SUMMARY
Systemic infections with Gram-negative bacteria are characterized by high mortality rates due to the “sepsis syndrome,” a widespread and uncontrolled inflammatory response. Though it is well recognized that the immune response during Gram-negative bacterial infection is initiated after the recognition of endotoxin by Toll-like receptor 4, the molecular mechanisms underlying the detrimental inflammatory response during Gram-negative bacteremia remain poorly defined. Here, we identify a TRIF pathway that licenses NLRP3 inflammasome activation by all Gram-negative bacteria. By engaging TRIF, Gram-negative bacteria activate caspase-11. TRIF activates caspase-11 via type I IFN signaling, an event that is both necessary and sufficient for caspase-11 induction and autoactivation. Caspase-11 subsequently synergizes with the assembled NLRP3 inflammasome to regulate caspase-1 activation and leads to caspase-1-independent cell death. These events occur specifically during infection with Gram-negative, but not Gram-positive, bacteria. The identification of TRIF as a regulator of caspase-11 underscores the importance of TLRs as master regulators of inflammasomes during Gram-negative bacterial infection.
INTRODUCTION
Germline-encoded receptors survey extracellular and intracellular compartments for signs of microbial infection. The cytosolic compartment in particular has emerged as a frontline of host defense, where distinct families of receptors recognize microbial products and initiate protective immune defenses (Franchi et al., 2012; Rathinam and Fitzgerald, 2011). A key component of cytosolic surveillance is the inflammasome, a multiprotein complex that controls the maturation of the proinflammatory cytokines interleukin-1β (IL-1β) and IL-18. Distinct inflammasomes have been identified that are differentiated by their protein constituents, activators, and effectors. In most cases, inflammasomes contain a nucleotide-binding and oligomerization leucine-rich repeat (NLR) protein, the best studied of which is NLRP3 (Franchi et al., 2012).
In response to diverse microbial, environmental, or endogenous danger signals, the NLRP3 inflammasome complex assembles, leading to the multimerization of the adaptor molecule ASC. Subsequently, procaspase-1 is recruited leading to caspase-1 autoactivation, which then cleaves IL-1β and IL-18 into biologically active cytokines. These cytokines have wide-ranging proinflammatory effects important in early control of microbial infection. Despite the identification of numerous triggers, direct binding of any ligands to NLRP3 has not been clearly demonstrated (Strowig et al., 2012). In the case of bacterial infection, pore-forming toxins and bacterial mRNA represent the major triggers of NLRP3 activation (Kanneganti et al., 2006; Sander et al., 2011).
Given the significant potential of IL-1 and related cytokines to cause detrimental inflammation, key regulatory checkpoints ensure that inflammasome-dependent production of these cytokines is tightly regulated (Rathinam et al., 2012). TLR signaling is one such checkpoint. TLRs control the expression of pro-IL-1β and of NLRP3 itself, events that depend predominantly on MyD88. TIR-domain-containing adaptor-inducing interferon-b (TRIF) has also been linked to NLRP3 inflammasome signaling in situations in which the autophagy machinery is depleted or blocked (Saitoh et al., 2008; Zhou et al., 2011). Depletion of the autophagic proteins Atg16L1, LC3B, or beclin 1 results in elevated activation of caspase-1 and secretion of IL-1β and IL-18 (Nakahira et al., 2011; Saitoh et al., 2008; Zhou et al., 2011). In the case of ATG16L1-deficiency, elevated caspase-1 activation and IL-1β production are dependent on TRIF (Saitoh et al., 2008). More recent studies have also linked TRIF to NLRP3 inflammasome activation in cells infected with avirulent Escherichia coli (Sander et al., 2011).
These observations suggest that TRIF is linked to NLRP3 inflammasome activation by as yet undefined mechanisms. Here, we identify a TRIF pathway that links TLR4 and NLRP3 signaling during the immune response to Gram-negative bacteria. This pathway is initiated by TLR4 and mediated by type I IFNs. Type I IFNs induce caspase-11 expression, an event that is both necessary and sufficient to promote caspase-11 autoprocessing in the absence of any other microbial trigger. Caspase-11 activation via the TLR4-TRIF-IFNβ pathway synergizes with the NLRP3 pathway to coordinate caspase-1-dependent IL-1β and IL-18 secretion and also leads to caspase-1-independent cell death. The identification of TRIF as a regulator of caspase-11 provides new insights into NLRP3 inflammasome activation during Gram-negative bacterial infection, highlights the central role of TLRs as master regulators of inflammasome signaling, and unveils new targets that might be manipulated to prevent uncontrolled inflammation during septic shock.
RESULTS AND DISCUSSION
TRIF Is Essential for NLRP3 Inflammasome Activation in Response to EHEC and Citrobacter rodentium
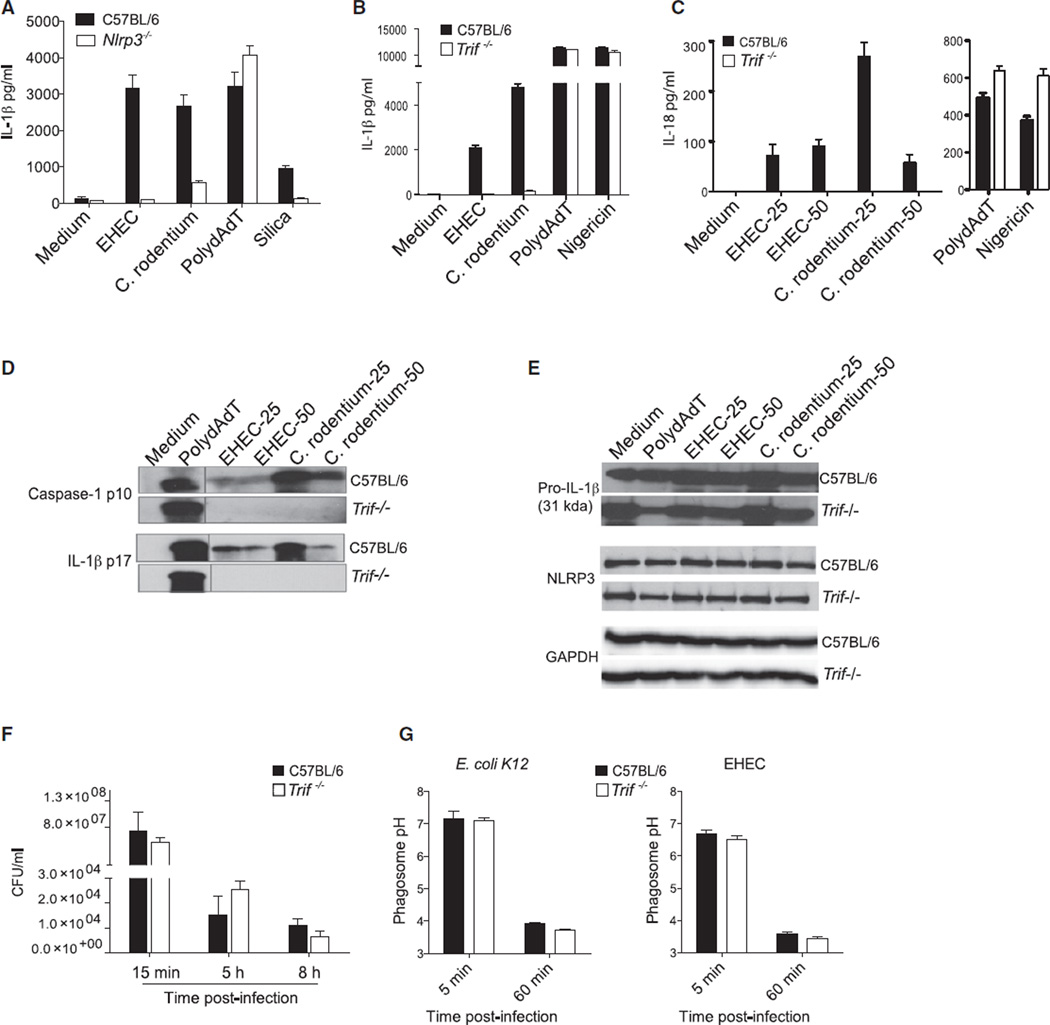
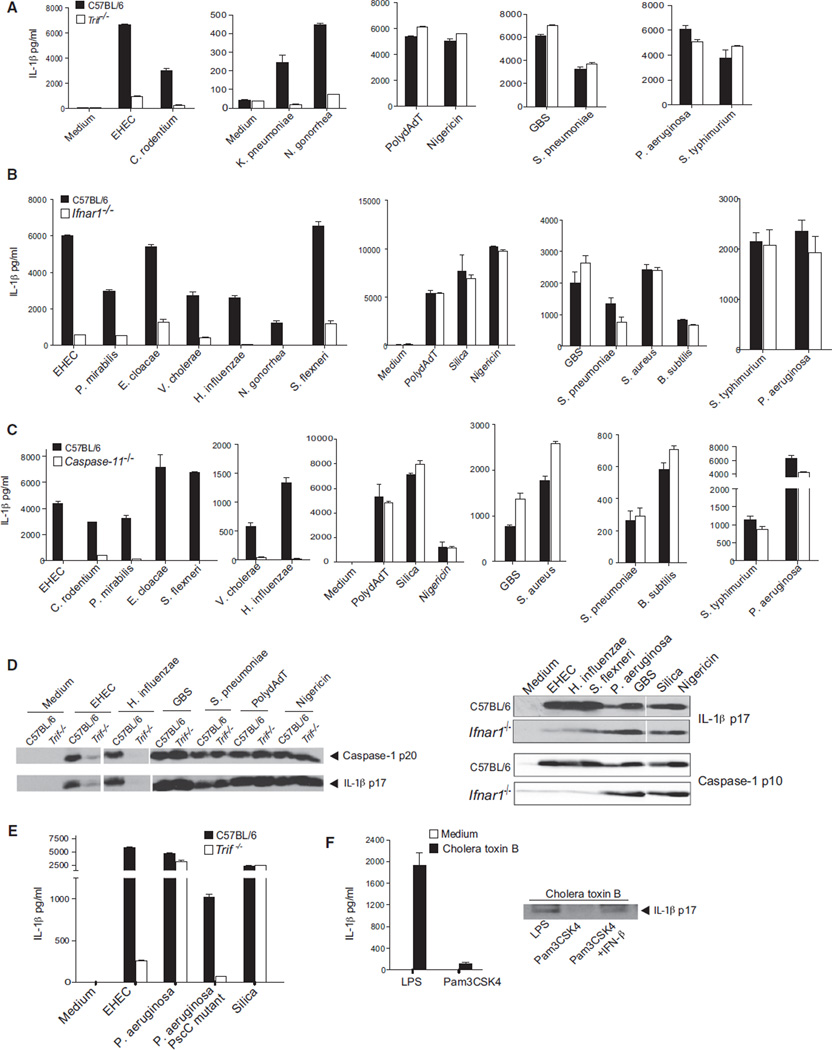
