Abstract
The discovery of cisplatin as a therapeutic agent stimulated a new era in the application of transition metal complexes for therapeutic design. Here we describe recent results on a variety of transition metal complexes targeted to DNA to illustrate many of the issues involved in new therapeutic design. We describe first structural studies of complexes bound covalently and non-covalently to DNA to identify potential lesions within the cell. We then review the biological fates of these complexes, illustrating the key elements in obtaining potent activity, the importance of uptake and subcellular localization of the complexes, as well as the techniques used to delineate these characteristics. Genomic DNA provides a challenging but valuable target for new transition metal-based therapeutics.
Introduction
Since the successful application of cisplatin as an anticancer drug, the field of inorganic medicinal chemistry has undergone a revolution.1 Several additional platinum complexes have achieved FDA approval for cancer treatment.2 Two ruthenium complexes are currently in clinical trials,3,4 and studies on the biological effects of potential metal-based therapeutics are being published at an increasing rate. Anticancer compounds have a myriad of targets (DNA, proteins, membranes, etc.), and in fact the true lesion responsible for the biological activity of a compound is difficult to determine.5 Nevertheless, this article focuses on compounds that are not unlike cisplatin, in that the complexes are thought to have DNA as their main target in cellulo.
Here we discuss studies of several metal complexes to explore their differences in structural interactions with DNA, their biological fates inside the cell, and the tools and techniques being used to probe the path taken by the small molecule in reaching its DNA target. Establishing cellular uptake and even the subcellular distributions of the metal complexes are critically important in understanding and optimizing their activity. While the subtle hydrolysis reactions associated with cisplatin uptake probably could not have been strategically designed, they are key to its mode of action. This review is illustrative rather than comprehensive in its approach, yet hopefully these illustrations provide a foundation for considering strategies for new design and for elucidating mechanisms of action.
DNA as a Target
DNA represents a fruitful target for metal complexes. DNA can function as a ligand either through interactions with the sugar-phosphate backbone or coordination to the bases. Moreover, non-covalent interactions with DNA lead to additional targets, and greater specificity, through an ensemble of interactions in the DNA grooves and base stack.
Covalent Interactions
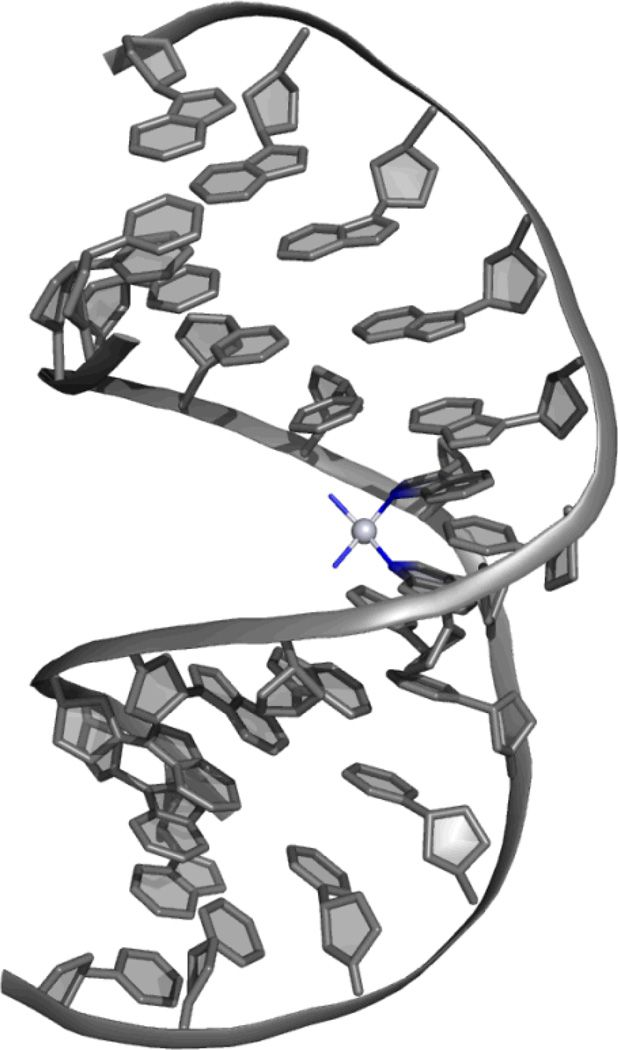
It has been widely accepted since the 1970’s that DNA is the biological target of cisplatin in vivo.6,7 However, the nature of the adduct formed between cisplatin and DNA was not determined until the 1980’s. This interaction was proposed to be an intrastrand cross-link between the N7 atoms of adjacent guanines and cisplatin based on the results of numerous biochemical studies.8,9 This adduct was further characteriz.10 However, this adduct was not fully structurally characterized by x-ray crystallography until 1995, when the Lippard group published the 2.6-Å resolution crystal structure of cisplatin bound to a double-stranded DNA dodecamer (Figure 1).11
Figure 1.
Crystal structure at 2.6-Å resolution of the intrastrand crosslink between cisplatin and the N7 atoms of adjacent guanines.11 The platinum center is shown as a light grey sphere, and its four nitrogen-based ligands are shown in blue. DNA is shown in grey. The dodecamer duplex is bent considerably towards the major groove, and the structure is no longer B-form DNA, but mostly A-form throughout the duplex.
In this structure, the duplex is bent considerably toward the major groove but without disruption of the Watson-Crick hydrogen bonding. In fact, the duplex is distorted to such a degree that the duplex changes conformation from B-DNA to A-DNA throughout most of the duplex. Such a significant distortion of the DNA is likely readily recognized by a host of cellular proteins. How efficiently such lesions are recognized and repaired rather than initiating a protein response and signaling cascade, is a question we still need to understand and which may make the difference between biological efficacy and little reaction.12
Intercalation
Metallointercalation is a DNA binding mode that has been extensively studied. The term was coined by Lippard and coworkers in studies of square planar Pt complexes with DNA.13 As with organic intercalators, these planar complexes containing aromatic heterocyclic ligands could stack among the DNA base pairs. However, the lack of site-specificity inherent in intercalation by a planar complex made detailed structural characterization difficult. The first structural characterization of metallointercalation was the 1.1-Å resolution crystal structure of the platinum complex [Pt(SEtOH)(terpy)]+ intercalated into the dinucleotide dimer deoxy CpG.14 Although not able to establish long range structural perturbations to DNA associated with intercalation, this structure did reveal that associated with intercalation, the DNA unwinds to accommodate the metal complex between bases and the pucker of the sugar rings changes geometry. This alternate sugar puckering was suggested as the basis for the “neighbor exclusion principle” associated with DNA intercalation, where at most intercalators bind in every other interbase-pair site.
