Abstract
Despite advances in our understanding of breast cancer, patients with metastatic disease have poor prognoses. GATA3 is a transcription factor that specifies and maintains mammary luminal epithelial cell fate, and its expression is lost in breast cancer, correlating with a worse prognosis in human patients. Here, we show that GATA3 promotes differentiation, suppresses metastasis and alters the tumour microenvironment in breast cancer by inducing microRNA-29b (miR-29b) expression. Accordingly, miR-29b is enriched in luminal breast cancers and loss of miR-29b, even in GATA3-expressing cells, increases metastasis and promotes a mesenchymal phenotype. Mechanistically, miR-29b inhibits metastasis by targeting a network of pro-metastatic regulators involved in angiogenesis, collagen remodelling and proteolysis, including VEGFA, ANGPTL4, PDGF, LOX and MMP9, and targeting ITGA6, ITGB1 and TGFB, thereby indirectly affecting differentiation and epithelial plasticity. The discovery that a GATA3-miR-29b axis regulates the tumour microenvironment and inhibits metastasis opens up possibilities for therapeutic intervention in breast cancer.
One of the classical hallmarks of cancer is the ability for tumour cells to invade and metastasize1. Metastasis is a multi-stage process that includes extracellular matrix remodelling, blood vessel recruitment, tumour cell entry and exit from circulation, and survival at a distant organ2. The tumour microenvironment is increasingly recognized as an important contributor to malignant progression and metastasis3,4. In addition to remodelling the microenvironment to facilitate metastasis, cancer cells also turn on embryonic morphogenesis regulators to undergo the epithelial-to-mesenchymal transition (EMT) and turn off differentiation programs5, allowing cancer cells to gain motility, modify cell adhesion and acquire stem-like properties6.
In contrast to EMT, differentiation is associated with less aggressive tumours and better prognosis. GATA3 is a transcription factor that specifies and maintains luminal epithelial cell differentiation in the mammary gland7–9. Loss of GATA3 is involved in breast cancer pathogenesis8–10 and a low GATA3 level is associated with poor prognosis11–13. Mutations in GATA3 that diminish or abolish its DNA-binding ability are commonly found in human breast cancers, and GATA3 was recently found to be one of three genes mutated in >10% of all breast cancers14,15. In several mouse models of breast cancer, Gata3 expression inversely correlates with tumour progression and metastasis: loss of GATA3 coincides with loss of differentiation, the transition from adenoma to carcinoma and the onset of tumour dissemination. Reintroduction of Gata3 into MMTV-PyMT carcinomas induces differentiation, suppresses dissemination10 and reduces tumour-initiating capacity16. However, how GATA3 induces tumour differentiation and inhibits dissemination, and what molecular and cellular events lie downstream of GATA3 are unknown.
MicroRNAs (miRNAs) are small, non-coding RNAs that modulate gene expression post-transcriptionally, either by inhibiting translation or by causing degradation through binding to the 3′ untranslated regions (UTRs) of target messenger RNAs (ref. 17). miRNAs are both positive and negative regulators of cancer metastasis18–20. Precedence for GATA-mediated miRNA regulation has recently been established: GATA1 promotes erythrocyte differentiation through miR-451, suggesting that GATA factors use miRNAs to make cell fate decisions21. In the mammary gland, miRNAs such as let-7 promote mammary differentiation and regulate self-renewal22,23. However, miRNAs downstream of GATA3 have yet to be investigated.
In this study, we predicted that GATA3 coordinates gene expression networks involved in metastasis through miRNA-mediated mechanisms. We investigated the molecular pathways by which GATA3 regulates differentiation and metastasis, and identified miR-29b, a miRNA downstream of GATA3 that modulates the tumour microenvironment, metastasis and epithelial plasticity.
RESULTS
GATA3 suppresses lung metastases from human and mouse breast cancer
Human breast cancers are classified into several subtypes that are prognostic of outcome24. Clinically, basal, triple-negative breast cancers are more aggressive and poorly differentiated25. In human breast cancer lines26, GATA3 is expressed at a higher level in luminal versus basal A and basal B subtypes (Fig. 1a), consistent with its luminal localization (Supplementary Fig. S1a).
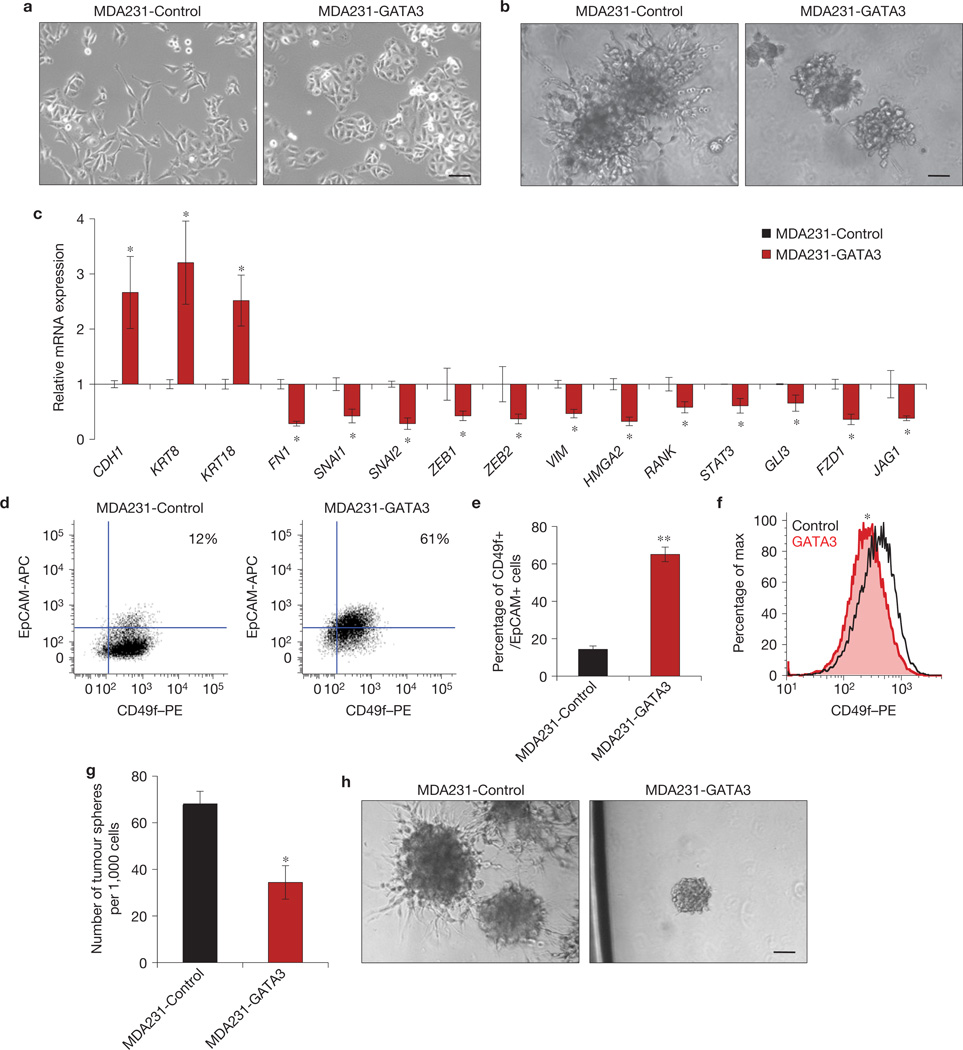
Figure 1.