While conducting a systematic investigation of inflammasome activation by enterohemorrhagic Escherichia coli (EHEC) and Citrobacter rodentium, two related Gram-negative enteropathogens, we revealed a requirement not only for NLRP3 (Figure 1A), but also for TRIF in the production of IL-1β or IL-18 at 16 hr postinfection (Figures 1B and 1C). Caspase-1 cleaves pro-IL-1β and pro-IL-18 into their biologically active forms. Secretion of the caspase-1 subunit p10, as well as of the mature IL-1β-p17, was abrogated in TRIF-deficient macrophages infected with EHEC and C. rodentium (Figure 1D). The requirement for TRIF was specific to EHEC and C. rodentium because normal processing and secretion of caspase-1 and IL-1β were observed in TRIF-deficient cells stimulated with polydAdT, which engages the AIM2 inflammasome (Rathinam et al., 2010), or nigericin, a canonical activator of the NLRP3 inflammasome. The requirement for TRIF in EHEC and C. rodentium infection was observed across a broad range of bacterial doses (MOI, 6, 12, 25, and 50) and was also seen at an earlier time point (8 hr postinfection) (Figures S1A and S1B available online). Whereas MyD88 has been linked to transcriptional regulation of pro-IL-1β and NLRP3 in LPS-primed cells, the role of TRIF in EHEC and C. rodentium infection was unrelated to these so-called “priming signals” because Pam3Csk4, which triggers TLR2 signaling, a TRIF-independent TLR, was used to ensure equivalent levels of pro-IL-1β and NLRP3 in all conditions. The protein levels of pro-IL-1β and NLRP3 were similar in wild-type and Trif−/− macrophages primed with Pam3Csk4 and infected with EHEC and C. rodentium (Figure 1E). EHEC and C. rodentium also triggered secretion of IL-18, which unlike IL-1β is synthesized constitutively in cells, and production of IL-18 was also dependent on TRIF. A role for TRIF in the regulation of IL-1β in infected cells was also observed in cells that were unprimed (data not shown), further validating a role for TRIF in inflammasome activation rather than priming.
Figure 1. TRIF Is Essential for NLRP3 Inflammasome Activation by EHEC and C. rodentium.
(A) IL-1β production by Pam3CSK4-primed BMDMs stimulated with EHEC (MOI of 25), C. rodentium (MOI of 25), or polydAdT for 16 hr or silica for 16 hr.
(B–E) ELISA for IL-1β (B) and IL-18 (C), immunoblots for cleaved caspase-1 and IL-1β in the supernatants (D), and immunoblots for proIL-1β, NLRP3, and GAPDH in the lysates (E) of Pam3CSK4-primed BMDMs stimulated with EHEC (MOI of 25 and/or 50), C. rodentium (MOI of 25 and/or 50), or polydAdT for 16 hr or nigericin for 1 hr.
(F) Intracellular bacterial numbers at various time points from EHEC-infected BMDMs.
(G) Phagosomal pH assessed by ratiometric analysis at 5 and 60 min postinfection in E. coli K12- or EHEC-infected BMDMs.
Data are presented as the mean ± SEM of one experiment representative of three (A–E) or two (F and G) experiments. See also Figure S1.
TRIF Does Not Control Bacterial Uptake, Killing, or Phagosomal Acidification
Nascent phagosomes mature by fusion with early and then late endosomes, ultimately fusing with lysosomes. The formation of an acidified mature phagolysosome is essential for NLRP3 inflammasome activation in response to extracellular bacteria such as E. coli, so we investigated whether TRIF regulates phagocytosis, phagosome maturation, or the destructive capacity of phagosomes during EHEC infection. In a gentamicin-killing assay, we observed no difference in internalization and killing between wild-type and Trif−/− cells (Figure 1F). Moreover, using a ratiometric fluorescence-based assay, we found that Trif−/− macrophages acidified EHEC-containing phagosomes with kinetics comparable to that of wild-type cells (Figure 1G). Similar observations were made by lysotracker staining analysis (data not shown). Phagosome membranes are permeant to certain ligands and can facilitate delivery of bacterial mRNA to the cytosol to engage NLRP3 (Sander et al., 2011). Phagolysosomal integrity after EHEC infection as monitored by acridine orange staining revealed no difference in the intrinsic permeability of phagolysosomes in TRIF-deficient cells (Figure S1C). Additionally, production of reactive oxygen species, which has been linked to NLRP3 activation, occurs independently of TRIF in E. coli-infected cells (Sander et al., 2011). Collectively, these observations indicated that TRIF does not influence bacterial uptake, killing, or phagosomal acidification and hence does not control egress of bacterial RNA into the cytosol during infection to trigger NLRP3.
TRIF Acts Upstream of Caspase-11
A recent study identified caspase-11 as a key regulator of caspase-1 activation in response to the enteric pathogens E. coli, C. rodentium, and Vibrio cholerae (Kayagaki et al., 2011). Early studies characterized caspase-11 as a critical component of the caspase-1 complex (Wang et al., 1996, 1998). Although caspase-11 does not process pro-IL-1β directly, it was shown to be required for the activation of caspase-1 (Wang et al., 1996). More recent studies have shown that secretion of the caspase-1 subunits p10 and p20 as well as IL-1β and IL-18 in response to enteropathogens required caspase-11, ASC, and NLRP3 (Kayagaki et al., 2011). Caspase-11 was dispensable, however, for caspase-1 activation in response to other NLRP3 agonists such as ATP. Caspase-1 activation by these atypical enteropathogenic activators required NLRP3 and ASC, but caspase-11 processing and cell death did not, suggesting that caspase-11 was activated by distinct mechanisms independent of NLRP3 and ASC. These data suggested that caspase-11 was activated by NLRP3-independent mechanisms but synergized with the assembled NLRP3 inflammasome to regulate caspase-1 activation in response to enteric pathogens.