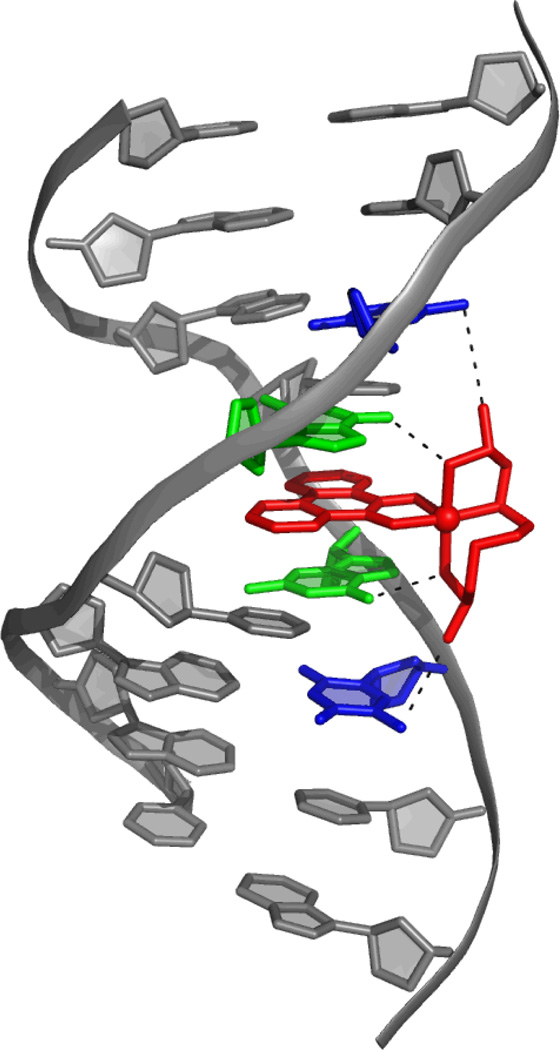
In succeeding years, our laboratory focused on intercalation by octahedral complexes containing at least one aromatic heterocyclic ligand for stacking, or partial intercalation, in between base pairs. The symmetry and functionality associated with the non-intercalated ligands could then provide the basis for highly specific interactions with several bases along the groove of DNA, once oriented by stacking in the helix of the intercalated ligand. In particular, we found the enantioselective intercalative binding of right-handed Δ-complexes into right-handed B-DNA.15 One of the first structural characterizations of metallointercalation into a long DNA duplex was thus provided from the 1.2-Å resolution crystal structure of the sequence-specific rhodium intercalator, Δ-α-[Rh[(R,R)-Me2trien]phi]3+, bound to a duplex octamer (Figure 2).16 The complex was designed to target the sequence 5’-TGCA-3’ through a mix of hydrogen bonding and methyl-methyl interactions in the DNA major groove. The complex was also shown as a result of its sequence-specificity to inhibit the binding of sequence-specific DNA-binding proteins, a first step in specifically inhibiting gene expression.17
Figure 2.
The 1.2-Å resolution crystal structure of the sequence-specific rhodium intercalator, Δ-α-[Rh[(R,R)-Me2trien]phi]3+, bound to a duplex octamer.16 The complex binds specifically to 5’-TGCA-3’. There are two Van der Waals Me-Me interactions between the metal complex ligand and T (T shown in blue, interaction shown with the black dotted line), and a hydrogen bond between the NH group on the metal complex ligand and the O6 of G (G shown in green, interaction shown with the black dotted line). This structure displays the doubling of the rise of the DNA, buckling of the adjacent base pairs, and a slight unwinding of the DNA upon intercalation.
Critically, the structure obtained provided detailed information on how metallointercalation in general modifies the conformation of DNA. In this structure, intercalation occurs from the major groove, with the aromatic intercalating phi ligand π-stacking with the π-orbitals of the flanking base pairs, similar to the stacking of consecutive base pairs in duplex DNA. This structure also confirmed the conformational changes revealed by shorter intercalated oligonucleotides such as doubling of the rise, buckling of the base pairs flanking the intercalation site, and a slight unwinding of the DNA localized at the site of intercalation. Interestingly, there were no long range effects on the DNA structure, no bending or kinking of the helix. Indeed, even the alternating sugar-pucker was not evident. The metallointercalator was simply like another base pair in the helical stack.
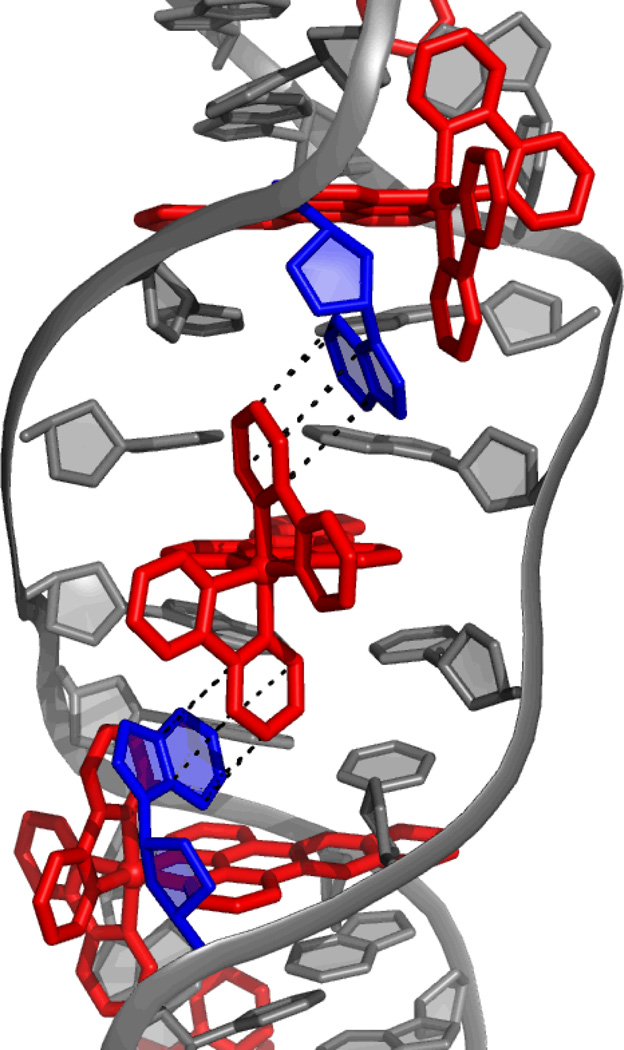
Structural characterization of intercalation from the minor groove was recently obtained from two independent crystal structures of dppz complexes of Ru(II) intercalated into duplex DNA. The first is the 0.92-Å resolution structure of two Δ-[Ru(bpy)2(dppz)]2+ complexes intercalated from the minor groove into a duplex 12-mer containing two mismatched sites.18 There are also two ruthenium complexes bound to the mismatched sites via insertion (vide infra), and the extruded adenosines π-stack with the bpy ligands of the intercalated complexes, serving to stabilize the intercalated complex in the minor groove (Figure 3). Intercalation of this complex from the minor groove was in contrast to NMR19,20 and competitive fluorescence studies21, both of which suggested that intercalation of the ruthenium complex occurred from the major groove. Crystal packing forces may play a role here in directing intercalation from the minor groove side; in any case it is clear that the energetic differences between intercalation from the major groove versus minor groove must be small. Furthermore, this structure showed the doubling of the rise of the DNA at each intercalation site, and an unwinding of the base pairs to accommodate the complexes. Interestingly, while the rise is doubled at each intercalation site, the rise between the base pairs with no metal complex bound is reduced from the 3.3Å that is expected, consistent with π-stacking interactions between the extruded mismatches and the ancillary bpy ligands being the dominating interaction that may be directing intercalation from the minor groove.
Figure 3.
The 0.92-Å resolution structure of two Δ-[Ru(bpy)2(dppz)]2+ complexes intercalated from the minor groove into a duplex 12-mer containing two AA mismatched sites.18 The mismatches are extruded from the base stack by two other Δ-[Ru(bpy)2(dppz)]2+ complexes that are metalloinserted at these sites. The extruded bases (shown in blue) π-stack with the bpy rings of the intercalated metal complexes (interactions shown in black, metal complexes shown in red).