GATA3 suppresses spontaneous and experimental breast cancer metastases to the lungs. (a) GATA3 expression levels from basal A, basal B and luminal breast cancer cell lines. Microarray data set is adapted from ref. 25. ** one-way analysis of variance P < 0.001. (b) Relative Gata3 levels in 4T1 cells ± Gata3 measured by qPCR (n = 8 independently obtained biological samples, **P < 0.001). (c–g) BALB/c mice were injected with 4T1 cells ± Gata3 into the inguinal mammary fat pad. Tumours were allowed to grow for three weeks and measured (c) and immunohistochemical staining for GATA3 in primary tumours was performed (d). The lungs were collected and examined for metastases (e). (n = 12 independent mice per group, *P < 0.01.) Representative H&E images (f) and immunohistochemical staining for GATA3 (g) in lung metastases are shown. (h,i) CD31 immunohistochemical mean intensity (h) and F4/80 immunohistochemical mean intensity (i) in primary 4T1 ± Gata3 tumours. (Values derived from n =8 independent tumours per group, and 10 random fields per tumour.) Representative images are shown below the graphs (*P < 0.05). (j) BALB/c mice were injected i.v. with 4T1 cells ± Gata3. Bioluminescence imaging was performed on day 14 post-injection and mice were euthanized immediately after imaging (n = 10 independent mice per group). (k) GATA3 immunohistochemical staining of 4T1 ± Gata3 experimental i.v. injected lung metastases. (l) BALB/c mice were co-injected i.v. 1:1 with control cells labelled with RFP (4T1-RFP) and Gata3-expressing cells labelled in GFP (4T1-Gata3–GFP). Mice were euthanized on day 12 post-injection and the percentages of RFP- and GFP-positive cells were determined by flow cytometry (n = 12 independent mice per group; **P < 0.002, paired t -test). Data are reported as mean±s.e.m. Scale bars, 200 µm (d,f,g,k) and 100 µm (h,i).
We overexpressed mouse Gata3 in basal 4T1 cells to evaluate metastasis in an immunocompetent model, and human GATA3 in basal MDA-MB-231 cells (referred to as MDA231), lines with low endogenous GATA3 (Fig. 1b and Supplementary Figs S1b,c and S2a,b). Orthotopic transplant of stably transduced 4T1-Gata3 cells into BALB/c mice gave rise to tumours similar in size to 4T1-Control cells, but showed a significant decrease in the number and size of spontaneous lung metastases (Fig. 1c–f). Interestingly, whereas primary 4T1-Gata3 tumours expressed GATA3, the lung metastases were mostly GATA3-low (Fig. 1g and Supplementary Fig. S1d), suggesting that metastatic cells lost GATA3 expression. We did not observe any difference in proliferation in vivo or in culture (Supplementary Fig. S1e,f), suggesting that GATA3 causes no intrinsic defects on viability, and that the difference in metastasis depends on in vivo microenvironment interactions. Indeed, 4T1-Gata3 tumours exhibited significant decreases in tumour vasculature and macrophage infiltrates (Fig. 1h,i). Consistent with this, we found a twofold reduction in serum VEGF-A levels in mice bearing orthotopic 4T1-Gata3 primary tumours (Supplementary Fig. S1g), suggesting that GATA3 regulates VEGF-A in vivo.
Human MDA231-GATA3 cells injected into nude mice formed smaller tumours than MDA231-Control cells, with decreased tumour vasculature and macrophage infiltrates and secreted less VEGF-A (Supplementary Fig. S2c–g). MDA231-GATA3 cells showed no difference in proliferation in culture or in viable tumour areas, but expressed higher levels of apoptotic markers and exhibited more necrosis in vivo (Supplementary Fig. S2h–k). These data support the concept that microenvironment interactions are critical for GATA3-mediated metastasis inhibition.
We next examined whether GATA3 only limits the ability of tumour cells to disseminate from the primary site, or if it also affects the later stages of metastasis, for example, colonization. Accordingly, we inoculated the cells directly into the circulation by intravenous (i.v.) injection (through the tail vein) to form experimental metastases. For both 4T1 and MDA231 cells, GATA3 decreased lung metastases (Fig. 1j and Supplementary Fig. S2l). We validated that these experimental metastases maintained GATA3 overexpression (Fig. 1k). Thus, sustained GATA3 expression also inhibits the late steps of metastasis.
To determine whether GATA3-expressing cells can compete with control cells, we labelled 4T1 cells with RFP (control) or Gata3–GFP, mixed them in a 1:1 ratio, and co-injected them i.v. After 2 weeks, we found that 4T1-Gata3–GFP cells accounted for only ~20% of the total metastatic cells in the lung, whereas 4T1-RFP cells accounted for ~80% (Fig. 1l). These results indicate that GATA3 confers a competitive disadvantage in vivo.
GATA3 promotes differentiation and limits cell migration
As GATA3 regulates luminal fate, we sought to determine whether GATA3-mediated metastasis suppression involves cell differentiation. In two-dimensional cell culture, MDA231-Control cells had a more spindle-shaped, mesenchymal morphology, whereas MDA231-GATA3 cells exhibited a more epithelial phenotype (Fig. 2a). In a wound-scratch assay, MDA231-Control cells migrated as single cells to close the wound, whereas MDA231-GATA3 cells migrated as a collective sheet (Supplementary Fig. S3a). In three-dimensional (3D) Matrigel culture, which better mimics physiological conditions27,28, GATA3-overexpressing MDA231 and Hs578T human basal cells were less invasive than their controls (Fig. 2b and Supplementary Fig. S3b–d and Videos S1 and S2).
Figure 2.
GATA3 induces a more luminal phenotype and suppresses cell migration. (a) Phase-contrast images of MDA231 cells ± GATA3 in 2D culture. (b) Phase-contrast images of MDA231 cells ± GATA3 embedded in 3D Matrigel. (Also see Supplementary Videos S1 and S2.) (c) Relative expression of epithelial markers (CDH1, KRT8, KRT18), mesenchymal markers (FN1, SNAI1, SNAI2, ZEB1, ZEB2, VIM, HMGA2, RANK) and major inflammatory and stemness-associated pathway genes including NF-ΚB (STAT3, RANK), Wnt (FZD1) Hedgehog (Gli3) and Notch (JAG1). (n =8 independently obtained biological samples, *P < 0.05 for all genes.) (d–f) Flow cytometry analysis of cell-surface markers EpCAM and CD49f in MDA231 cells ± GATA3 (d) and quantification (e) of n = 8 independent experiments, *P < 0.001. A representative histogram of CD49f expression is shown in (f). (*P < 0.01 by probability binning χ2 test.) (g,h) Number of tumour spheres from single MDA231 cells ± GATA3 (*P < 0.02; g), and representative phase-contrast images (h) of n = 3 independent experiments performed in triplicate. Data are reported as mean±s.e.m. All scale bars, 100 µm.
Using quantitative PCR (qPCR), we found that GATA3 increased the level of expression of luminal genes and decreased the level of expression of basal, EMT, stemness and inflammatory genes (Fig. 2c and Supplementary Fig. S3e,f). Despite these gene expression changes, 4T1-Gata3 cells were morphologically similar to 4T1-Control cells (Supplementary Fig. S3g). GATA3 also induced basal cells to adopt a luminal cell surface phenotype characterized by decreased CD49f (integrin α6) and increased EpCAM expression29 (Fig. 2d–f). Furthermore, MDA231-GATA3 and 4T1-Gata3 cells formed fewer, smaller and less invasive tumour spheres from single cells (Fig. 2g,h and Supplementary Fig. S3h). Taken together, these results indicate that GATA3 promotes a luminal differentiation program and opposes the basal/EMT state.