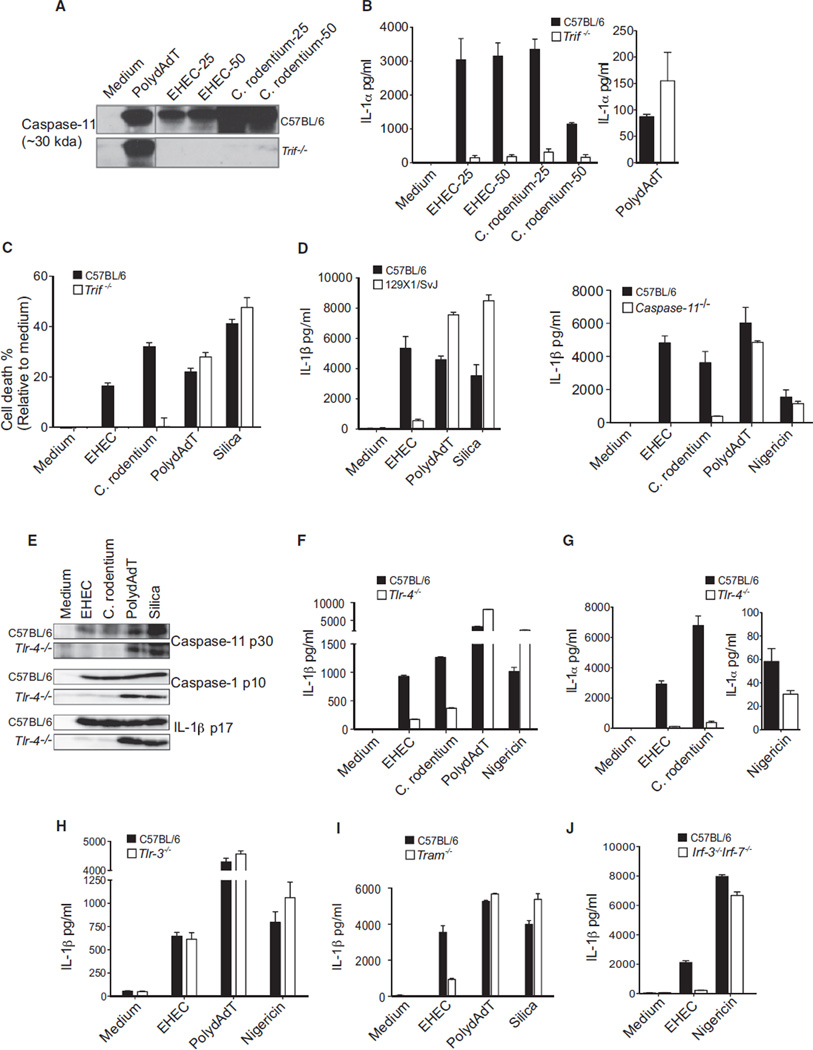
We hypothesized that TRIF might lie upstream of caspase-11 in order to regulate caspase-1 activation during infection with EHEC and C. rodentium. We therefore monitored the activation status of caspase-11 in wild-type and TRIF-deficient cells. The caspase-11 locus encodes two isoforms of 43 and 38 kDa, and the processed form of caspase-11 is a ~26–30 kDa form. Infection of wild-type BMDMs with EHEC or C. rodentium resulted in the generation of the cleaved caspase-11 p30 subunit, and this response was completely absent in TRIF-deficient cells (Figure 2A). Caspase-11 has also been shown to regulate the secretion of IL-1α and high-mobility group box protein 1 (HMGB1), as well as cell death, in an NLRP3-caspase-1-independent manner (Kayagaki et al., 2011). Consistent with a role for TRIF upstream of caspase-11, secretion of IL-1α as well as cell death were markedly impaired in Trif−/− cells infected with EHEC or C. rodentium (Figures 2B and 2C). PolydAdT also stimulated formation of the caspase-11 p30 subunit. However, caspase-11 was dispensable for polydAdT-induced processing and secretion of IL-1β by macrophages from 129X1/SvJ mice (which are mutant for caspase-11) or caspase-11-deficient mice (Figure 2D). This was in contrast to EHEC and C. rodentium, both of which induced IL-1β release in a manner strictly dependent on caspase-11. Caspase-11 processing after polydAdT treatment is due to the promiscuous nature of caspase-1 activity in the in vitro assay conditions and is not physiologically relevant. This processing of caspase-11 (which occurred with delayed kinetics) was suggested to be a consequence of caspase-1 activation in vitro (Kayagaki et al., 2011). These data reveal a central requirement for TRIF in caspase-11 activation, caspase-11-dependent caspase-1 activation, and caspase-11-dependent IL-1α release and cell death during infection with EHEC and C. rodentium, but not other triggers of the NLRP3 inflammasome.
Figure 2. TRIF Signaling Downstream of TLR4-TRAM Mediates Inflammasome Activation by EHEC and C. rodentium.
(A) Cleaved caspase-11 in the supernatants of Pam3CSK4-primed BMDMs stimulated for 16 hr with EHEC (MOI of 25 and 50), C. rodentium (MOI of 25 and 50), or polydAdT.
(B, D, and F–J) IL-1β or IL-1α production by Pam3CSK4-primed BMDMs from C57BL/6 wild-type and various knockout mice stimulated with EHEC, C. rodentium, polydAdT, or silica for 16 hr or nigericin for 1 hr.
(C) Cell death in BMDMs stimulated with EHEC, C. rodentium, polydAdT, or silica for 16 hr.
(E) Immunoblots for cleaved caspase-11, caspase-1, and IL-1β in the supernatants of Pam3CSK4-primed BMDMs stimulated with EHEC, C. rodentium, polydAdT, or silica for 16 hr.
Data are presented as the mean ± SEM of one experiment representative of three experiments. See also Figure S2.
TRIF Signaling Downstream of TLR4 Is Essential for Caspase-11-Dependent Inflammasome Activation in Response to EHEC and C. rodentium
We next wanted to understand how TRIF could facilitate caspase-11 activation. TRIF is an integral component of TLR3 and TLR4 signaling (Kawai and Akira, 2010) and also functions in an RNA-sensing pathway involving DDX1, DDX21, and DHX6 (Zhang et al., 2011). TRIF is recruited to TLR4 via TRAM or directly to the TIR domain of TLR3 (Kagan et al., 2008; Yamamoto et al., 2003). Like TRIF-deficient cells, TLR4-deficient cells were impaired in their ability to induce proteolytic processing of caspase-11 (Figure 2E) and, as a consequence, failed to induce processing of caspase-1 and IL-1β in response to EHEC and C. rodentium (Figures 2E and 2F). TLR4-deficient cells also failed to secrete IL-1α (Figure 2G). Caspase-1 activation and IL-1β, as well as IL-1α release in response to polydAdT, nigericin, or silica, were all unaffected in Tlr4−/− cells. EHEC-induced IL-1β secretion was normal in Tlr3−/− macrophages (Figure 2H). We also observed a defect in IL-1β secretion in Tram−/− macrophages infected with EHEC (Figure 2I). TRIF signaling downstream of TLR4-TRAM leads to activation of the interferon regulatory factors IRF3/7 (Fitzgerald et al., 2003), NFκB, or apoptosis (Cusson-Hermance et al., 2005; Han et al., 2004; Kaiser and Offermann, 2005; Weber et al., 2010). TRIF interacts with TBK1 to activate IRF3/7, with RIP kinases to mediate NFκB activation, or with caspase-8 to induce apoptosis. IL-1β production as well as cell death in EHEC- and C. rodentium-infected macrophages from mice lacking RIP3 or RIP3 and caspase-8 were unaffected (Figure S2). This was in contrast to macrophages from mice lacking IRF3/7 that were severely impaired in their ability to mediate IL-1β release (Figure 2J). These data indicate that caspase-11 activation and caspase-11-dependent caspase-1 activation are downstream of a pathway involving TLR4, TRAM, TRIF, and IRF3/7 in response to a subset of NLRP3 activators, including EHEC and C. rodentium.