The second set of structures show the binding of Λ-[Ru(phen)2(dppz)]2+ to two different duplex 10-mers.22 These structures reveal three different conformations of intercalation for the same complex. Namely, when intercalated at the TA/TA central step of the oligonucleotide d(CCGGTACCGG)2, the complex is intercalated deeply, perpendicularly, and symmetrically into the base stack from the minor groove. However, when the complex intercalates at the terminal GG/CC step, the intercalation geometry is shallower and angled. Finally, this angled intercalation allows for the phenanthroline ligands to semi-intercalate into the neighboring duplex. This “semi-intercalation” was also seen in a structure of Λ-[Ru(TAP)2(dppz)]2+ bound to a duplex 10-mer.23
These various structural characterizations of metallointercalation highlight the versatility of this binding mode, and likely also the shallow energy profile among different intercalative binding modes. Likely the structural diversity reflects the sequence selectivity associated with the different complexes, where the non-intercalating interactions in the DNA groove lead to some structural variations. But none of these conformational changes yield dramatic changes in DNA structure, like the bend in DNA generated by a Pt crosslink. In the context of therapeutic design, the fact that metallointercalators cause no major structural distortions in DNA needs to be considered. Intercalators generally, especially those that have no sequence-specificity, have little therapeutic applicability. Perhaps it is the fact that a strongly defined DNA lesion is not produced with intercalation that limits the biological consequences for metallointercalators.
Insertion
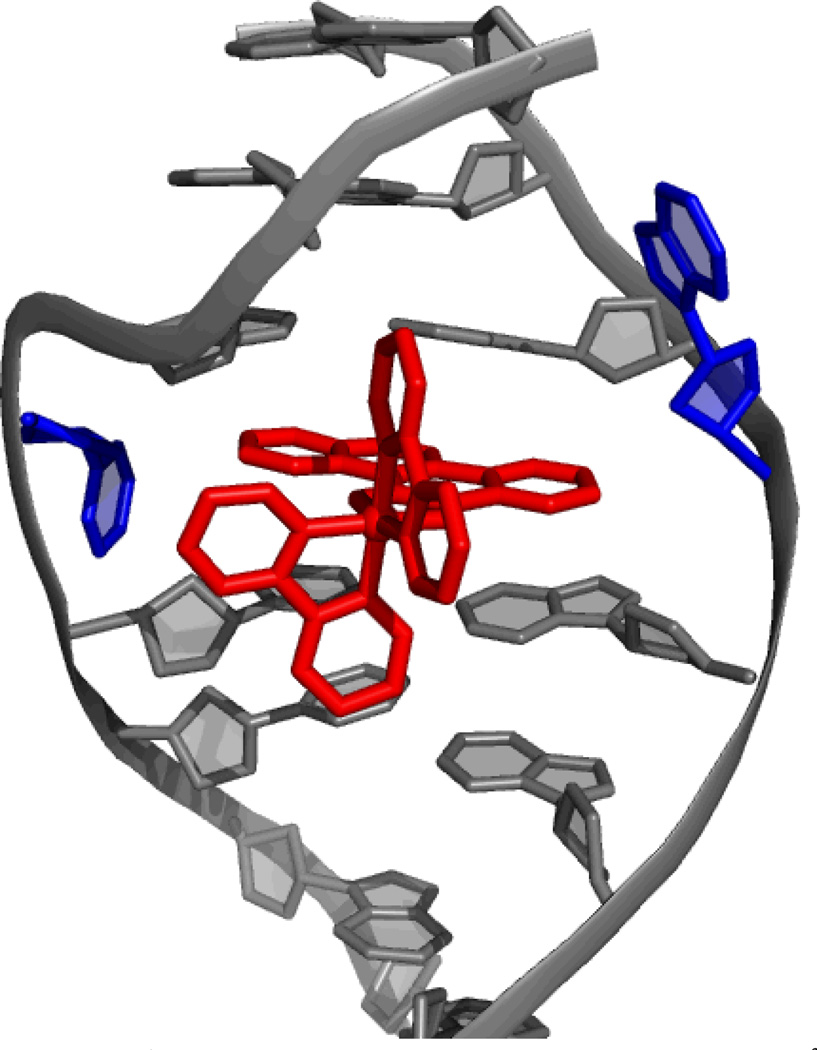
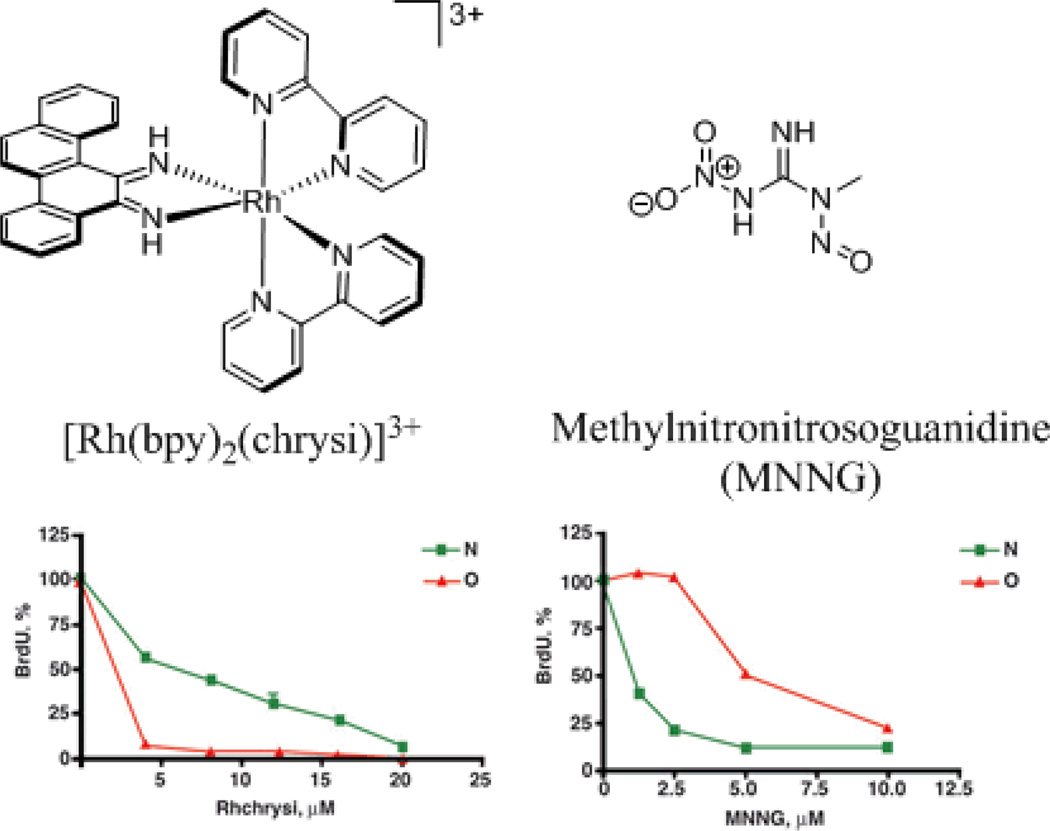
Rhodium metalloinsertors, as described in our laboratory, bind to mismatches in DNA with high affinity and specificity.24,25 While it was known that these compounds could preferentially target thermodynamically destabilized mismatches in DNA over matched base pairs by a factor of over 1000, 26 for ten years there was no structural information on the interaction between these metal complexes and mismatched DNA. The 1.1-Å resolution crystal structure of [Rh(bpy)2(chrysi)]3+ bound to an AC mismatch revealed the binding mode to be metalloinsertion, where the chrysi ligand inserts into the base stack via the minor groove and ejects both mismatches bases (Figure 4).27
Figure 4.
The 1.1-Å resolution structure of Δ-[Rh(bpy)2(chrysi)]3+ bound to an AC mismatch.27 The rhodium complex, shown in red, inserts into the DNA from the minor groove and completely ejects the mismatches bases, shown in blue.