GATA3 promotes miR-29b expression
In addition to the cell-intrinsic effects of GATA3 on differentiation, we were intrigued by the observation that GATA3-expressing tumours had reduced serum VEGF-A and tumour vasculature. However, the VEGFA promoter lacked clear GATA3-binding sites, and GATA3 did not decrease the activity of a luciferase reporter containing the VEGFA promoter (Supplementary Fig. S3i). We therefore reasoned that GATA3 regulates VEGFA and perhaps other pro-metastatic genes indirectly through miRNAs and conducted a screen in MDA231 cells ± GATA3 using qPCR miRNA arrays. Of 88 miRNAs evaluated, miR-29b was the most upregulated miRNA in MDA231-GATA3 cells (Fig. 3a,b and Supplementary Table S1), and the expression level of miR-29b was also increased in Hs578T-GATA3 cells (Supplementary Fig. S4a). The miR-29 family consists of three members with the same seed sequence: miR-29a, miR-29b and miR-29c (Supplementary Fig. S4b). We also found that miR-29a and miR-29c were increased by GATA3 (Supplementary Fig. S4c), with miR-29a being ~400-fold more abundant than miR-29c at basal levels.
Figure 3.
miR-29b is induced by GATA3, enriched in luminal, good prognostic breast cancers, and associated with reduced metastatic potential. (a,b) Eighty-eight miRNAs were screened using qPCR miRNA arrays in MDA231 cells ± GATA3 (a) and miR-29b expression was further validated by TaqMan qPCR (b). (n = 8 independently obtained biological samples, *P < 0.05.) (c) MDA231 cells were co-transfected with an miR-29a/b1–Luc reporter and pcDNA-EGFP (control) or increasing amounts of pcDNA-GATA3. Firefly luciferase was normalized to Renilla luciferase and plotted relative to the control. (n = 5 independent experiments performed in triplicate, *P < 0.05.) (d) MDA231 cells were co-transfected with pcDNA-eGFP or pcDNA- GATA3 and the miR-29a/b1–Luc reporters containing GATA site deletions. The non-mutated (WT) reporter was used as the control. Firefly luciferase was normalized to Renilla luciferase and plotted relative to the control. (n =3 independent experiments performed in triplicate, *P < 0.05.) (e,f) Relative TGF-β–Luc reporter activity (e) and NFκB–Luc reporter activity (f) in MDA231 cells ± GATA3, with and without TGF-β1, sRANKL or TNF-α stimulation. (n =5 independent experiments performed in triplicate, *P < 0.05.) (g,h) Relative miR-29a, miR-29b and miR-29c expression in primary human basal-like, HER2, luminal A and luminal B breast cancers (g), and in primary human oestrogen-receptor (ER)-positive and negative tumours (h). Microarray data set is adapted from ref. 32. (n = 93 primary breast tumour samples, * Kruskal–Wallis test statistic, P < 0.05.) (i) Relative miR-29a, miR-29b and miR-29c expression in primary mouse basal and luminal breast cancers. Microarray data set is adapted from ref. 36. (n = 41 primary tumours from individual mice and 5 normal mammary glands, ***P < 0.001.) (j) Relative miR-29b expression in 67NR, 168FARN, 4TO7 and 4T1 cells, with metastatic capability shown below. Microarray data set adapted from ref. 37. (k) Relative Gata3 and miR-29b expression in primary normal mammary epithelial cells, adenoma and carcinoma cells from MMTV-PyMT mice, with metastatic capability shown below. (n = 6 independently obtained biological samples per group, *P < 0.05.) Data are reported as mean±s.e.m.
The miR29a/b1 promoter, which was previously identified30, contains three GATA3-binding sites (Supplementary Fig. S4d). GATA3 increased the activity of the miR29a/b1-promoter reporter (Fig. 3c), and deleting the GATA sites diminished GATA3-mediated reporter induction, demonstrating that these sites are necessary and functional (Fig. 3d).
As previous studies showed that miR-29b is negatively regulated by TGF-β and NF-κB (refs 30–32) we also examined whether GATA3 inhibits the TGF-β and NF-κB pathways, thereby regulating miR-29b indirectly. Indeed, GATA3 inhibited TGF-β and NF-κB reporter activities and suppressed TGF-β-induced EMT and components of the TGF-β pathway transcriptionally (Fig. 3e,f and Supplementary Fig. S4e–g). Stimulation with recombinant TGF-β, TNF-Α or sRANKL decreased miR-29a and miR-29b levels in control, but not GATA3, cells (Supplementary Fig. S4h,i). Thus, GATA3 induces miR-29b expression directly (by binding the GATA sites on the promoter) and indirectly (by inhibiting the TGF-β and NF-ΚB pathways).
miR-29 expression is enriched in more differentiated breast cancer and normal luminal epithelial cells
We examined whether miR-29 correlates with more differentiated breast cancers, which are associated with better patient outcomes12. In an miRNA data set of 99 primary human breast tumours33, the levels of all three miR29 members were higher in luminal and oestrogen-receptor-positive (ER+) tumours (Fig. 3g,h). Moreover, miR-29c was associated with more favourable prognoses in a meta-analysis of over 1,000 breast cancers34 and with luminal differentiation in an independent data set of over 100 human breast tumours35.
We then used a miRNA data set of mouse breast tumours36 and found that miR-29 members were expressed at higher levels in luminal when compared with basal models (Fig. 3i). In normal epithelial cells isolated from the mouse mammary gland, all miR-29 members were expressed at higher levels in the luminal fraction (Supplementary Fig. S5a–c).
We next examined whether miR-29b is inversely related to metastatic ability. Using a data set of syngeneic cell lines with varying metastatic capabilities37,38, we found that expression of miR-29b was lowest in cells with the highest metastatic capacities (Fig. 3j and Supplementary Fig. S5d). In the MMTV-PyMT model, which mimics progressive stages of human luminal breast cancer39, adenoma and carcinoma cells had decreased Gata3 and miR-29b expression levels compared with normal epithelium (Fig. 3k), with the lowest levels at the carcinoma stage coinciding with metastasis. Together, these results demonstrate that miR-29 expression correlates with more favourable outcomes, more differentiated phenotypes in normal and cancer cells and reduced metastatic potential.
miR-29b promotes and maintains luminal differentiation
We next examined whether miR-29b promotes luminal characteristics. MMTV-PyMT tumours become less differentiated during tumour progression39. Stable overexpression of miR-29b in a primary cell line derived from a late-stage MMTV-PyMT carcinoma resulted in a more epithelial phenotype, higher luminal marker and lower basal marker expression levels (Fig. 4a,b). In addition, miR-29b promoted branching of PyMT cell aggregates in 3D Matrigel (Fig. 4c), a feature of normal branching morphogenesis40.
Figure 4.
miR-29b promotes luminal characteristics and loss of miR-29b induces a de-differentiated, mesenchymal phenotype. (a) Phase-contrast images of PyMT cells ± miR-29b in 2D culture. (b) Relative expression of luminal (Krt8, Esr1 and Gata3) and basal (Krt14) epithelial markers in PyMT cells ± miR-29b by qPCR (n =6 independently obtained biological samples, *P < 0.05 for all genes.) (c) Phase-contrast images of PyMT ± miR-29b aggregates embedded in 3D Matrigel. (d) Phase-contrast images of MDA231 cells ± Zip29 knockdown. (e) Flow cytometry analysis of CD49f (integrin α6) and CD29 (integrin β1) cell-surface expression in MDA231 cells ± Zip29. Representative histograms of n = 5 independent experiments (*P < 0.01 by probability binning χ2 test). (f) Phase-contrast images of HMLE cells ± Zip29. (g) Representative flow cytometry analysis of cell-surface CD24 and CD44 expression of HMLE cells ± Zip29. (n = 5 independent experiments.) (h) Relative expression of epithelial and mesenchymal markers in HMLE cells ± Zip29 by qPCR. (n =6 independently obtained biological samples, *P <0.05.) (i) Western blot analysis of phospho-Smad3, vimentin and actin in HMLE cells ± Zip29. (j) Flow cytometry analysis of the CD44hi/CD24low population in HMLE cells ± Zip29 and ± LY-364947. (n =4 independent experiments, *P < 0.05.) Data are reported as mean±s.e.m. All scale bars, 100 µm. Uncropped images of blots are shown in Supplementary Fig. S9a.