TLR4, TRIF, and IFNAR Signaling Are Essential for EHEC- and C. rodentium-Induced Caspase-11 Activation and Caspase-11 Effector Function
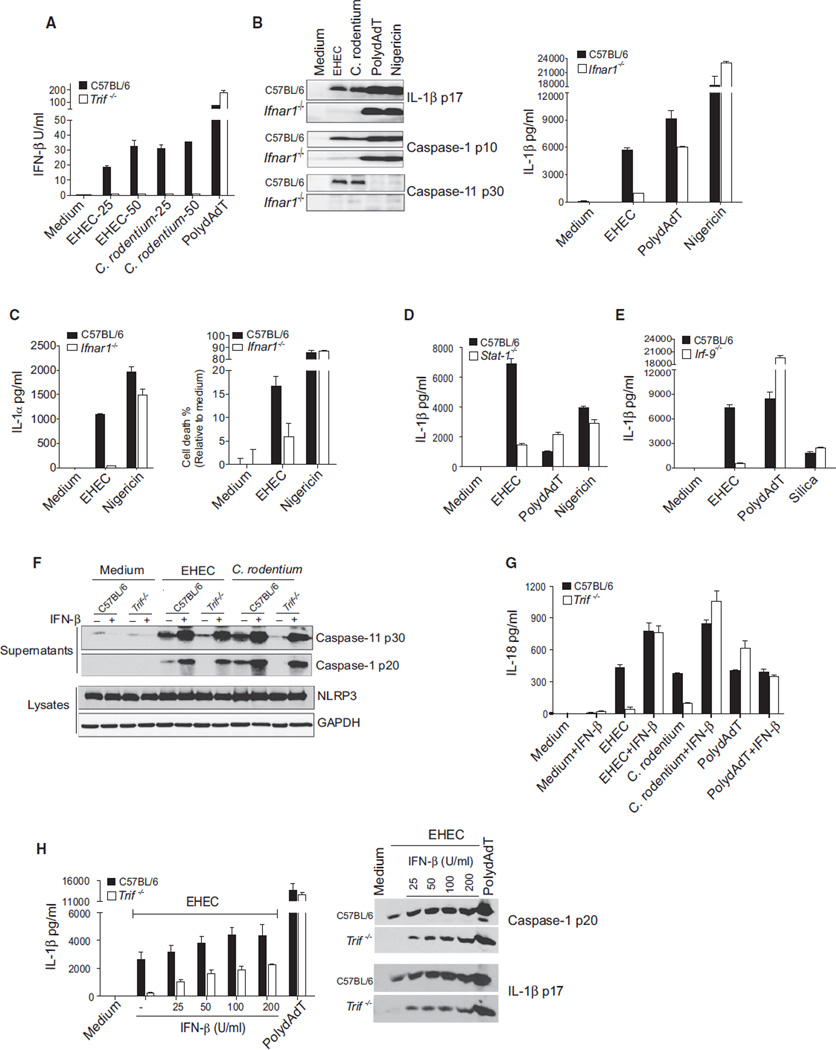
A prominent feature of TRIF-dependent IRF signaling is the production of type I interferons. We examined IFNβ production in macrophages from mice lacking either TRIF or, as a control, STING, a key mediator of cytosolic pathways (Ishikawa et al., 2009). EHEC and C. rodentium both induced IFNβ in a TRIF-dependent but STING-independent manner (Figures 3A and S3A). Consistent with a role for TRIF rather than STING in IFNβ induction, EHEC and C. rodentium induced normal levels of IL-1β in macrophages from STING-deficient mice (Figure S3A). IFNβ mediates TRIF-dependent caspase-11-inflammasome activation during EHEC infection because processing of caspase-11 was markedly impaired in IFNAR1-deficient cells infected with EHEC (Figure 3B). Consequently, EHEC- or C. rodentium-infected IFNAR1-deficient cells were extremely defective in their ability to cleave caspase-1 and IL-1β, as well as in their ability to secrete IL-1β (Figure 3B). The requirement for IFNAR1 was seen regardless of the dose of bacteria used (Figure S1A) Furthermore, the secretion of IL-1α, cell death, and HMGB1 release—events unique to the caspase-11 pathway—were also defective in EHEC-infected Ifnar1−/− cells (Figures 3C and S3B). In contrast, IFNAR1 was dispensable for these events in cells stimulated with polydAdT and nigericin. Similar to TRIF, IFNAR1 signaling did not control bacterial phagocytosis, phagosomal acidification, or bacterial killing by macrophages (Figures S3C and S3D).
Figure 3. Type I Interferon Response Triggered by TLR4-TRIF Mediates Caspase-11-Dependent Inflammasome Activation by EHEC and C. rodentium.
(A) IFN-β production by BMDMs stimulated with EHEC (MOI of 25 and 50), C. rodentium (MOI of 25 and 50), or polydAdT for 16 hr.
(B) ELISA for IL-1β and immunoblots for cleaved caspase-1, caspase-11, and IL-1β in the supernatants of BMDMs stimulated with EHEC, C. rodentium, or polydAdT for 16 hr or nigericin for 1 hr.
(C) IL-1α secretion by and cell death in BMDMs stimulated with EHEC for 16 hr or nigericin for 1 hr.
(D and E) IL-1β production by Pam3CSK4-primed C57BL/6 wild-type and Stat1−/− (D) or Irf9−/− (E) BMDMs stimulated with EHEC, polydAdT, or silica for 16 hr or nigericin for 1 hr.
(F and G) Cleaved caspase-1 and caspase-11 in the supernatants and NLRP3 in the lysates (F) and IL-18 in the supernatants (G) of Pam3CSK4-primed BMDMs stimulated with EHEC, C. rodentium, or polydAdT for 16 hr. Cells were subjected to 1,000 U/ml IFN-β treatment at the time of infection as indicated.
(H) Secreted IL-1β and cleaved caspase-1 and IL-1β in the supernatants of Pam3CSK4-primed BMDMs treated with indicated doses of IFN-β and stimulated with EHEC or polydAdT for 16 hr.
Data are presented as the mean ± SEM of one experiment representative of 2–3 experiments. See also Figures S1A and S3.
Type I IFNs signal in an autocrine/paracrine manner via IFNα/βR1 and IFNα/βR2. Dimerization of the receptor chains causes the receptor-associated JAKs to become activated, resulting in the engagement of signal transducer and activator of transcription (STAT)-1 and STAT-2, which associate with IRF9 to form the ISGF3 complex. ISGF3 translocates to the nucleus and initiates transcription of multiple IFN-stimulated genes (ISGs) (González-Navajas et al., 2012). Whereas STAT-1 and IRF9 were dispensable for polydAdT and nigericin or silica-induced production of IL-1β, both factors were essential for IL-1β production in EHEC-infected macrophages (Figures 3D and 3E). We also tested the ability of exogenous type I IFN to rescue the defect in inflammasome-dependent responses in Trif−/− cells. Exogenous IFNβ restored caspase-11 processing, caspase-1 processing, and IL-18 production in TRIF-deficient cells infected with EHEC and C. rodentium (Figures 3F and 3G). Type I IFN has been shown to impair transcription of pro-IL-1β, thereby reducing the pool of precursor levels (Guarda et al., 2011). Indeed, exogenous treatment with 1000 U/ml IFNβ, the dose used in previous experiments, led to a significant reduction in pro-IL-1β levels and failed to rescue EHEC- or C. rodentium-induced IL-1β production (Figure S3E). However, treatment of TRIF-deficient macrophages with 25–200 U/ml of IFNβ rescued caspase-1 and IL-1β processing and IL-1β secretion in a dose-dependent manner (Figure 3H). Neutralization of IFN-β also led to a dose-dependent attenuation of EHEC-induced, but not polydAdT-induced, IL-1β production (Figure S3F). These data indicate that the processing of caspase-11 is dependent on TLR4, TRAM, TRIF, and the type I IFN-STAT1-signaling pathway.