Metalloinsertion of this complex results in only small conformational changes in the duplex near the binding site but a large perturbation is associated with the ejection of the mismatched bases into the DNA groove. The structure also explains the enantiospecificity of binding of [Rh(bpy)2(chrysi)]3+; the deep insertion of the complex within the minor groove with no increase in base pair rise results in a steric clash between the ancillary ligands and the sugar phosphate backbone if the left-handed isomer were to be bound. For intercalation, in contrast, where there is an increase in rise at the binding site, enantioselective intercalation requires a much bulkier ancillary ligand than bpy.28 As with the case of cisplatin, this structure may also suggest the basis for the biological activity of these complexes. Ejection of the mismatched bases results in a large lesion that could be easily recognized in vivo. This lesion likely is responsible for the selective cell death of MMR-deficient cells over MMR-proficient cells following rhodium treatment, as there are 1000 times as many mismatches in the MMR-deficient cells. This binding conformation has been corroborated by additional crystal structures of this same compound bound to different mismatches.29 Furthermore, the generality of metalloinsertion as a binding mode for different bulky metal complexes has been established by the crystal structure mentioned earlier, that of Δ-[Ru(bpy)2(dppz)]2+ bound by insertion at two mismatched sites.18 Again, the information garnered from these structures is instrumental in unraveling the mechanism of action of these therapeutic agents, and thus in the development of future agents with improved biological activity.
Biological Activities of Metal Complexes
Undeniably, optimizing a compound as a therapeutic requires the complete assessment of its biological activity in vitro. Here we focus on the biological activity in cellulo of several classes of transition metal complexes that are thought to target DNA. As the activities of cisplatin and other platinum compounds have been extensively discussed elsewhere,12,30,31 we describe here the characterization of non-platinum-based therapeutics.
Polypyridyl Complexes
The biological activities of countless ruthenium polypyridyl complexes have been reported in the literature and have been the subject of many reviews.32 Indeed, the exploration of biological activities of polypyridyl complexes began more than fifty years ago in classic studies by Dwyer and coworkers.33 At that stage, no biological target was identified, but the more recent studies on coordinatively saturated metal complexes suggest that DNA was likely the target for the full family of complexes examined earlier.
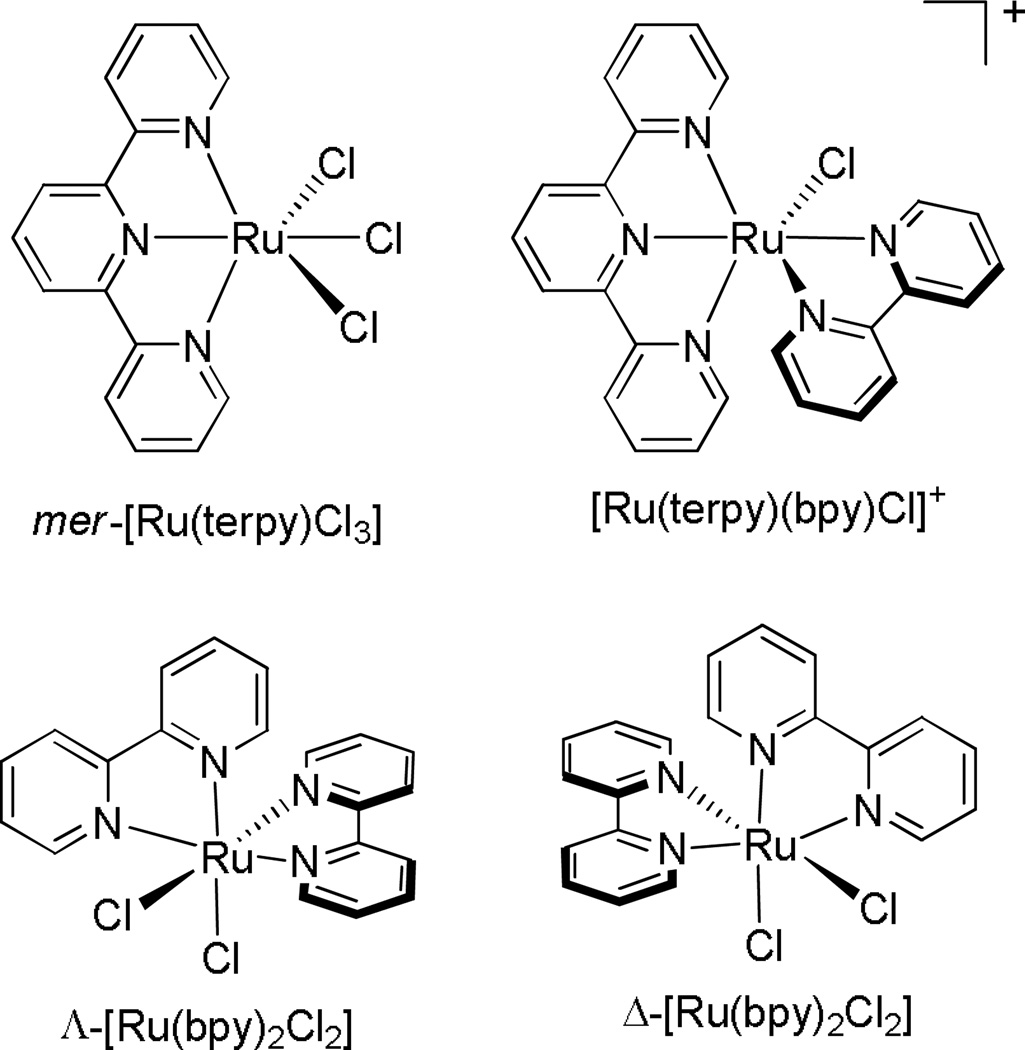
Many more studies have been conducted on ruthenium complexes that bind DNA covalently, by analogy to cisplatin, than those which are coordinatively saturated and inert to substitution, binding non-covalently. In one of the earliest studies, Novakova and coworkers studied four chloropolypyridyl ruthenium complexes (Figure 5) in murine and human tumor cell lines. Interestingly, only the complex with three leaving chloride ligands, mer-[Ru(terpy)Cl3], displayed significant cytotoxicity. While the binding affinities of the different complexes could not explain this discrepancy, it was discovered that only the mer- [Ru(terpy)Cl3] complex had the ability to form interstrand crosslinks in the DNA, thus harkening on the importance of specific lesions and structural distortion in determining biological activities, not simply avidity for binding DNA.34
Figure 5.
Chemical structures of four ruthenium polypyridyl complexes studied by Novakova and coworkers.34 Out of all four complexes, only the mer-[Ru(terpy)Cl3] complex displays significant cytotoxicity in human and murine tumor cell lines.