We then examined whether loss of miR-29b induces a de-differentiated, mesenchymal phenotype. We generated miR-29b-knockdown cells using miR-Zip29b lentiviruses, which stably knockdown endogenous miRNAs. Although miR-29b levels decreased as expected, we also observed decreases in miR-29a and miR-29c levels, probably owing to their similar sequences (Supplementary Fig. S6a,d). MDA231-Zip29 cells were more elongated, protrusive and spindle-like, expressed lower GATA3 and had increased levels of CD49f and CD29 (Fig. 4d,e and Supplementary Fig. S6b), two markers of the basal/stem cell population that have miR-29b-binding sites. Loss of miR-29b in mouse 4TO7 cells also caused a spindle-like morphology and increased mesenchymal marker levels (Supplementary Fig. S6c,d).
We extended our findings to normal human mammary cells (HMLE), which exhibit phenotypic plasticity in vitro6. HMLE-Zip29 cells were more elongated and spindle-like and had increased mesenchymal and decreased epithelial marker expression levels. The EMT program has previously been linked to stem-like traits6. Loss of miR-29b increased the CD44hi/CD24low stem-cell-enriched compartment and increased the level of stem cell marker expression (Fig. 4f–i and Supplementary Fig. S6e,f). Loss of miR-29 also increased basal/stem cell markers in PyMT and 4T1 cells (Supplementary Fig. S6g–j).
HMLE-Zip29 cells had higher basal levels of phospho-Smad3 (Fig. 4i), suggesting that miR-29b knockdown increases unchecked TGF-β signalling, which promotes EMT. Consistent with this, inhibiting TGF-β signalling using LY-364947 in HMLE-Zip29 cells reversed the increased CD44hi/CD24low population (Fig. 4j). Taken together, these results indicate that miR-29b promotes differentiation, and loss of miR-29b causes de-differentiation and increases the level of mesenchymal marker expression characteristic of a progenitor-like state, which is mediated, at least in part, by an increased level of TGF-β signalling.
miR-29b downregulates pro-metastatic genes that modify the tumour microenvironment and indirectly regulate epithelial plasticity
We used prediction algorithms to generate a list of candidate miR-29b targets41–43. Although transcription factors associated with EMT (for example, SNAI1, TWIST or ZEB1) did not have miR-29b-binding sites in their 3′ UTRs, TGFB, a potent inducer of EMT and de-differentiation contained miR-29b-binding sites (Fig. 5a). In addition, ITGA6 (CD49f) and ITGB1 (CD29), which maintain and enhance stemness44 and are commonly used markers to identify the basal population, contained miR-29b-binding sites in their 3′ UTRs. Interestingly, we identified miR-29b sites in many microenvironmental genes involved in angiogenesis, collagen remodelling and matrix degradation, including ANGPTL4, LOX, MMP2, MMP9, PDGF and VEGF (Fig. 5a), which have been implicated in promoting metastasis4,45. To determine whether miR-29b modulates these genes directly, we cloned these 3′ UTRs into luciferase reporters and co-transfected them with either miR-29b or control mimic. For all 3′ UTRs tested, miR-29b decreased luciferase activity by 40–80% (Fig. 5b), which was partially relieved when the miR-29b sites were mutated (Fig. 5c), indicating that these sites are functional and specific for miR-29b.
Figure 5.
miR-29b targets pro-metastatic genes involved in remodelling the tumour microenvironment and tumour differentiation. (a) Computationally predicted interactions between miR-29b and the 3′ UTRs of several mRNAs involved in differentiation, EMT, angiogenesis, ECM crosslinking and ECM proteolysis. The miR-29b seed sequence is in red and the complementary binding sites are in green. The mutations generated within the 3′ UTRs for c are in purple. (b,c) The wild-type (b) and mutant (c) 3′ UTRs of the indicated miR-29b targets were cloned into dual luciferase reporters and co-transfected with miR-29b or cel-67 control mimic. Renilla luciferase activity was measured 48 h post-transfection and normalized to firefly luciferase (n =5 independent experiments performed in triplicate, *P < 0.05). (d,e) Relative miR-29b expression (d) and mRNA expression of indicated miR-29b targets (e) in stably transduced 4T1 cells ± miR-29b (n =8 independently obtained biological samples, *P < 0.05.) (f,g) Western blot (f) and flow cytometer analysis (g) of 4T1 cells ± miR-29b for ANGPTL4, LOX, VEGF-A and ITGA6 (CD49f). (*P < 0.01 by probability binning χ2 test). Data are reported as mean±s.e.m. Uncropped images of blots are shown in Supplementary Fig. S9b.
We stably transduced 4T1 and MDA231 cells with miR-29b and confirmed its overexpression (Fig. 5d). In both 4T1 and MDA231 cells, miR-29b repressed endogenous mRNA levels of target genes (Fig. 5e and Supplementary Fig. S7a,b). We validated a subset of targets at the protein level by western blot and cell surface staining, and found that miR-29b decreased the levels of ANGPTL4, ITGA6 (CD49f), LOX and VEGF-A (Fig. 5f,g). These results suggest that miR-29b downregulates a cohort of pro-metastatic genes involved in differentiation and modifying the tumour microenvironment.
miR-29b expression suppresses lung metastasis
To determine whether miR-29b suppresses metastasis, we injected mice with 4T1-Control or 4T1-miR-29b cells orthotopically. Although there was no difference in primary tumour size, 4T1-miR-29b tumours had significantly fewer blood vessels and a decreased level of fibrillar collagen (Fig. 6a–c). Importantly, mice with 4T1-miR-29b tumours had fewer and smaller metastases (Fig. 6d). miR-29b also significantly reduced the size and number of experimental lung metastases in both 4T1 and MDA231 cells (Fig. 6e and Supplementary Fig. S7c–e).
Figure 6.
miR-29b inhibits lung metastasis and loss of miR-29b increases lung metastasis. (a) BALB/c mice were injected with 4T1 cells ± miR-29b into the mammary fat pad to form primary orthotopic tumours. Tumours were allowed to grow for three weeks and measured (n =8 independent mice per group). (b,c) Representative images of 4T1 ± miR-29b primary orthotopic tumours stained for CD31 to evaluate tumour vasculature (b) or picrosirius red to evaluate fibrillar collagen (c). (d) Representative H&E images of lung metastases from mice injected with primary 4T1 ± miR-29b tumours. (e) BALB/c mice were injected i.v. with 4T1 cells ± miR-29b and bioluminescence imaging was performed on day 14 post-injection. Mice were euthanized immediately after imaging (n = 10 independent mice per group.) Representative H&E images of the lung metastases are shown below. (f) Relative expression of miR-29b targets in primary normal mammary epithelial cells and primary MMTV-PyMT adenoma and carcinoma cells measured by qPCR (n = 6 independently obtained biological samples, *P < 0.05). (g,h) FVB/n mice were injected i.v. with PyMT cells±miR-29b, and euthanized at six weeks. A representative set of gross lungs (g) and H&E images of the lung metastases (h) are shown (n=8 independent mice per group). (i,j) FVB/n mice were injected i.v. with PyMT cells ± Zip29 to knockdown endogenous miR-29b, and bioluminescent imaging was performed on week three (i). The graph depicts the number of surface lung metastases per lung lobe (j; n =8 independent mice per group, *P < 0.05). Data are reported as mean±s.e.m. Scale bars, 100 µm (b–d), 200 µm (e), 1 cm (g) and 1mm (h).