Transcriptional Induction of Caspase-11 Is Coupled to Caspase-11 Autoprocessing
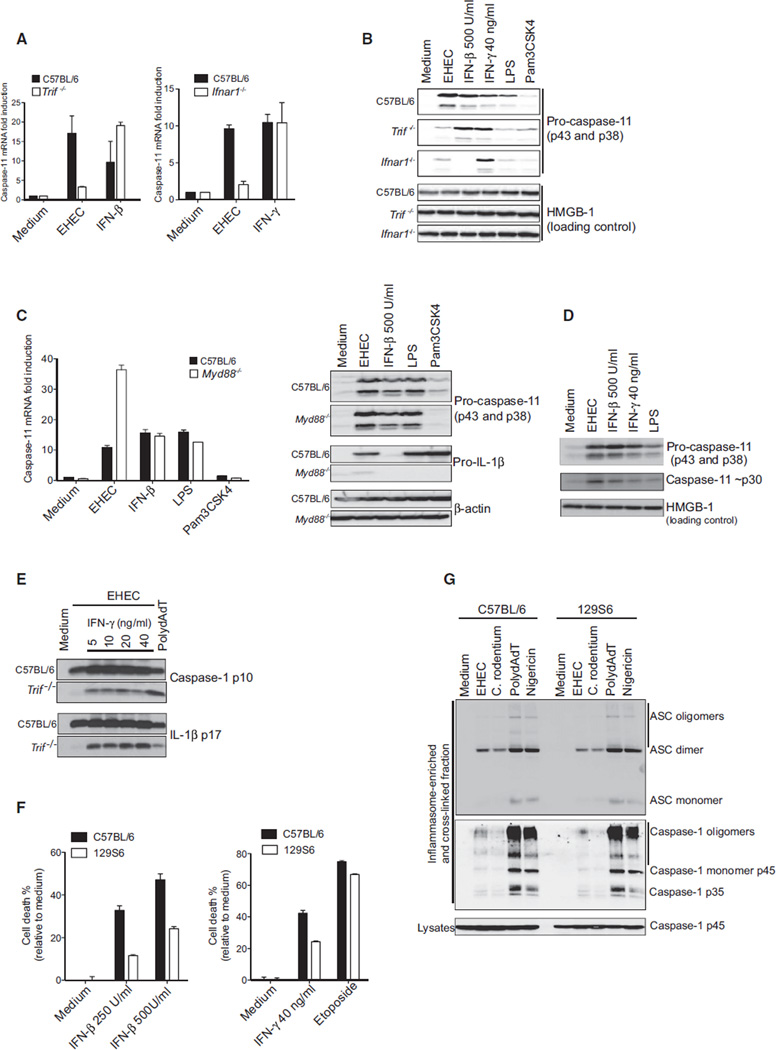
Caspase-11 is unique among caspases in its regulation at the transcriptional level (Wang et al., 1996, 1998). Previous work has shown that macrophages express very low levels of the procaspase-11 isoforms p43 and p38, the expression of which increases following LPS (Wang et al., 1996, 1998) or type I IFN stimulation (Schauvliege et al., 2002; Yen and Ganea, 2009). Similar findings have been reported in vivo (Wang et al., 1996, 1998). Consistent with these published data, we found that caspase-11 was transcriptionally regulated during EHEC infection in a manner dependent on TRIF and IFNAR signaling (Figure 4A). Procaspase-11 protein levels accumulated in EHEC-infected macrophages in a manner dependent on TRIF and IFNAR (Figure 4B). This was in contrast to MyD88-deficient macrophages, in which expression of procaspase-11 was intact (Figure 4C). As expected, pro-IL-1β levels in EHEC-infected macrophages were impaired in the absence of MyD88. IFNβ and IFNγ also turn on procaspase-11 expression (Schauvliege et al., 2002; Yen and Ganea, 2009) (Figures 4A and 4B). In the case of IFNγ treatment, procaspase-11 mRNA induction occurred independently of IFNAR signaling.
Figure 4. EHEC-Induced Caspase-11 Transcriptional Induction Is TRIF and IFNAR Dependent.
(A) Caspase-11 transcript levels in BMDMs infected with EHEC for 12 hr or stimulated with IFN-β (500 units/ml) or IFN-γ (40 ng/ml).
(B) Procaspase-11 and HMGB-1 (as a loading control) in the lysates of BMDMs stimulated with indicated treatments for 16 hr.
(C) Caspase-11 transcript and protein levels and pro-IL-1β protein levels in C57BL/6 and MyD88−/− BMDMs stimulated with EHEC, IFN-β, LPS (200 ng/ml), or Pam3CSK4 (400 ng/ml) for 12 hr (transcript) or 16 hr (protein).
(D) Pro- and processed caspase-11 in the lysates of BMDMs treated with EHEC, IFN-β, IFN-γ, or LPS (200 ng/ml) for 16 hr.
(E) Cleaved caspase-1 and IL-1β in the supernatants of Pam3CSK4-primed BMDMs treated with indicated doses of IFN-γ and stimulated with EHEC or polydAdT for 16 hr.
(F) Cell death in immortalized C57BL/6 and 129S6 BMDMs stimulated with IFN-β (250 U/ml and 500 U/ml) or IFN-γ (40 ng/ml) for 40 hr or etoposide (50 µM) for 24 hr.
(G) Oligomerization of ASC and caspase-1 in the inflammasome-enriched and cross-linked lysates of immortalized C57BL/6 and 129S6 BMDMs treated with EHEC or C. rodentium for 6 hr or polydAdT for 3 hr or nigericin for 30 min. Monomers, dimers, and oligomers of ASC and caspase-1 are indicated accordingly.
Data are presented as the mean ± SEM of one experiment representative of three (A–D) or two (E–G) experiments. See also Figure S4.
Based on the unique inducibility of caspase-11 and the ability of IFNβ to restore caspase-11-dependent activation of caspase-1 in TRIF-deficient cells, we hypothesized that induction of procaspase-11 during infection with EHEC was sufficient to facilitate caspase-11 autoactivation. To test this hypothesis directly, we monitored procaspase-11 induction and processing in macrophages stimulated with LPS, IFNβ, or IFNγ alone in the absence of any other microbial trigger. Consistent with a model whereby transcriptional induction of procaspase-11 is coupled to autoprocessing of caspase-11, LPS treatment alone led not only to transcriptional induction of both procaspase-11 isoforms p43 and p38, but also to the appearance of processed caspase-11 (Figure 4D). Similarly, IFNβ or IFNγ alone induced both transcriptional induction and processing of caspase-11. Ectopic expression of caspase-11 in HEK293 cells resulted in production of the autoprocessed form (Figure S4A), and in-vitro-transcribed/-ranslated caspase-11 generated in a rabbit reticulocyte system also resulted in production of the autoprocessed form (Figure S4B). These findings are consistent with published studies (Kang et al., 2000; Kayagaki et al., 2011). Further support for this model is based on the fact that exogenous IFNγ could also induce procaspase-11 expression, autoprocessing, and restoration of caspase-1 and IL-1β processing in TRIF-deficient macrophages after EHEC infection (Figure 4E). Finally, IFNβ or IFNγ alone induced cell death in a caspase-11-dependent manner (Figure 4F). Collectively, these data indicate that elevated caspase-11 expression can result in autoprocessing and activation. Though these findings do not exclude the possibility of a distinct molecular scaffold regulating caspase-11 processing, they indicate that the regulation of caspase-11 at the transcriptional level is sufficient for activation of caspase-11 and suggest that it is the critical regulated step for its downstream function. This mechanism is distinct from that regulating all other inflammatory and initiator apoptotic caspases in which a molecular scaffold promotes activation. It is important to note here that active caspase-11 only leads to caspase-1 activation when the NLRP3-ASC scaffold has been assembled.