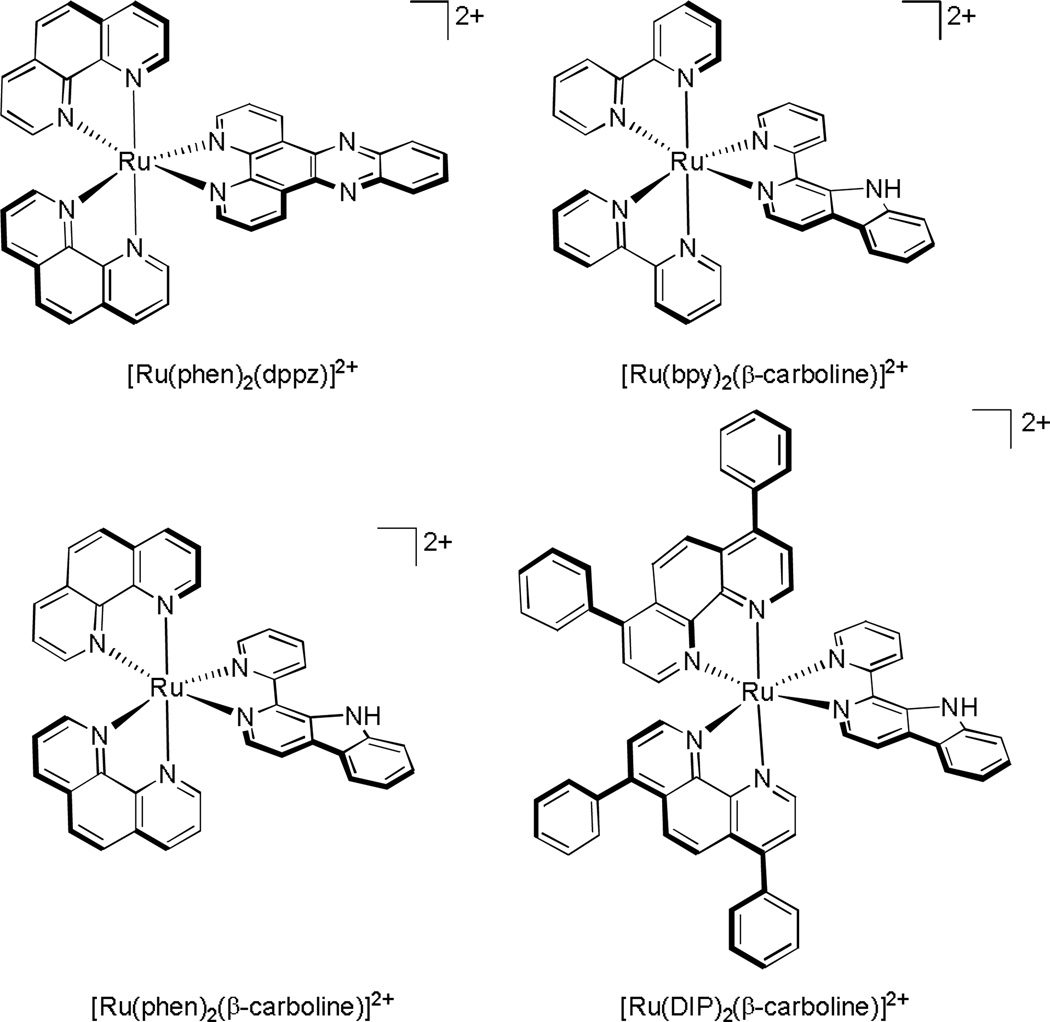
In a more recent study conducted by Tan and coworkers, several inert ruthenium polypyridyl complexes were studied, three ruthenium complexes containing a β–carboline ligand, along with the control compound [Ru(phen)2(dppz)]2+ (Figure 6). They observed that upon substitution of the dppz ligand for the β–carboline ligand, the complex now accumulates in the nucleus as well as the cytoplasm. Furthermore, the complexes with β–carboline ligands are significantly more cytotoxic towards HeLa cells, inducing apoptosis and autophagy, while [Ru(phen)2(dppz)]2+ does not. Since the cytotoxicities of the β–carboline complexes correlated well with their DNA binding affinities, they concluded that genomic DNA may be their primary target in cellulo.35 The comparative results with these complexes, however, also suggest that the differential activities of the complexes may depend on more than just affinities. Both cellular uptake and localization may be issues. Moreover the bulkiness and geometry of the coordinated carboline ligand may indicate that metalloinsertion plays some role in the binding interaction.
Figure 6.
Four Ru(II) polypyridyl complexes studied by Tan and coworkers.35 All three complexes with β-carboline as a ligand were cytotoxic towards HeLa cells, whereas the control [Ru(phen)2(dppz)]2+ complex was not.
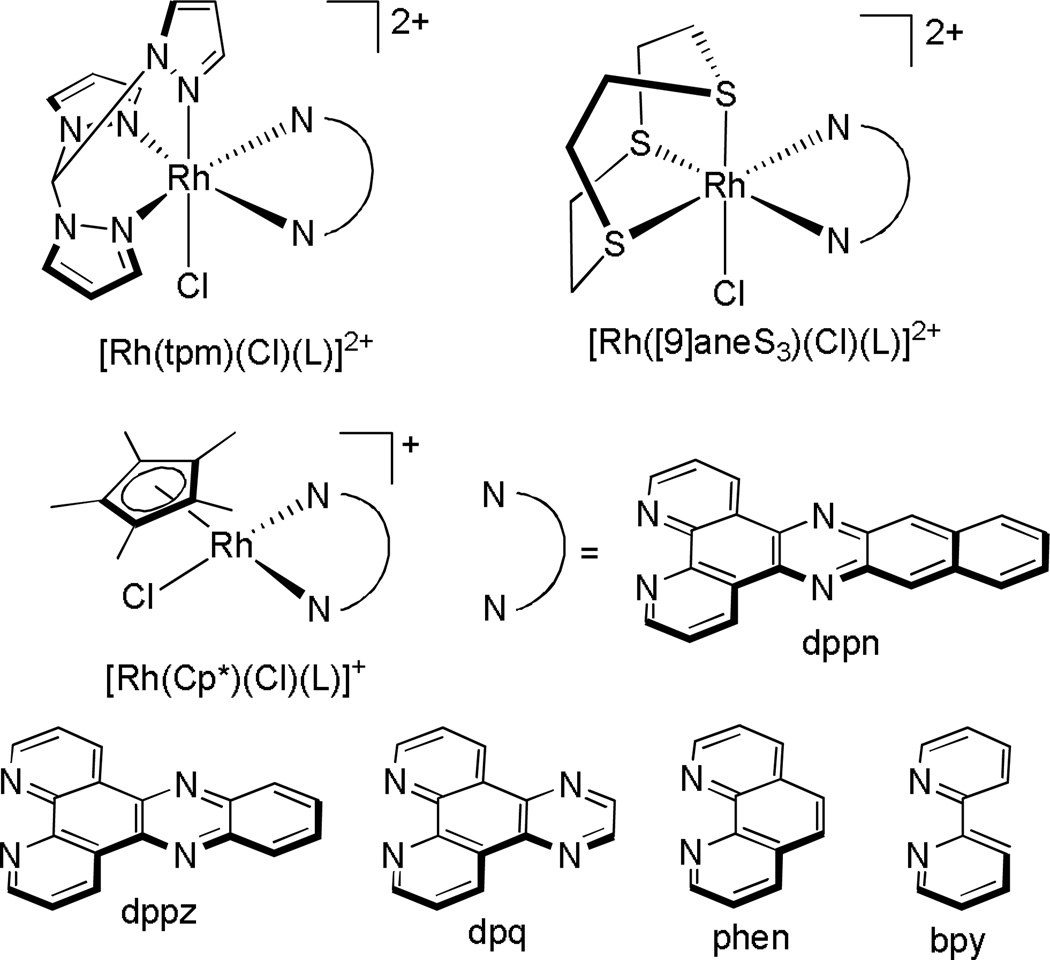
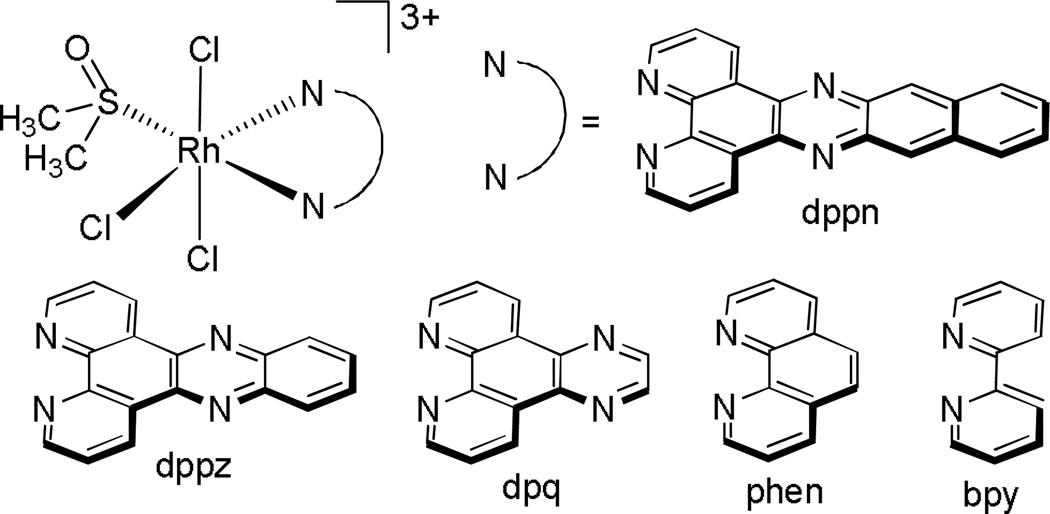
The Sheldrick laboratory has focused on studies of a variety of rhodium polypyridyl complexes that can bind both covalently and non-covalently with DNA.36,37 A series of polypyridyl rhodium complexes containing a facial tripodal (tpm), Cp*, or thioether ([9]aneS3) ligand (Figure 7) were synthesized and their cytotoxicities towards the MCF-7 and HT-29 cell lines examined. They observed, in general, while keeping the facial ligand constant, an increase in potency with increasing surface area and hydrophobicity of the polypyridyl ligand. Furthermore, it was found that for the polypyridyl ligands phen, dpq, and dppz, the facial ligands had the effect of increasing the cytotoxicity in the order [Cp*]−<[9]aneS3<tpm. This trend was found to be consistent not with binding affinity, but with the magnitude of rhodium uptake into the cells.38–40 Some of these complexes were furthermore found to selectively target lymphoma (BJAB) cells over healthy leukocytes.41
Figure 7.