As miR-29b levels decrease during tumour progression in MMTV-PyMT mice, we examined whether any miR-29b targets increase in parallel. In carcinomas, where miR-29b levels were lowest, the expression level of many miR-29b targets increased (Fig. 6f). Moreover, PyMT-miR-29b cells showed decreased target expression levels (Supplementary Fig. S7f–h) and significantly reduced lung metastasis (Fig. 6g,h). Conversely, loss of miR-29b in PyMT, 4T1 and MDA231 cells increased spontaneous and experimental lung metastases and EMT markers in vivo (Fig. 6i,j and Supplementary Fig. S8a–f). Taken together, these results show that miR-29b functions as a metastasis suppressor in human and mouse models of breast cancer.
miR-29b suppresses metastasis by repressing microenvironmental targets
To gain further insight into how miR-29b suppresses metastasis, we examined whether the levels of miR-29b targets increase after miR-29 knockdown. Many miR-29b targets we identified were upregulated after miR-29 loss, including ANGPTL4, LOX, MMP9, VEGFA, ITGA6 and ITGB1 (Figs 7a and 4e). Moreover, miR-29 knockdown increased the level of 3′ UTR luciferase reporters of miR-29b targets (Fig. 7b), further validating these as bona fide targets.
Figure 7.
miR-29b knockdown increases the level of expression of its target genes and miR-29b suppresses metastasis by repressing four microenvironmental targets. (a) Relative expression of miR-29b targets in Zip29-knockdown cells by qPCR (n = 5 independently obtained biological samples, *P < 0.05). (b) The 3′UTR reporters of the indicated miR-29b targets were co-transfected with anti-miR-29b or control inhibitor into HEK293T cells. (n =3 independent experiments performed in triplicate, *P <0.05.) (c,d) BALB/c mice were injected i.v. with 4T1 cells ± miR-29b re-expressing ANGPTL4, LOX, MMP9 or VEGF-A, and euthanized at two weeks. Representative fluorescence micrographs of lungs were taken on a dissection microscope (c) and GFP fluorescence intensity was quantified (d). (n =8 independent mice per group, *P < 0.01.) Data are reported as mean±s.e.m. Scale bar, 1 mm.
We examined whether restoring the expression of four miR-29b targets, with known roles in angiogenesis and ECM remodelling, would increase metastasis. miR-29b alone inhibited metastasis, and re-expression of ANGPTL4, LOX, MMP9 or VEGF-A in 4T1-miR-29b cells restored lung metastasis (Fig. 7c,d), indicating that miR-29b suppresses metastasis, at least in part, by regulating these microenvironmental genes.
Regulation of miR-29b mediates the ability of GATA3 to suppress metastasis and promote differentiation
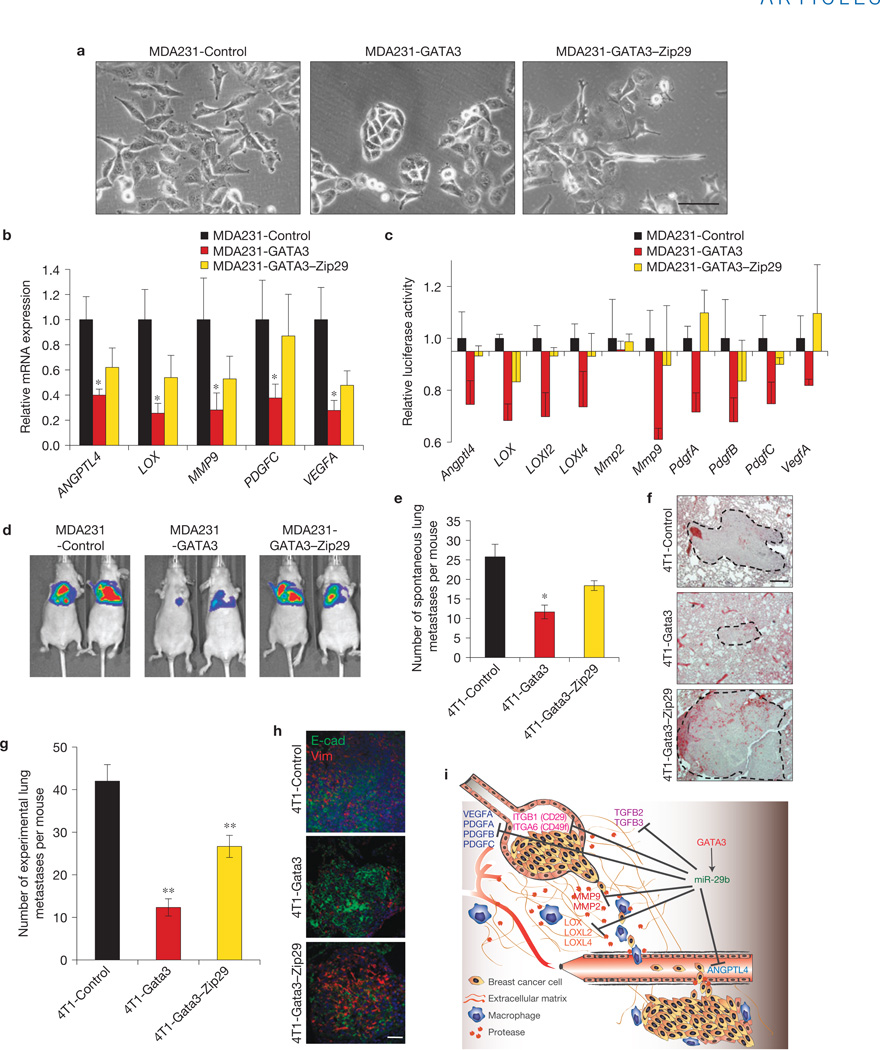
To determine whether GATA3-mediated metastasis suppression and differentiation requires miR-29b, we knocked down miR-29b in MDA231-GATA3 cells. GATA3 overexpression promoted epithelial clustering, and concomitant loss of miR-29b abrogated the effects of GATA3, resulting in elongated, spindle-like cells (Fig. 8a). Similarly, loss of miR-29 in T47D luminal breast cancer cells, which highly express endogenous GATA3, caused a mesenchymal morphology accompanied by an increase in the levels of EMT markers and target expression (Supplementary Fig. S8g–i). Thus, tumour cells acquire mesenchymal characteristics in the absence of miR-29b, even with high levels of GATA3 expression.
Figure 8.
miR-29b is an important downstream target of GATA3 that mediates its ability to promote luminal differentiation and suppress metastasis. (a) Phase-contrast images of MDA231 cells ± GATA3 ± Zip29. (b) Relative expression of miR-29b targets ANGPTL4, LOX, MMP9, PDGFC and VEGFA in MDA231 cells ± GATA3 ± Zip29 by qPCR (n =6 independently obtained biological samples, *P <0.05). (c) The 3′UTR reporters of the indicated miR-29b targets were transfected into MDA231 cells ± GATA3 ± Zip29 and the luciferase activity was measured. (n =3 independent experiments performed in triplicate.) (d) Bioluminescent imaging of mice injected i.v. with MDA231 cells ± GATA3 ± Zip29at four weeks post-injection (n =8 independent mice per group). (e,f) BALB/c mice were injected with 4T1 cells ± Gata3 ± Zip29 into the mammary fat pad. Tumours were allowed to grow for four weeks and lungs were examined for spontaneous metastases (e). Representative H&E images are shown (f). (n =10 independent mice per group, *P < 0.05.) (g,h) BALB/c mice were injected i.v. with 4T1 cells ± Gata3 ± Zip29 knockdown and euthanized two weeks post injection. The number of lung surface metastases was quantified (g) and immunofluorescence staining was performed for E-cadherin (green) and vimentin (red; h; n =8 independent mice per group, **P <0.01). (i) Proposed model of how GATA3 promotes differentiation and suppresses breast cancer metastasis through regulation of miR-29b. Data are reported as mean±s.e.m. Scale bars, 100 µm (a), 200 µm (f) and 50 µm (h).