Caspase-11 Does Not Regulate Assembly of the NLRP3 Inflammasome
A key question that arises from these studies is how caspase-11 impacts NLRP3 inflammasome activation. Caspase-11 does not process pro-IL-1β directly; however, it is required for the activation of caspase-1 (Kang et al., 2000; Wang et al., 1996, 1998). One possibility is that caspase-11 promotes the assembly of the NLRP3 inflammasome complex. To test this, we examined the assembly of the NLRP3-ASC complex in EHEC- and C. rodentium-infected macrophages by examining the oligomerization status of ASC in wild-type and 129S6 macrophages (which lack caspase-11). EHEC and C. rodentium induced oligomerization of ASC in wild-type macrophages, and these events were unaffected in macrophages from 129S6 mice, which lack caspase-11 (Figure 4G), or in caspase-11-deficient macrophages (data not shown). These data suggest that formation of the NLRP3-ASC complex is not dependent on the presence of caspase-11. We also tested the possibility that caspase-11 promotes the recruitment of caspase-1 to the assembled NLRP3-ASC complex but found no difference in the recruitment of caspase-1 to the oligomerized ASC complex in cells lacking caspase-11 (Figure 4G). What was striking, however, was the considerably weaker oligomerization of both ASC and caspase-1 in EHEC- and C. rodentium-infected macrophages compared to that observed with PolydAdT or canonical activators of the NLRP3 inflammasome, such as nigericin. This weaker response was particularly surprising in light of the mostly comparable degree of IL-1β production seen in all of these experimental conditions (Figure 2D). Previous studies have shown that caspase-11 interacts with caspase-1 and forms a heterodimeric complex in infected cells (Kayagaki et al., 2011; Wang et al., 1998). It is likely, therefore, that caspase-11 amplifies caspase-1 activation by processing caspase-1 itself or by enabling caspase-1 autoprocessing through heterodimerization.
NLRP3-Dependent Caspase-1 Activation and IL-1β Production by All Gram-Negative Bacteria Is Mediated by the TRIF-IFN-Caspase-11 Pathway
We next wanted to test whether this TRIF-IFNAR-caspase-11 pathway represented a fundamental mechanism by which a broader range of bacterial pathogens activated the NLRP3 inflammasome. We chose to study three distinct classes of bacterial pathogens. The first of these were Gram-negative bacteria known to engage the NLRP3 inflammasome and included Hemophilus influenzae, Klebsiella pneumoniae, Neisseria gonorrhea, Shigella flexneri, Enterobacter cloacae, Vibrio cholerae, and Proteus mirabilis (Duncan et al., 2009; Willingham et al., 2009; Willingham et al., 2007). In all cases, production of IL-1β and/or IL-18 was strictly dependent on TRIF, IFNAR, STAT1, and caspase-11 (Figures 5A, 5B, 5C, S5A, and S5B). Moreover, caspase-1 and IL-1β processing triggered by these bacterial pathogens were impaired in macrophages from mice lacking TRIF or IFNAR1 (Figure 5D). Interestingly, the TRIF-IFN-caspase-11 pathway was dispensable for processing of caspase-1 and/or IL-1β production in macrophages infected with Salmonella typhimurium and Pseudomonas aeruginosa, which predominantly engage the NLRC4 rather than the NLRP3 inflammasome (Figures 5A, 5B, and 5C) (Mariathasan et al., 2004; Miao et al., 2006, 2010; Sutterwala et al., 2007).
Figure 5. NLRP3 Inflammasome Activation by All Gram-Negative Bacteria Requires TRIF-IFNβ-Caspase-11 Axis.
(A–C) IL-1β production by Pam3CSK4-primed C57BL/6 wild-type and Trif−/− (A) or Ifnar1−/− (B) or caspase-11−/− (C) BMDMs stimulated with indicated bacteria for 16 hr.
(D) Cleaved caspase-1 and IL-1β in the supernatants of Pam3CSK4-primed BMDMs stimulated with indicated bacteria for 16 hr.
(E) IL-1β secretion by Pam3CSK4-primed C57BL/6 wild-type and Trif−/− BMDMs stimulated with P. aeruginosa PAK wild-type strain or pscC mutant strain (that lacks the type III secretion system) for 16 hr.
(F) Secreted and cleaved IL-1β in the supernatants of BMDMs primed as indicated and stimulated with cholera toxin B for 16 hr.
Data are presented as the mean ± SEM of one experiment representative of 2–3 experiments. See also Figure S5 and Table S1.
We also examined a set of Gram-positive bacteria that engage the NLRP3 inflammasome (Costa et al., 2012; Fang et al., 2011; Franchi et al., 2007; Mariathasan et al., 2006). Inflammasome activation in response to Group B Streptococcus (GBS), Streptococcus pneumoniae (Pneumococcus), Staphylococcus aureus, and Bacillus subtilis, however, proceeded independently of the IFNAR-STAT-1-caspase-11 pathway (Figures 5B, 5C, S5A and, S5B). Listeria monocytogenes also induced IL-1β production in a manner that was TRIF and IFNAR independent (Figure S5C). Presumably the highly potent pore-forming cytotoxins such as hemolysins expressed by these Gram-positive bacteria facilitate direct activation of the NLRP3 inflammasome without a requirement for the amplifying type I IFN-caspase-11 pathway.
We speculated that, in the absence of NLRC4 activation by a functional T3SS, Gram-negative bacteria such as S. typhimurium or P. aeruginosa might also engage the TRIF and caspase-11 pathway. We compared IL-1β production in macrophages infected with wild-type P. aeruginosa and a mutant lacking a T3SS (pscC). Whereas wild-type P. aeruginosa induced normal levels of IL-1β in TRIF-deficient macrophages, the pscC mutant became dependent on the TRIF-induced caspase-11 pathway (Figure 5E). Similar findings were obtained using a T3SS mutant of S. typhimurium (data not shown). Cholera toxin B (CTB) has also been shown to engage the caspase-11-dependent NLRP3 pathway (Kayagaki et al., 2011). We speculated that CTB would only engage the caspase-11 pathway in cells that were primed with LPS to provide the TRIF-IFNβ-caspase-11 response to synergize with CTB-induced NLRP3 activation. Indeed, CTB treatment of macrophages elicited IL-1β release only in macrophages that were first primed with LPS and not those primed with the TLR2 ligand Pam3Csk4 (Figure 5F). In contrast to LPS, priming with the TLR2 ligand Pam3Csk4 failed to induce substantial caspase-11 expression (Figures 4B and 4C). Exogenous treatment of TLR2-primed macrophages with IFNβ restored CTB-induced IL-1β processing.