Chemical structures of the compounds studied by the Sheldrick laboratory.38–41 An extensive structure-activity study was undertaken on all complexes in the MCF-7 and HT-29 cell lines. Upon increasing the surface area of the “L” ligand, the cellular uptake of the drug increased, resulting in an increased potency of the drug.
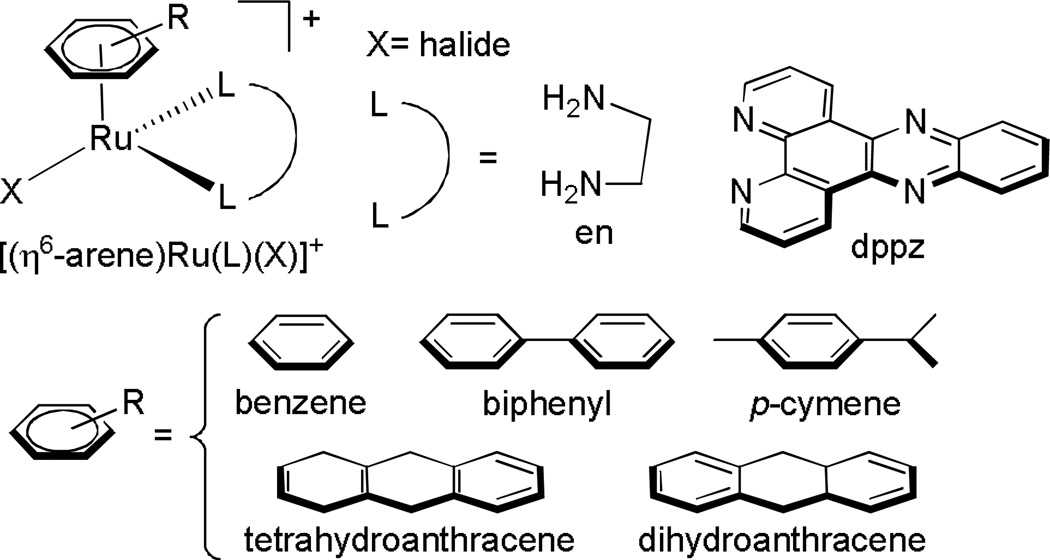
Ruthenium Arene Complexes
There have been many studies exploring the cytotoxicities of “piano stool” complexes.42 Compounds of the type [(η6-arene)Ru(L)(X)]+ (where L is a bidentate ligand and X is a halide, Figure 8) have been shown to exhibit anticancer activity,43 while their analogs with three monodentate ligands are completely inactive towards A2780 human ovarian cancer cells.44 In particular, the results from a few studies by the Sadler laboratory revealed that while the identity of the halide and chelating ligand had minor effects on the biological activities of these complexes towards A2780 cells, the size of the arene ligand had a major effect. The potency of the drug followed the trend benzene < p-cymene < biphenyl < dihydroanthracene < tetrahydroanthracene, which suggests increased cellular accumulation enhances their activity. 44,45 It was also noted that the arene complexes of the form type [(η6-arene)Ru(en)(Cl)]+ (en=ethylenediamine) did not display cross-resistance with cisplatin, suggesting an entirely different mechanism of action of these types of compounds.
Figure 8.
Compounds studied by the Sadler laboratory.44–48 It was discovered that as the lipophilicity of the arene ligand increased, the potency of the drug towards A2780 cells increased. It was also found that complexes with L=en have a mechanism of action distinct from cisplatin.
Further studies confirmed that these complexes bind to DNA and induce structural distortions to the DNA that are distinct from those induced by cisplatin.46 Specifically, they have been found to bind selectively to G bases in DNA oligonucleotides, regardless of the presence of other biologically relevant binding sites,47 along with partial intercalation of the arene ligand into the base stack.48
Rhodium Metalloinsertors
The biological activities of rhodium metalloinsertors, showing a preferential activity in mismatch repair (MMR) –deficient cells, reflect well on the initial strategy used for their design, namely targeting DNA mismatches and therefore cells that have a higher frequency of DNA mismatches, the MMR-deficient cells. First studies were designed and carried out to establish in vitro that the complexes bind to DNA mismatches with high affinity and specificity.24,25 Several years later, their unique biological activity was characterized. Two metalloinsertors were found to preferentially inhibit growth in MMR-deficient cells over MMR-proficient cells (Figure 9).49 Furthermore, while only the Δ-[Rh(bpy)2(chrysi)]3+ enantiomer binds to mismatches in vitro, likewise only this enantiomer was found to possess this biological activity, implying that the biological activity of these complexes originates from binding to mismatches in cellulo. Further evidence to support this notion was achieved in a succeeding study in which the ability of several different metalloinsertors with varying ancillary ligands to target MMR-deficient cells preferentially was directly correlated with their mismatch binding affinities.50 A subsequent study on the mechanism of activity of these metalloinsertors revealed that not only do they selectively inhibit growth of MMR-deficient cells, but after longer incubation times, they are also selectively cytotoxic, inducing necrosis in the MMR-deficient cells.51 The ability of these complexes to target MMR-deficient cells over MMR-proficient cells with this selectivity is distinctive. Commonly used chemotherapeutics, alkylators, DNA damaging agents as well as cisplatin, all suffer from a selective toxicity instead with MMR-proficient cells, leading to a build-up in resistance to MMR-deficient cancers (Figure 9).52 Moreover it appears that the selective activity of metalloinsertors in MMR-deficient cells is not only unique but also general to the full family of metalloinsertors. The further development of this class of compounds, then, might help overcome one of the largest issues we have with current platinum-based therapeutics: acquired or inherent resistance.