We examined whether GATA3 downregulated miR-29b targets, and found that the levels of expression and 3′ UTR reporters of miR-29b targets, including ANGPTL4, LOX, MMP9 and VEGFA, were decreased by GATA3; this inhibition was partially reversed in GATA3-Zip29 cells (Fig. 8b,c). Significantly, we found that miR-29 knockdown reversed GATA3-mediated suppression of experimental and spontaneous metastases and increased the vimentin expression level (Fig. 8d–h and Supplementary Fig. S8j), indicating that perturbation of miR-29b attenuates the anti-metastatic and pro-differentiation functions of GATA3. Together, our results demonstrate that miR-29b is an important node downstream of GATA3 that controls differentiation and the expression of pro-metastatic genes involved in modifying the tumour microenvironment, ultimately leading to metastasis suppression (Fig. 8i).
DISCUSSION
In this study, we show that GATA3 promotes luminal differentiation and suppresses lung metastasis through miR-29b, an anti-metastatic microRNA that promotes differentiation and regulates the tumour microenvironment (Fig. 8g). We demonstrate that GATA3 increases the level of expression of miR-29b, which is exquisitely positioned to inhibit several steps required for metastasis: a network of mRNAs involved in stemness/EMT, angiogenesis, proteolysis, ECM signalling and ECM remodelling have miR-29b-binding sites and are bona fide miR-29b targets. Although we did not find miR-29b sites in classical EMT-promoting transcription factors, ITGA6 (CD49f) and ITGB1 (CD29), markers of the basal/stem cell population previously implicated in maintaining and enhancing stemness44, and TGFB2 and TGFB3, all had miR-29b-binding sites. As miRNAs act pleiotropically and modulate a range of biological processes, they represent an ideal set of targets through which transcription factors such as GATA3 can operate.
The loss of GATA3 triggers fibroblastic transformation and cell invasion46, and coincides with the onset of angiogenesis, inflammatory cell recruitment, and dissemination10. It has been shown that GATA3 reduces tumour vasculature and macrophage infiltrates, components of the tumour microenvironment that promote metastasis16,47. In human breast cancer, GATA3 correlates with higher E-cadherin and ER expression levels13. These observations suggest that promoting differentiation in primary tumours limits metastasis by both non-cell-autonomous mechanisms (for example, angiogenesis, inflammation, ECM remodelling) and cell-autonomous mechanisms (for example, increased cell adhesion).
In breast cancer, the level of miR-29b expression is highest in good prognostic, well-differentiated, luminal-type cancers and inhibits metastasis, the main cause of cancer-related deaths. miR-29b promoted luminal differentiation; conversely, loss of miR-29b promoted mesenchymal traits and metastasis. Concomitant loss of miR-29b in GATA3-expressing cells restored metastasis and abrogated the ability of GATA3 to promote differentiation, indicating that miR-29b is an important node that exerts its anti-metastatic effects by cell-intrinsic and cell-extrinsic mechanisms. We found that miR-29b-mediated metastasis inhibition depended on repression of its targets. Of these, ANGPTL4 affects lung seeding by disrupting endothelial cell junctions45; LOX increases tissue fibrosis and integrinmediated survival signalling48; MMP9 remodels the ECM and releases sequestered VEGF-A (ref. 49); and VEGF-A promotes angiogenesis. Interestingly, dysregulation of the miR-29 family has been implicated in tissue fibrosis50–52 and many cancers, including leukaemia, lung cancer, liver cancer, rhabdomyosarcoma and melanoma32,53–55.
TGF-β signalling activates the promoters of many of the same genes that are miR-29b targets, including LOX, MMPs and VEGFA (refs 56–58), and is a potent inducer of EMT. We found that GATA3 inhibited TGF-β and that TGFB2 and TGFB3 are miR-29b targets. Therefore, to repress TGF-β-induced activation of its target genes, GATA3 inhibits the signalling network directly at the transcriptional level and indirectly at the post-transcriptional level through miR-29b (that is, degradation and translational inhibition of already-made TGFB mRNAs). This multi-tiered system ensures efficient GATA3-mediated inhibition of TGF-β signalling. Interestingly, miR-29b also increased GATA3, and loss of miR-29b decreased GATA3, suggesting that GATA3 and miR-29b form a positive feedback loop and act collaboratively to reinforce fate decisions. The exact mechanisms of how GATA3 and miR-29b function in cell-type-specific manners remain to be further investigated.
Our work on miR-29b adds to the growing body of evidence implicating miRNA-mediated regulation of cancer and the tumour microenvironment. Previous studies have shown that many microRNAs play important pro- and anti-metastatic roles by regulating diverse cellular processes used during metastasis20,59,60. Future work aimed at identifying other GATA3 targets and determining their function will allow us to further understand the role of GATA3 in development and cancer.
Rather than functioning as a classical tumour suppressor, GATA3 defines a distinct class of pro-differentiation factors capable of also modifying the tumour microenvironment. The identification of one GATA3 target, miR-29b, illustrates how epithelial plasticity, the tumour microenvironment and metastasis are linked. Finally, miRNAs are being pursued as potential anti-cancer agents61,62. Whether increasing miR-29b levels in primary breast tumours improves patient survival will ultimately determine the therapeutic use of miR-29b mimics.
METHODS
Animal studies
All animal experiments were performed at UCSF, and reviewed and approved by the UCSF IACUC. Mice were housed under pathogen-free conditions in the UCSF barrier facility. BALB/c and nude mice were purchased from Simonsen Laboratories. FVB/n mice, originally from Jackson Laboratories, were bred in-house. For experimental metastasis experiments, age-matched female mice were injected i.v. (through the tail vein) with 1 × 105 cells (4T1), 5 × 105 cells (MDA-MB-231) or 5–10 × 105 cells (PyMT) in PBS. For primary tumours and spontaneous metastasis assays, age-matched female mice were injected with the indicated number of cells in 1:1 DMEM/Matrigel (BD Biosciences) into the fourth mammary fat pad without clearing. Tumour measurements were made blinded, using a caliper at least once per week, and volumes were calculated using the formula: V=0.52 × (length)2 × width. Bioluminescence imaging was performed using an IVIS Spectrum and image radiance values were normalized using Living Image (Caliper LifeScience).
Cell culture
MDA-MB-231, T47D, Hs578T, HEK293T, GP2, 4T1 and 4TO7 cells were obtained from the ATCC, LBNL or UCSF Cell Culture Facility, and grown in standard conditions (DMEM with 10% FBS). The immortalized human mammary epithelial cells (HMLE) were obtained from J. Debnath (UCSF, San Francisco, CA) and W. Hahn (Harvard University, USA) and maintained in MEGM (Lonza) as previously described63. The PyMT cell line was generated by isolating a late-stage MMTV-PyMT/FVB tumour, dissociating the cells in collagenase, and culturing in DMEM/F12 media supplemented with 5% FBS, insulin and hydrocortisone. For TGF-β stimulation, cells were serum-starved for 18–24 h before adding TGF-β1 or TGF-β2 (R&D Systems) at 5 ng ml−1. For TGF-βR inhibition, cells were grown in standard conditions with 10 µm LY-364947 (Calbiochem) or dimethylsulphoxide (Sigma). For cells embedded in Matrigel, cells were aggregated overnight on ultralow attachment plates (Corning) and then embedded into growth-factor-reduced Matrigel (BD Biosciences) the next day. Cells were grown in serum-free media supplemented with insulin–transferrin (Invitrogen) and 50 ng ml−1 of EGF (Invitrogen) or 2.5nM FGF2 (Sigma).
