng and colleagues propose that the detection of c-di-GMP via DDX41 promote enhanced DDX41-STING interactions, leading to an increase in the binding affinity of STING for c-di-GMP, which ultimately drives downstream signaling events. This model is consistent with the possibility that STING functions as a secondary receptor or coactivator in the cyclic di-nucleotide signaling pathway.
Recognition of endogenous DNA and the autoimmune diseases
While DNA-induced immune responses are central to immunity, inappropriate recognition of self-DNA can lead to deleterious consequences to the host. Systemic lupus erythematosus (SLE) is one such disease, where type I IFN and autoantibodies directed against dsDNA, RNA and nucleosomes are implicated in diseases pathogenesis (83). Multiple fail-safe mechanisms deployed by the host subvert endogenous DNA-induced immune responses. One level of regulation is provided by cellular endonucleases such as DNase I, DNase II and DNase III (also known as Trex1) that are involved in the clearance of extracellular, lysosomal and cytosolic DNA, respectively. Recent evidence clearly indicates that the functional defects in these enzymes are associated with SLE and other human diseases. For example, DNase I gene mutations have been identified in subgroups of SLE patients (84), a clinical association further supported by lupus-like syndrome in DNase I deficient mice (85). Defects in DNase I lead to the accumulation of extracellular DNA released by apoptotic/necrotic cells, which is highly immunostimulatory (86). DNase II is expressed in lysosomes where it degrades DNA from phagocytosed apoptotic and necrotic cells. Interestingly, DNase II knockout mice are embryonic lethal, however they are viable on the IFNR1-knockout background indicating that type I IFNs mediate the lethality of DNase II genetic deficiency (87). Here, type I IFN responses are mediated in a TLR-independent, but IRF3/7 dependent manner. Recent evidence indicates that DNaseII/STING deficient mice are also viable indicating the importance of STING in this model (88). The DNase II/ IFNR1 mice develop autoimmune polyarthritis within 2 months of age with features reminiscent of human rheumatoid arthritis further highlighting a central role for DNA-induced immune responses in autoimmune diseases. The sensors of DNase I and DNase II substrates are unclear, but it is likely that one or more of the aforementioned cytosolic DNA sensor(s) recognize and respond to this accumulated dsDNA. The DNase III/Trex1 is another nuclease, which is normally involved in the clearance of cell-intrinsic ssDNA (89). Trex1 is the most abundant 3’-5’ exonuclease, and is localized to the endoplasmic reticulum. Recent studies in Trex1 knockout mice provide great insight into the regulation of endogenous DNA and its role in autoimmunity. Trex1−/− mice are viable, however they exhibit reduced life span (2–4 months), and manifest inflammatory myocarditis (90). In the absence of Trex1, there is an accumulation of ~60 bp ssDNA believed to be produced during replication, which leads to the activation of DNA-damage associated signaling pathways. Further work by Stetson and colleagues revealed a role for Trex1 in preventing cell-intrinsic initiation of autoimmunity (91). Trex1 substrates are ssDNA, which are either the by products of replication and/or reverse transcribed from endogenous retroelements. As has been observed in DNaseII deficient mice, inflammatory myocarditis associated with Trex1-deficiency is rescued by crossing Trex-1 deficient animals to STING-deficient mice. Together, these studies highlight the important role of strict regulation of the DNA sensing pathway, and highlight the importance of DNA sensing in both protective and pathological immune responses.
Conclusions
In recent years there has been tremendous progress in understanding how cells recognize and respond to microbial threats via cytosolic DNA recognition. Studies from several groups have clearly indicated that multiple sensors exist in the cytosol to trigger inflammatory responses to DNA. These responses are central to antimicrobial immunity and therefore it is not surprising that multiple DNA sensors exist and operate in different cell-types. However, as with all areas of progress, many new questions arise and key aspects of DNA recognition remain to be better understood. The potential functional overlap and redundancy of key sensors and signaling intermediates as well as a better clarification of the relative importance of DDX41 versus STING in sensing cyclic di-nucleotides will likely be assisted following the generation and characterization of DDX41-deficient cells and mice. A better understanding of the molecular mechanisms of how cells generate inflammation in response to DNA could provide new targets that could be manipulated for the treatment of infectious as well as autoimmune disease.
Acknowledgements
We apologize to colleagues whose work could not be cited due to space limitations.
This work is supported by NIH grants (AI093752, AI067497 and AI083713) awarded to KAF.