Figure 9.
Chemical structures (top) and biological activities (bottom) of the rhodium metalloinsertor [Rh(bpy)2(chrysi)]3+ (left), and the DNA alkylating agent MNNG (right) studied in our laboratory.49 MMR-proficient (green) and MMR-deficient (red) cells were treated with varying concentrations of each compound and the proliferation of growth was quantified via a BrdU incorporation assay. With the rhodium metalloinsertor, the MMR-deficient cells are preferentially targeted over MMR-proficient cells, whereas the DNA alkylating agent targets the MMR-proficient cells, a trend seen with many commonly used DNA-targeted therapeutics.
Cellular Uptake of Metal Complexes
Establishing biological activity of small molecules and complexes in the cellular milieu is clearly more complicated than establishing chemical targets and structures in a test tube. A key element underlying this complexity is whether and how the complex enters the cell. Does it ever make it to its test tube target?
Methods to monitor Cellular Uptake
Because transition metals like Rh, Ru, and Pt are not inherently found in the cell, techniques that focus on detecting metals, such atomic absorption spectroscopy (AAS) or inductively coupled plasma mass spectrometry (ICP-MS) are invaluable to monitor cellular uptake. The foundation of AAS is the aspect that metal ions absorb strongly at discrete, characteristic wavelengths. In this technique, the sample is suctioned into an atomizer, which reduces everything in the sample to its atomic states. The atoms are then irradiated, during which time they will absorb light of a certain energy which equals a specific electronic transition of a particular element. The remaining radiation then passes through a monochromator which picks out the wavelength of light specific to the metal of interest, and sends it to the detector. Quantitative information is obtained by taking the ratio of the flux without a sample to the flux with the sample and converting to concentration using the Beer-Lambert Law.53,54
ICP-MS is highly sensitive and is applicable to a wide range of metals. Furthermore, it is compatible with a wide range of sample matrices, including many biologically relevant ones. The sample is introduced into a spray chamber, where it is nebulized into an aerosol. These small droplets are then transferred to the inductively coupled plasma. The sample droplets are atomized and ionized by the plasma, leading to atomic, singly charged ions which are subsequently transferred to the mass spectrometer, which is used to determine the concentration of the metals of interest.55
Confocal fluorescence microscopy allows for the acquiring of high-resolution 3-D images with low background interference, and can be used to observe intracellular fluorescent compounds.56 As such, confocal fluorescence microscopy has been utilized in countless studies to probe the cellular uptake of metal complexes, but only those that are luminescent.57 However, due to differences in the quantum yields among different luminescent compounds, this technique cannot be utilized to quantify the amount of drug localized inside the cell. Another technique that can be used to monitor uptake of luminescent molecules is flow cytometry. In this technique, cells are individually counted based on the amount of luminescence inside of them. A stream of cells is passed through a laser beam, and the instrument records their light scatter and luminescence. The readout is a histogram showing the number of cells versus luminescence intensity. Importantly, combining either of these techniques with ICP-MS or AAS can give much more information than either technique alone.
Relationships Between Drug Uptake and Activity
One major disadvantage to cisplatin treatment is acquired resistance.58 While there are several different explanations that can account for this resistance, one important mechanism of cisplatin resistance is decreased cellular accumulation of the drug.59 Specifically, in an early study by Andrews and coworkers, AAS was used to quantify cisplatin accumulation into parent and cisplatin-resistant 2008 human ovarian carcinoma cells. The resistant cells, which exhibited a 3.3-fold resistance, displayed 50% less intracellular platinum than the parental cell line. 60 Since this study, there have been numerous investigations on various different cell lines, many of which employed AAS or ICP-MS, to corroborate this observation.
In a study done by the Sheldrick laboratory, a series of rhodium(III) polypyridyl complexes (Figure 10) was synthesized and their cytotoxicities towards MCF-7 and HT-29 cancer cells determined. It was noted that as the lipophilicity of the polypyridyl ligand was increased, the cytotoxicity increased as well, and they therefore measured the cellular accumulation of each compound using AAS. As with the cisplatin example, the most potent complexes exhibited the greatest amount of cellular rhodium accumulation.61
Figure 10.
Chemical structures of Rh(III) polypyridyl complexes synthesized and studied to monitor uptake. As complexes increased in lipophilicity (bpy to phen to dpq to dppz to dppn derivatives), their cellular uptake into MCF-7 and HT-29 cells increased, resulting in increased potency.61
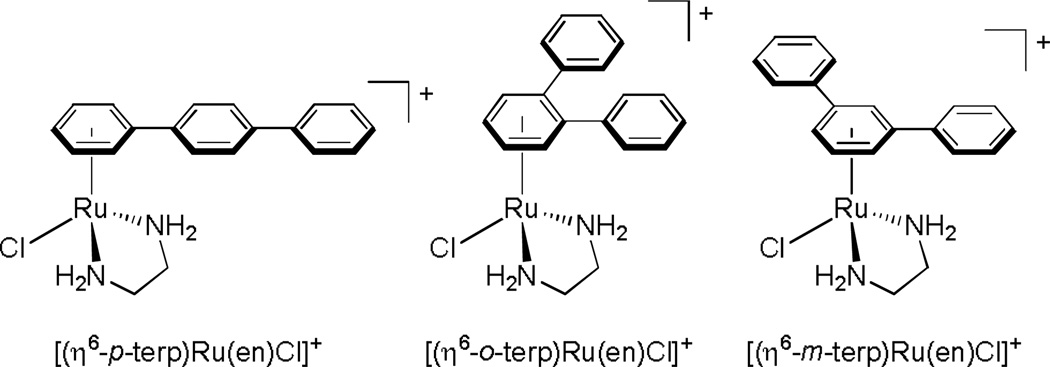
The relationship between intracellular drug concentration and efficacy is not always as straightforward as in the above studies, however. For example, in a study conducted by Bugarcic and coworkers, three isomeric terphenyl Ru(II) piano-stool complexes (Figure 11) were examined. Their cytotoxicities against two cisplatin sensitive and two cisplatin resistant cancer cell lines were determined, and their intracellular ruthenium concentrations were determined by AAS. Surprisingly, the extent of ruthenium uptake of the three complexes did not correlate at all with their different cytotoxicities. In fact, the most potent of the three complexes (the p-terp complex) displayed the least amount of ruthenium uptake into the cells. After numerous DNA binding studies, the authors attributed the enhanced cytotoxicity of this complex to its ability to not only covalently interact with DNA (as the other two isomers could) but also intercalate into DNA.62 Perhaps the structural distortions generated in this bound lesion produced a great cellular response. In any case, this study nicely highlights the fact that in many cases more information than just intracellular drug concentration is necessary in order to explain biological activity.
Figure 11.
Chemical structures of three isomeric terphenyl Ru(II) piano-stool complexes studied by Bugarcic and coworkers.62 Uptake of the complexes into two cisplatin resistant and two cisplatin sensitive cell lines was found not to correlate with their potencies.