Lentiviral and retroviral production
Viral production was carried out using calcium-phosphate-mediated transfection of HEK293T or GP2 cells. Virus was concentrated by ultracentrifugation, and added to cells with Polybrene. Stably transduced cells were selected in puromycin, G418 or hygromycin for at least 5 days or selected by FACS.
Plasmids
Several plasmids were obtained from Addgene including: pcDNA-miR-29b from J. Mendell (Johns Hopkins University School of Medicine, Baltimore, MD; plasmid 21121; ref. 64), pcDNA-GATA3 from G. Hotamisligil (Harvard School of Public Health, Boston, MA; plasmid 1332; ref. 65), p3TP-Lux from J. Massagué (Memorial Sloan-Kettering Cancer Center, New York, NY; plasmid 11767; ref. 66), SBE4-Luc from B. Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD; plasmid 16495; ref. 67), pWZL Blast VEGF-A from R. Weinberg (MIT, Cambridge, MA; plasmid 10909; ref. 68) and pBabe Angptl4 from J. Massagué (Memorial Sloan-Kettering Cancer Center, New York, NY; plasmid 19156; ref. 45). Several plasmids were gifts, including: pBM-IRES–puro and pBM-Mmp9 from E. Raines69 (University of Washington, Seattle, WA) and G. Bergers (UCSF, San Francisco, CA), pMSCV-miR-29b from A. Goga (UCSF, San Francisco, CA), pLV-LOX from V. Weaver (UCSF, San Francisco, CA), pGL3-V2774 (containing the VEGFA promoter) from K. Xie (MD Anderson, Houston, TX) and pGL3-miR29–Luc (containing the miR29a/b1 promoter) from J. Mott30 (University of Nebraska, Omaha, NE). The pMSCV-Luciferase retrovirus was generated using XhoI and EcoRI from pGL3 (Promega). The pmiRZip-29b (System Biosciences) to stably knockdown miR-29b expression was used following the manufacturer’s instructions and contained the following shRNA sequence: 5′-GTAGCACCGTTTGAAATCAATGCTCTTCCTGTCAGAACACTGATTTCAAATGGTGCTATTTTT-3′ .
For the 3′ UTR luciferase reporters, the 3′ UTRs were PCR amplified from genomic DNA, cloned into pCR2.1 by TOPO cloning (Invitrogen) and verified by sequencing. Fragments were then digested with XhoI, SgfI and/or NotI and cloned into the psiCheck2 reporter (Promega). The psicheck2-ITGB1 3′ UTR plasmid was a gift from P. Gonzalez44 (Duke University, Durham, NC). Site-directed mutagenesis to mutate the miR-29b seed in the 3′ UTR reporters or to delete the GATA sites in the pGL3-miR29–Luc reporter was performed according to the manufacturer’s instructions (Stratagene). All mutants were verified by sequencing. Primer sequences used to generate the wild-type and mutant 3′ UTRs and the GATA mutants are detailed in Supplementary Tables S3–S5.
miRNA PCR arrays and qPCR
Total RNA was isolated from cells using the miRNeasy Mini Kit (Qiagen). For miRNA PCR arrays (SABiosciences), MDA231 cells ± GATA3were screened according to the manufacturer’s instructions. Complementary DNA was synthesized using the RT2 miRNA First Strand Kit (SABiosciences). Data were analysed using SABiosciences software. For qPCR, cDNA was synthesized using the Superscript III RT First Strand Kit (Invitrogen). qPCR was performed using FastStart Universal SYBR Green master mix (Roche) in an Eppendorf Mastercycler realplex machine. Ct values were normalized to actin and GAPDH, and relative expression was calculated using the 2ΔΔCt method. For quantification of miRNA expression, TaqMan probes were used according to the manufacturer’s protocol (Applied Biosystems). Ct values were normalized to RNU48 (human samples) or snoRNA202 (mouse samples). Primer sequences for qPCR were found using the Harvard Primer Bank and are detailed in Supplementary Table S2.
Serum cytokine analysis and ELISA
Sera from mice bearing 4T1 primary tumours were collected when the mice were euthanized. Samples (n=5 per group) were analysed in duplicate by Eve Technologies, using a multiplex bead platform. For analysis of secreted VEGF-A from cells in vitro, supernatants were collected in triplicate 48 h after serum-starvation and analysed by ELISA (enzyme-linked immunosorbent assay) according to manufacturer’s instructions (R&D Systems).
Cell viability assays
Cell viability over several days was measured using the CellTiter MTT Assay according to the manufacturer’s instructions (Promega). Cells were seeded in triplicate at the same initial density, and attenuance at 590nm was read on sequential days using a plate reader (Bio-Rad).
Luciferase assays
For 3′ UTR assays, HEK293T or MDA231 cells were co-transfected with the indicated sicheck2 wild-type or mutant reporter and either miR-29b, control cel-67, anti-miR-29b or anti-control mimic (100nM final) using Dharmafect Duo (Dharmacon).
For the TGF-β and NF-ΚB reporter assays, the SBE4-Luc, p3TP-Lux or pGL-NF κB-Luc reporter was co-transfected with Renilla luciferase (pRL-TK) using Lipofectamine 2000 (Invitrogen) into MDA231 cells. Cells were serum-starved overnight, and then stimulated with 5 ng ml−1 TGF-β (R&D), 500 ng ml−1 sRANKL (Invitrogen) or 100,ng ml−1 TNF-Α (Peprotech) for 24 h.
For the miR-29a/b1 promoter reporter assays, the pGL3-miR-29–Luc wild-type or mutant reporter was co-transfected with pRL-TK and either pcDNA-eGFP (control) or pcDNA–GATA3 into MDA231 cells. Lysates were collected 48 h post-transfection. Renilla and firefly luciferase activities were measured using the Dual-Luciferase Reporter System and a GloMax luminometer (Promega). Transfection efficiency was normalized to the control luciferase.
Immunostaining and histology
Tissues were fixed in 4% PFA overnight, and paraffin processed or embedded into OCT for frozen sections. Standard haematoxylin and eosin (H&E) staining was performed for routine histology. Picrosirius red staining was performed as previously described48 and fibrillar collagen visualized using crossed polarizers. Standard antigen retrieval was performed using citrate or proteinase K for immunohistochemistry9. The TSA Amplification Kit (Perkin Elmer #NEL700A001KT) was used according to the manufacturer’s instructions. Primary antibodies were incubated overnight, and secondary antibodies were incubated for 1 h. The following antibodies were used at the indicated concentrations: CD31 (BD Pharmingen #553370, 1:50), F4/80 (Invitrogen #MF48000, 1:100), phospho-histone H3 (Cell Signaling #9701, 1:100), cleaved caspase-3 (Cell Signaling #9661, 1:100), GATA3 (R&D #AF2605 1:50 and Abcam #ab32858, 1:100), E-cadherin (Zymed #13-1900, 1:500), biotinylated anti-rat (Jackson #112-067-003, 1:300), biotinylated anti-rabbit (Dako #E0431, 1:300) and biotinylated anti-goat (Jackson #305-067-003, 1:300). Fluorescent antibodies 488-anti-rabbit and 568-anti-rat were from Molecular Probes (Invitrogen #A11008 and #A11077, 1:600), and confocal microscopy was performed on a Nikon C1si confocal microscope. Image analysis was performed using ImageJ or Nikon software.