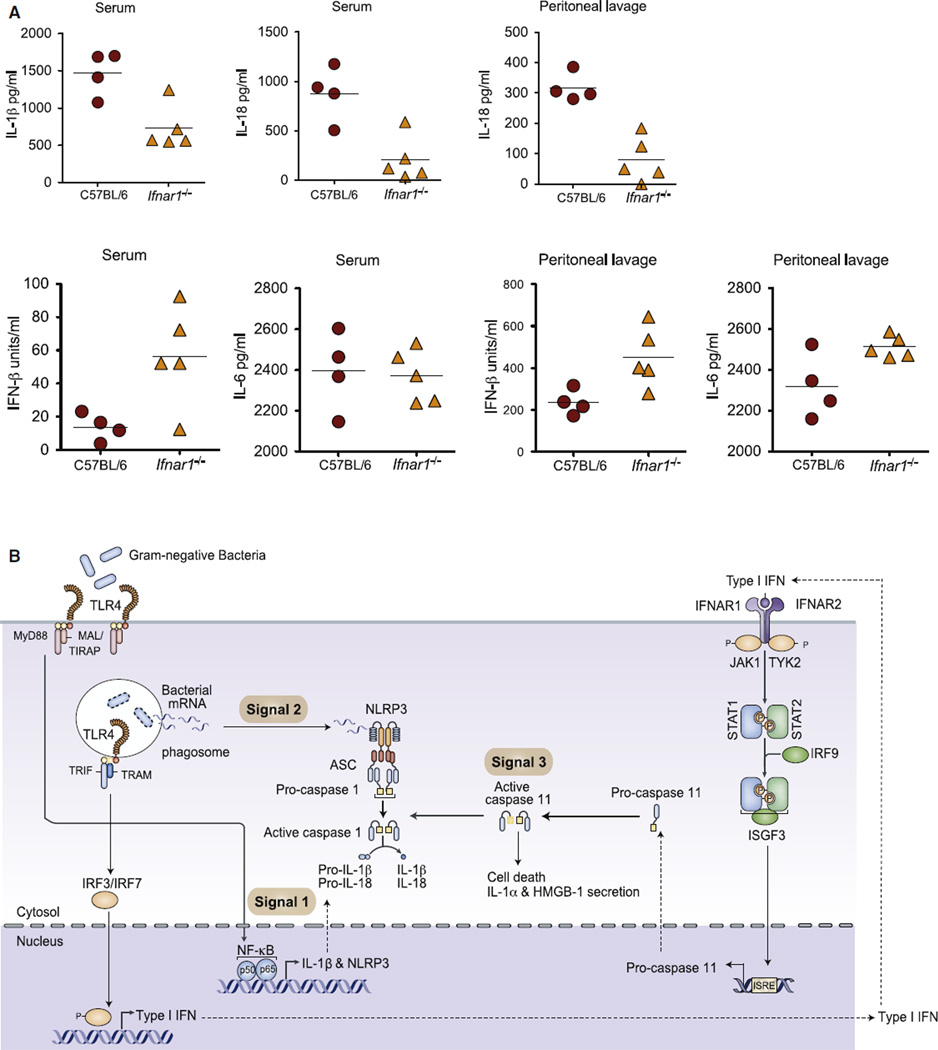
Finally, we wanted to address the role of the TRIF-IFNAR pathway in regulating inflammasome activation in vivo. Wildtype and Ifnar1−/− mice were infected intraperitoneally (i.p.) with E. coli, and cytokine levels were measured in both the serum and peritoneal lavage. Whereas E. coli induced high levels of IL-1β in serum and IL-18 in serum and the peritoneal lavage of wild-type mice, both of these responses were significantly impaired in IFNAR1-deficient mice (Figure 6A). In contrast, induction of IL-6 and IFNβ were not affected by IFNAR1 deficiency. Moreover, we observed enhanced survival of TRIF-deficient mice following E. coli infection in vivo (Figure S6). These data indicate the importance of the TRIF-IFNAR-caspase-11 pathway in E. coli-induced IL-1β and IL-18 production not only in vitro, but also in vivo.
Figure 6. Type I Interferon Signaling Is Essential for Inflammasome Activation in an E. coli-Induced Acute Peritonitis Model.
(A) Cytokines in the serum and peritoneal lavages of C57BL/6 and Ifnar1−/− mice infected with E. coli BL21 for 6 hr (n = 4–5). Data are presented as the mean ± SEM of one experiment representative of two experiments.
(B) Integrative model of TLR4-TRIF-IFN and NLRP3 signaling to activate caspase-11-dependent caspase-1, IL-1β, as well as IL-18 processing.
See also Figure S6.
The integration of TLR and NLR pathways during the inflammatory response is well documented. Here, we have identified a TRIF-dependent pathway that integrates TLR and NLRP3 signaling during the immune response to all Gram-negative bacteria. TRIF licenses NLRP3 inflammasome activation by all Gram-negative bacteria (Figure 6B) via the type I IFN pathway. Type I IFNs upregulate caspase-11 expression, an event that is both necessary and sufficient to enable caspase-11 autoactivation. At the same time, by a mechanism still not understood in detail, bacterial mRNA from viable bacteria that have been phagocytosed access the cytosolic compartment, leading to assembly of the NLRP3 inflammasome (Sander et al., 2011).Caspase-11 activation via the TLR4-TRIF-IFN-β pathway then synergizes with this bacterial mRNA-activated NLRP3 platform to orchestrate caspase-1-dependent IL-1β and IL-18 processing and secretion (Figure 6B). Caspase-11 also leads to cell death and release of endogenous alarmins, such as IL-1α and HMGB1. The identification of TRIF signaling as a key determinant of caspase-11 induction and activation and the crosstalk revealed between TLR and NLR pathways provide important insights into the integration of signaling events during Gram-negative bacterial infection. This study establishes TLRs as “master” regulators of inflammasome activation by revealing the utilization of distinct modules of TLR signaling to orchestrate IL-1β-driven inflammation. By engaging MyD88 downstream of TLR4, Gram-negative bacteria turn on transcription of pro-IL-1β and of Nlrp3 mRNA (signal 1). Phagocytosis of bacteria and the destructive environment of the phagolysosomal compartment then lead to the release of bacterial mRNA into the cytosolic compartment, allowing assembly of the NLRP3 inflammasome (signal 2). Simultaneously, engagement of TRIF downstream of TLR4 couples transcription of caspase-11 to its autoactivation via type I interferons (signal 3). Activated caspase-11 synergizes with the bacterial mRNA-assembled NLRP3 complex to coordinate caspase-1 activation and maturation of IL-1β and IL18. These studies support the rational use of neutralizing type I IFN antibodies and JAK inhibitors, as well as the design of additional therapies targeting this caspase-11 pathway for the treatment of detrimental inflammation associated with infectious diseases caused by a range of Gram-negative bacterial pathogens.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice from The Jackson Laboratory (Bar Harbor, ME) were bred at UMASS Medical School. Trif−/−, Nlrp3−/−, Tlr4−/−, Tlr3−/−, Tram−/−, Irf3−/−Irf7−/−, Ifnar1−/−, Stat1−/− (kind gift of Christine Biron, Brown University), Irf9−/− (kind gift of Karen Mossman, McMaster University), Myd88−/−, and caspase-11−/− (kind gift of Vishva Dixit, Genentech) were all on C57BL/6 background. 129X1/SvJ mice were purchased from The Jackson Laboratory. Sting−/− mice (Sauer et al., 2011) and caspase-8−/− Rip3−/− and caspase-8+/− Rip3−/− mice have been described previously (Kaiser et al., 2011). Mouse strains were maintained in specific pathogen-free conditions in UMASS Medical School, and the animal protocols were carried out in accordance with the guidelines set forth by UMASS Medical School Institutional Animal Care and Use Committee.
Bacteria
EHEC (EDL933 strain) and C. rodentium (ATCC 51459) were grown overnight in LB broth, and the macrophages were infected at an MOI of 25 unless otherwise indicated. Strain names and MOI for additional bacteria used in this study are given in Table S1.
BMDM Culture and Stimulations
Bone marrow cells from wild-type and various knockout mice were cultured in DMEM with 10% fetal bovine serum and 20% L929 supernatants. BMDMs were primed with 400 ng/ml Pam3CSK4 (Invivogen) unless otherwise indicated for 4 hr and then stimulated with bacteria at indicated MOIs for 1 hr, and then medium was replaced with medium containing gentamicin (100 µg/ ml). The supernatants were collected 16 hr postinfection. In certain experiments, the cells were also stimulated with indicated doses of recombinant murine IFN-β (PBL Interferon Source) or IFN-γ (R & D systems) at the time of infection. For control purposes, the cells were transfected with polydAdT (1 µg/106 cells; Sigma-Aldrich) or stimulated with silica (200 µg/ml) or nigericin (10 µM; Sigma-Aldrich). Additionally, BMDMs immortalized with J2 virus have been used where indicated.
ELISA and Cell Death Assay
Cell culture supernatants, serum, and peritoneal lavages were assayed by ELISA for IL-1β (BD Biosciences), IL-1α (BD Biosciences), IL-18 (eBioscience), and IL-6 (eBioscence). A sandwich ELISA for mouse IFN-β was used as described (Roberts et al., 2007). Cell death was assessed by CellTiter-Glo luminescent cell viability assay (Promega).
Immunoblotting
Proteins from the cell culture supernatants were precipitated by methanolchloroform extraction method. Cells were lysed with 1% NP-40 lysis buffer. Immunoblot analysis was done with antibodies to mouse caspase-1 p10 (sc-514; Santa Cruz Biotechnology), mouse caspase-1 p20 (clone 5B10; eBioscience), mouse IL-1β (AF-401-NA; R&D Systems), mouse caspase-11 (clone 17D9; Sigma-Aldrich), mouse HMGB-1 (clone 3E8; BioLegend), mouse NLRP3 (clone cryo-2, Enzo Life Sciences), ASC (sc-22514-R; Santa Cruz Biotechnology), b-actin, and GAPDH (clone 71.1, Sigma).