Abbreviations
- PRR
Pattern recognition receptor
- TLR
Toll-like receptor
- NLR
NOD-like receptor
- RLR
RIG-I-like receptor
- MAMP
Microbe-associated molecular patterns
- TIR
Toll/Interleukin-1 receptor
- TRIF
TIR-domain-containing adaptor inducing IFN-β
- RIG-I
Retinoic acid inducible-I
- MDA5
Melanoma differentiation-associated protein 5
- MAVS
Mitochondrial antiviral signaling
- IRF
IFN regulatory factor
- ISD
Immunostimulatory DNA
- TBK1
Tank-binding kinase 1
- IFN
Interferon
- PYD
Pyrin domain
- CARD
Caspase-1 activation and recruitment domain
- AIM2
Absent in melanoma 2
- IFI16
Interferon inducible protein 16
- STING
Stimulator of type I IFN gene
- ISG
Interferon-stimulated genes
- PYHIN
Pyrin and HIN200 domain-containing
- ODN
Oligodeoxynucleotides
- LRRFIP1
Leucine-rich repeat in Flightless-I interacting protein 1
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Microbes and infection / Institut Pasteur. 2004;6:1382–1387. doi: 10.1016/j.micinf.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 4.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 6.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 7.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 8.Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunological reviews. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 11.Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:27–31. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- 12.Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Current opinion in immunology. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 14.Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. The Journal of experimental medicine. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature immunology. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 16.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Dinarello CA. The IL-1 family and inflammatory diseases. Clinical and experimental rheumatology. 2002;20:S1–S13. [PubMed] [Google Scholar]
- 18.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annual review of immunology. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 19.Gracie JA, Robertson SE, McInnes IB. Interleukin-18. Journal of leukocyte biology. 2003;73:213–224. doi: 10.1189/jlb.0602313. [DOI] [PubMed] [Google Scholar]
- 20.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 21.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 22.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nature immunology. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nature immunology. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 25.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS pathogens. 2009;5 doi: 10.1371/journal.ppat.1000480. e1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 27.Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. Adenovirus membrane penetration activates the NLRP3 inflammasome. Journal of virology. 2011;85:146–155. doi: 10.1128/JVI.01265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. The Journal of experimental medicine. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belhocine K, Monack DM. Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cell Microbiol. 2012;14:71–80. doi: 10.1111/j.1462-5822.2011.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. European journal of immunology. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren SE, Armstrong A, Hamilton MK, Mao DP, Leaf IA, Miao EA, Aderem A. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. Journal of immunology. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, Yamamoto M, Takeda K. Critical role of AIM2 in Mycobacterium tuberculosis infection. International immunology. 2012;24(10):637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- 38.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13:1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell host & microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 41.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Molecular and cellular biology. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. Journal of immunology. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, Dixit VM, Monack DM. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Almeida LA, Carvalho NB, Oliveira FS, Lacerda TL, Vasconcelos AC, Nogueira L, Bafica A, Silva AM, Oliveira SC. MyD88 and STING signaling pathways are required for IRF3-mediated IFN-beta induction in response to Brucella abortus infection. PloS one. 2011;6:e23135. doi: 10.1371/journal.pone.0023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koppe U, Hogner K, Doehn JM, Muller HC, Witzenrath M, Gutbier B, Bauer S, Pribyl T, Hammerschmidt S, Lohmeyer J, Suttorp N, Herold S, Opitz B. Streptococcus pneumoniae stimulates a STING- and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. Journal of immunology. 2012;188:811–817. doi: 10.4049/jimmunol.1004143. [DOI] [PubMed] [Google Scholar]
- 51.Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li XD, Knapp S, Kovarik P. Type I interferon production induced by Streptococcus pyogenes-derived nucleic acids is required for host protection. PLoS pathogens. 2011;7 doi: 10.1371/journal.ppat.1001345. e1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 53.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 56.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature immunology. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 59.Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS pathogens. 2009;5 doi: 10.1371/journal.ppat.1000665. e1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melchjorsen J, Rintahaka J, Soby S, Horan KA, Poltajainen A, Ostergaard L, Paludan SR, Matikainen S. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. Journal of virology. 2010;84:11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orzalli MH, Deluca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunological reviews. 2011;243:109–118. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 65.Conrady CD, Zheng M, Fitzgerald KA, Liu C, Carr DJ. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal immunology. 2012;5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annual review of immunology. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. Journal of immunology. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. Journal of immunology. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: a novel double stranded RNA binding protein which interacts with the leucine rich repeat of flightless I. Nucleic Acids Res. 1998;26:3460–3467. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nature immunology. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 74.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Molecular cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature structural & molecular biology. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 78.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Romling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Science signaling. 2008;1:pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 80.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2012 doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2012 doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature immunology. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ronnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasutomo K, Horiuchi T, Kagami S, Tsukamoto H, Hashimura C, Urushihara M, Kuroda Y. Mutation of DNASE1 in people with systemic lupus erythematosus. Nature genetics. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 85.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nature genetics. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 86.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nature immunology. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 87.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, Nagata S. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 88.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19386–19391. doi: 10.1073/pnas.1215006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3' -->5' exonuclease genes. The Journal of biological chemistry. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 90.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis. Molecular and cellular biology. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]