Time-lapse microscopy
Bright-field time-lapse videos were collected on a Zeiss Axiovert S-100 microscope. The temperature was held at 37 °C and CO2 was held at 5% by using a CTI Controller 3700 and Temperature Control combination. Images were acquired every 20 min and assembled into videos using MetaMorph (Molecular Devices).
Western blotting
Cells were lysed in RIPA buffer plus protease inhibitors (Roche) or directly in Lamelli Buffer with dithiothreitol. Protein concentration was measured using the BCA Protein Assay Kit (Thermo Scientific). Lysates were subjected to SDS–PAGE, transferred to PVDF membranes, blocked in 5% BSA, incubated with primary antibody overnight and visualized using ECL Detection Reagents (Pierce). Exposures were acquired using a LAS-4000 Imager (Fuji). Antibodies used include: actin (Santa Cruz #sc-47778, 1:1,000), LOX (Novus #NB100-2527, 1:500), VEGF-A (Abcam #ab46154, 1:1,000), ANGPTL4 (Invitrogen # 40-9800, 1:250), GATA3 (R&D #AF2605 1:1,000), phospho-Smad3 (Cell Signaling #9520, 1:1,000), vimentin (Cell Signaling #5741, 1:1,000), nucleolin (Abcam #ab22758, 1:1,000), HRP anti-rabbit and HRP anti-mouse (GE Healthcare #NA9340 and #NXA931, 1:5000) and HRP anti-goat (Invitrogen #811620, 1:5000).
Flow cytometry
To sort primary mouse mammary epithelial cells into basal and luminal fractions, mammary glands from adult virgin females were digested with collagenase. Organoids were collected by brief centrifugation and digested with trypsin to dissociate into single cells. The cells were stained with antibodies against CD49f, CD24 and lineage markers (CD45, CD31, Ter119) (eBioscience #17-0495-82, 48-0242-82, 12-0451, 11-0311-85, 11-5921-82, respectively; all used at 1:150), as described previously70. Antibodies against CD29, GATA3, CD44, CD24 and EpCAM (eBioscience #12-0299-71, 12-0241-81, 50-9966-41, 12-0441, 17-0247-41 and 50-9326-41, respectively; all used at 1:150) were used to stain cell lines. For the GATA3 stain, cells were permeabilized using 0.2% Triton X-100. Analysis and cell sorting were performed on an LSRII or FACS Aria II (Becton Dickinson), and analysed using FlowJo (TreeStar) or FACSDiva software (BD Biosciences).
Statistical analysis
Statistical analysis was performed using Prism 4 software (GraphPad Software). All data are presented as mean±s.e.m., unless otherwise stated. When two groups were compared, the two-tailed Student t -test was used, unless otherwise stated. When three or more groups were compared, the one-way analysis of variance test was used, followed by Tukey’s test to determine significance between groups. To compare histograms collected from the flow cytometer, we used the probability binning χ2 test on FlowJo (TreeStar). We considered P < 0.05 as significant.
Accession numbers for data sets
The data set generated has been deposited to GEO under the primary accession number GSE42468. Eight reference accession data sets were reanalysed in the study, including six from the GEO database (GSE7842, GSE19783, GSE22220, GSE23938, GSE23977 and GSE23978) and two from the ArrayExpress database (E-MEXP-2289 and E-TABM-157).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Werb laboratory for discussions, P. Shahi, J. Dai and J. Tai for experimental assistance and E. Atamaniuc, Y. Yu, and H. Capili for technical assistance. We thank T. Rambaldo and M. Kissner for flow cytometer assistance, the UCSF Biological Imaging Development Center for microscopy assistance, J. Debnath, G. Bergers, and D. Sheppard for discussions, and A. Goga, V. Weaver, J. Mott, P. Gonzalez, K. Xie and E. Raines for reagents. We also thank C. Choi for discussion and support. This research was supported by funds from the National Cancer Institute (R01 CA129523 to Z.W.), a Developmental Research grant from the Bay Area Breast Cancer SPORE (P50 CA058207 to Z.W.), a Department of Defense Predoctoral Fellowship (W81×WH-10-1-0168 to J.C.) and the UCSF Medical Scientist Training Program (J.C.). We dedicate this work to the memory of L. Verber.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
J.H.L. and A.B. contributed equally to this work. J.C. designed and performed experiments, with assistance from J.H.L., A.B., J-w.K. and S.P. Z.W. designed experiments and supervised research. J.C. and Z.W. wrote the manuscript, and all authors discussed the results and provided comments and feedback.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J. Cell Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 9.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouros-Mehr H, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon NK, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum. Pathol. 2010;41:1794–1801. doi: 10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehra R, et al. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res. 2005;65:11259–11264. doi: 10.1158/0008-5472.CAN-05-2495. [DOI] [PubMed] [Google Scholar]
- 13.Jacquemier J, et al. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 2009;11:R23. doi: 10.1186/bcr2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Usary J, et al. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 15.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asselin-Labat ML, et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol. Cell Biol. 2011;31:4609–4622. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 19.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Dore LC, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 24.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. New Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 26.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debnath J, Brugge JS. Modelling glandular epithelial cancers in threedimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 28.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mott JL, et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-κB. J. Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winbanks CE, et al. TGF-β regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buffa FM, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 35.Enerly E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu M, et al. Integrated miRNA and mRNA expression profiling of mouse mammary tumor models identifies miRNA signatures associated with mammary tumor lineage. Genome Biol. 2011;12:R77. doi: 10.1186/gb-2011-12-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 38.Dykxhoorn DM, et al. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucl. Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 44.Yu KR, et al. CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells. 2012;30:876–887. doi: 10.1002/stem.1052. [DOI] [PubMed] [Google Scholar]
- 45.Padua D, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J. Biol. Chem. 2010;285:14042–14051. doi: 10.1074/jbc.M110.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu IM, et al. GATA3 inhibits lysyl oxidase-mediated metastases of human basal triple-negative breast cancer cells. Oncogene. 2012;31:2017–2027. doi: 10.1038/onc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du R, et al. HIF1–induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roderburg C, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2010;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 52.Wang B, et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 2011;2:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen T, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2010;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong Y, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2009;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 56.Sethi A, Mao W, Wordinger RJ, Clark AF. Transforming growth factor-β induces extracellular matrix protein cross-linking lysyl oxidase (LOX) genes in human trabecular meshwork cells. Invest. Ophthalmol. Vis. Sci. 2011;52:5240–5250. doi: 10.1167/iovs.11-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dumont N, Arteaga CL. Transforming growth factor-β and breast cancer: tumor promoting effects of transforming growth factor-β. Breast Cancer Res. 2000;2:125–132. doi: 10.1186/bcr44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Elsner T, et al. Synergistic cooperation between hypoxia and transforming growth factor-β pathways on human vascular endothelial growth factor gene expression. J. Biol. Chem. 2001;276:38527–38535. doi: 10.1074/jbc.M104536200. [DOI] [PubMed] [Google Scholar]
- 59.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasisregulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 65.Tong Q, et al. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 66.Wrana JL, et al. TGF β signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 67.Zawel L, et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 68.Watnick RS, Cheng YN, Rangarajan A, Ince TA, Weinberg RA. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell. 2003;3:219–231. doi: 10.1016/s1535-6108(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 69.Gough PJ, Gomez IG, Wille PT, Raines EW. Macrophage expression of active MMP-9 induces acute plaque disruption in apoE-deficient mice. J. Clin. Invest. 2006;116:59–69. doi: 10.1172/JCI25074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.