Gentamicin-Killing Assay
This assay was performed as described previously (Rathinam et al., 2008). In brief, BMDMs were infected with EHEC at an MOI of 25. After 1 hr of infection, nonphagocytosed bacteria were removed by treating the cells with 100 µg/ml of gentamicin. At specified time points, intracellular viable bacteria were counted by lysing the cells with 0.1% Triton X-100 and spreading serial dilutions of lysates on LB agar plates. Colonies were counted after overnight incubation at 37°C.
Quantitative RT-PCR
RNA was extracted from infected BMDMs at indicated time points using RNeasy kit (QIAGEN). cDNA was synthesized from total RNA using the iScript Select cDNA synthesis kit (Bio-Rad). Quantitative RT-PCR for caspase-11 was performed by using iQ SYBR green supermix (Bio-Rad) and the primers (forward, 5′-ACAATGCTGAACGCAGTGAC-3′; reverse, 5′-CTGGTTCCTCCATTTCCAGA-3′; Kayagaki et al., 2011). Caspase-11 mRNA in the samples was normalized to that of β-actin or GAPDH, and the fold difference in caspase-11 transcript levels was analyzed by Livak’s method (Livak and Schmittgen, 2001).
Phagosome Acidification Assay
Ratiometric acidification assay was done as previously described (Ip et al., 2010). In brief, immortalized or primary BMDMs were incubated with heat-inactivated (65°C for 30 min) E. coli K12 or EHEC at low MOI (≤5) for 30 min on ice to synchronize phagocytosis. In all cases, cells were prechilled for 15 min on ice before adding bacteria. Bacteria were labeled with FITC (pH-sensitive) and Alexa Fluor 647 (pH-insensitive) fluorescent dyes, and bacterial clusters were disrupted by repeated passage through a 30 gauge needle before incubation with macrophages. Cells were further incubated at 37°C for 5, 30, or 60 min. Next, cells were washed twice with ice-cold PBS with 5 mM EDTA, detached, and resuspended in PBS. Cells were analyzed by flow cytometry to determine the MFI emission ratio between FITC and Alexa Fluor 647 of the bacteria inside phagosomes. To calculate the pH using the ratiometric assay, values were compared with a standard curve. For standard curve, cells after 2 hr of phagocytosis were permeabilized for 10 min at room temperature in buffers with a fixed pH (ranging from pH 3.5 to 8) containing 0.05% Triton X-100. The cells were analyzed by flow cytometry to determine the emission ratio of the two fluorescent dyes at each pH.
ASC Oligomerization Assay
ASC oligomerization assay was performed as described with minor modifications (Fernandes-Alnemri et al., 2007). In brief, BMDMs were primed with Pam3CSK4 for 3 hr and stimulated with EHEC or C. rodentium for 6 hr or polydAdT for 3 hr or nigericin for 30 min. Cytosolic lysates from the cells were enriched for inflammasome fractions by low-speed centrifugation and subjected to cross-linking with disuccinimidyl suberate (2 mM). The cross-linked samples were analyzed for ASC and caspase-1 oligomerization by immunoblotting.
In Vivo Infection
Age- and sex-matched C57BL/6 and Ifnar1−/− mice were infected with 109 CFUof E. coli BL21 strain to induce acute peritonitis and shock. Cytokine levels in the serum and peritoneal lavage were analyzed at 6 hr postinfection. Data from in vivo experiments were analyzed by unpaired two-tailed Student’s t test with Prism software. p values of less than 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Anna Cerny for animal husbandry and genotyping; Sanjay Ram, Douglas Golenbock, Rahul Gupta, Victor Boyartchuk, Brian Akerley, Sandy Wong, Beth McCormick, and Stephen Lory for bacterial strains; Christine Biron and Delia Demers for Stat-1−/− femurs; Karen Mossman and Derek Cummings for Irf9−/− femurs; Vishva Dixit and Nobuhiko Kayagaki for caspase-11−/− mice and additional reagents; Bill Kaiser for caspase-8−/−Rip3−/− and caspase-8+/−Rip3−/− femurs; Jakob von Moltke and Russell Vance for immortalized BMDMs; Egil Lien, Douglas Golenbock, and Ann Rothstein for critical reading of the manuscript; and all members of the Fitzgerald lab for helpful discussions. This work is supported by NIH grants AI083713 (to K.A.F.), AI046454 (to J.M.L.), and AI079198 (to L.M.S.) and NERCE Post-Doctoral Fellowship Awards NIH/NIAID U54 AI057159 to V.A.K.R and S.K.V.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.07.007.
REFERENCES
- Costa A, Gupta R, Signorino G, Malara A, Cardile F, Biondo C, Midiri A, Galbo R, Trieu-Cuot P, Papasergi S, et al. Activation of the NLRP3 inflammasome by group B streptococci. J. Immunol. 2012;188:1953–1960. doi: 10.4049/jimmunol.1102543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-kappaB activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gao X, Huang MT, O’Connor BP, Thomas CE, Willingham SB, Bergstralh DT, Jarvis GA, Sparling PF, Ting JP. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J. Immunol. 2009;182:6460–6469. doi: 10.4049/jimmunol.0802696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Tsuchiya K, Kawamura I, Shen Y, Hara H, Sakai S, Yamamoto T, Fernandes-Alnemri T, Yang R, Hernandez-Cuellar E, et al. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J. Immunol. 2011;187:4890–4899. doi: 10.4049/jimmunol.1100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kanneganti TD, Dubyak GR, Núñez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J. Biol. Chem. 2007;282:18810–18818. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J. Biol. Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- Ip WK, Sokolovska A, Charriere GM, Boyer L, Dejardin S, Cappillino MP, Yantosca LM, Takahashi K, Moore KJ, Lacy-Hulbert A, Stuart LM. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J. Immunol. 2010;184:7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Wang S, Hara H, Peterson EP, Namura S, Amin-Hanjani S, Huang Z, Srinivasan A, Tomaselli KJ, Thornberry NA, et al. Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 2000;149:613–622. doi: 10.1083/jcb.149.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Ozören N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Hoag KA, Mansfield LS. Dendritic cells from C57BL/6 mice undergo activation and induce Th1-effector cell responses against Campylobacter jejuni. Microbes Infect. 2008;10:1316–1324. doi: 10.1016/j.micinf.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat. Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ZJ, Goutagny N, Perera P-Y, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J. Exp. Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Müller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J-D, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauvliege R, Vanrobaeys J, Schotte P, Beyaert R. Caspase-11 gene expression in response to lipopolysaccharide and interferon-gamma requires nuclear factor-kappa B and signal transducer and activator of transcription (STAT) 1. J. Biol. Chem. 2002;277:41624–41630. doi: 10.1074/jbc.M207852200. [DOI] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miura M, Jung Y, Zhu H, Gagliardini V, Shi L, Greenberg AH, Yuan J. Identification and characterization of Ich-3, a member of the interleukin-1beta converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J. Biol. Chem. 1996;271:20580–20587. doi: 10.1074/jbc.271.34.20580. [DOI] [PubMed] [Google Scholar]
- Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- Weber A, Kirejczyk Z, Besch R, Potthoff S, Leverkus M, Häcker G. Proapoptotic signalling through Toll-like receptor-3 involves TRIF-dependent activation of caspase-8 and is under the control of inhibitor of apoptosis proteins in melanoma cells. Cell Death Differ. 2010;17:942–951. doi: 10.1038/cdd.2009.190. [DOI] [PubMed] [Google Scholar]
- Willingham SB, Bergstralh DT, O’Connor W, Morrison AC, Taxman DJ, Duncan JA, Barnoy S, Venkatesan MM, Flavell RA, Deshmukh M, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Allen IC, Bergstralh DT, Brickey WJ, Huang MT, Taxman DJ, Duncan JA, Ting JP. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J. Immunol. 2009;183:2008–2015. doi: 10.4049/jimmunol.0900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- Yen JH, Ganea D. Interferon beta induces mature dendritic cell apoptosis through caspase-11/caspase-3 activation. Blood. 2009;114:1344–1354. doi: 10.1182/blood-2008-12-196592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.