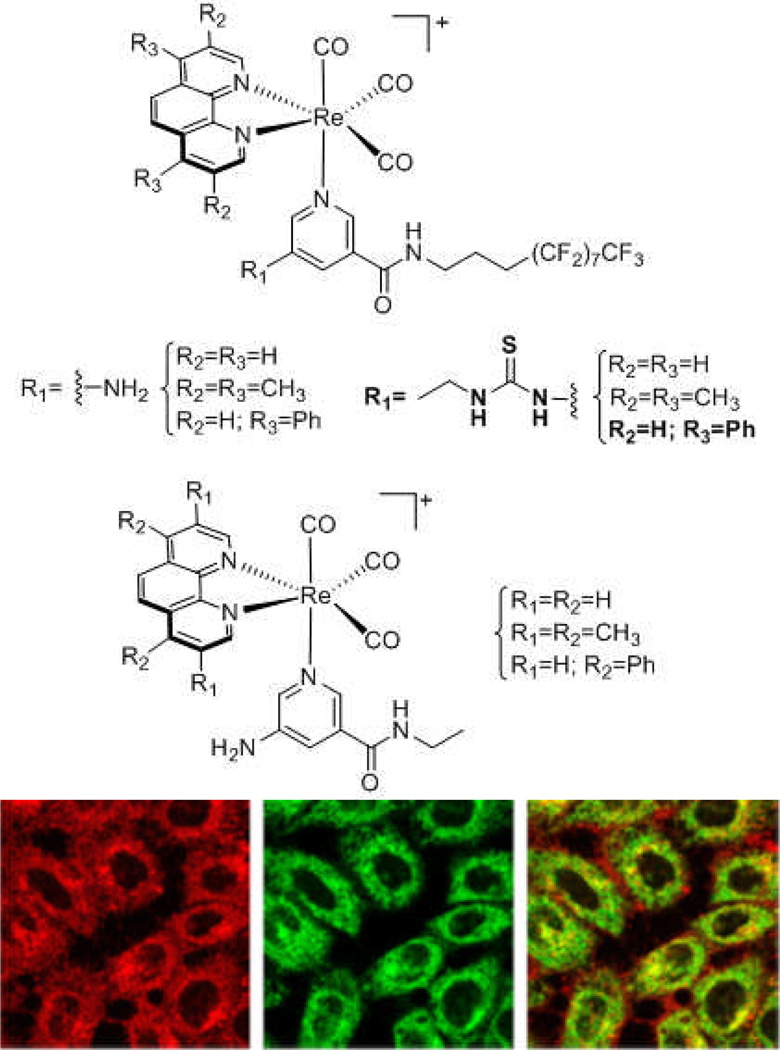
One study that combines ICP-MS and fluorescence microscopy to study the relationship among structure, activity, and uptake was done by Louie and coworkers. In this work, a series of luminescent rhenium(I) polypyridyl was examined (Figure 12). Here there was a direct correlation found between intracellular rhenium concentrations, detrmined by ICP-MS, and cytotoxicities in HeLa cells. Due to the luminescent nature of the complexes, the authors were also able to monitor uptake in live cells via confocal microscopy, and even observe the localization of the complexes in the mitochondria of the cells.63
Figure 12.
Chemical structures (top) of a series of luminescent rhenium(I) polypyridyl complexes by Louie and coworkers.63 Using ICP-MS, the authors were able to correlate the cytotoxicities of these complexes in HeLa cells with intracellular rhenium concentrations. Using fluorescence microscopy (bottom), the authors were able to observe the localization of one such complex (structure shown in bold, fluorescence shown on bottom left in red) in HeLa cells. The authors then used MitoTracker (bottom middle in green) to show that the complex localizes mainly in the mitochondria (bottom right, yellow shows the overlap). Copyright (2012) American Chemical Society
Mechanisms of Uptake
While knowing the relationship between the amount of a therapeutic taken into a cell and the activity of the drug is important, understanding how the compound gains entry into the cell is likewise crucial for optimization of next-generation complexes. The mechanism of cellular uptake of a drug can direct its localization within the cell as well as the specificity of a given compound for one cell type versus others.
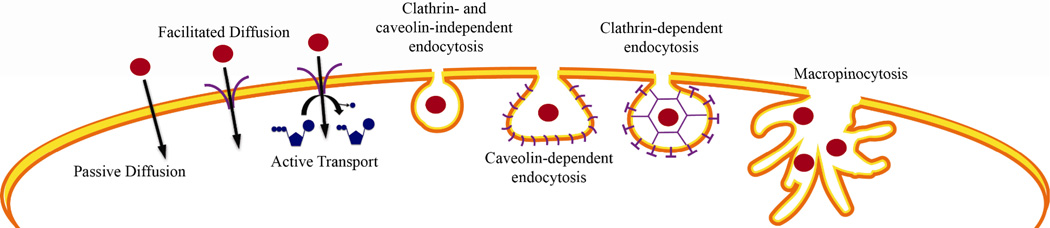
The different routes of entry into the cell include passive diffusion, facilitated diffusion, active protein transport, and endocytosis (Figure 13). Passive diffusion is the movement of the molecule of interest across the cell’s lipid bilayer, facilitated by the concentration gradient. Facilitated diffusion is the transport of the molecule of interest across the cell’s lipid bilayer, facilitated by a membrane-bound transport protein such as a channel or a passive carrier. Active transport is very similar, but the proteins involved in this type of transport are membrane-bound ATPases, meaning the process of moving a substance from the outside of the cell to the inside uses ATP. Endocytosis is a general term for the process by which the cell will “engulf” a molecule using a vesicle formed from the plasma membrane.64 Endocytosis can be broken down into five different categories: macropinocytosis, clathrin-mediated endocytosis, caveolin-mediated endocytosis, clathrin- and caveolin-independent endocytosis, and phagocytosis. All forms of endocytosis involve the formation of a membrane compartment, and simply differ in the size and composition of the compartments involved. Phagocytosis involves the uptake of particles larger than 0.5 µm in diameter, and thus is not applicable to small transition metal complexes.65 Macropinocytosis involves “ruffling” of the membrane to form large pockets greater than 1 µm in diameter, or endocytic vesicles, which are filled with both extracellular solvent and solute molecules. These vesicles are then broken down by endosomes or lysosomes. Clathrin-mediated endocytosis is mediated by “clathrin-coated pits”, which are about 100 nm in diameter, and have a crystalline coat made up of transmembrane receptors associated with the protein clathrin. These receptors bind their respective ligands (or a therapeutic agent that resembles their ligand) and then pinch off to form clathrin-coated vesicles (CCVs) which are internalized into the cell. In caveolin-mediated endocytosis, flask-shaped pits in the plasma membrane, about 60 to 80 nm in diameter, are shaped by caveolin, a protein that binds cholesterol. Finally, in clathrin and caveolinin-dependent endocytosis, small structures that are 40 to 50 nm in diameter act as “rafts”, freely diffusing along the cell surface. These rafts will then be captured and internalized within any endocytic vesicle.66,67
Figure 13.
Schematic diagram of the different possible routes of entry into the cell taken by small complexes.
However, just as membrane-bound proteins can facilitate the entry of a complex into the cell, certain proteins, called efflux transporters, can facilitate the extrusion of such compounds from the cell. In fact, multidrug efflux pumps, which can recognize multiple structurally dissimilar compounds, are often responsible for chemotherapeutic resistance.68 The most well-studied of such mammalian efflux transporters is the ATP-binding cassette (ABC) family. These efflux transporters use ATP hydrolysis to drive the extrusion of drugs from the cell, and can do so against significant concentration gradients.69 The reason behind the broad substrate specificity of these efflux transporters is the presence of a large, flexible hydrophobic biding pocket which allows for substrate binding via hydrophobic and electrostatic interactions, rather than the specific hydrogen-bonding networks present in less promiscuous transport proteins.70
A basic experiment in determining the mechanism of uptake is determining whether the mechanism of uptake is energy-dependent or –independent. Both passive and active diffusion are energy-independent, while active transport and endocytosis are energy-dependent. By incubating cells at low temperature (4° C) or in the presence of metabolic inhibitors (2-deoxyglucose and olygomycin), processes that require energy will be blocked. If the drug of interest has decreased uptake under these conditions, the mechanism of uptake involves an energy-dependent process, while if uptake is unchanged the mechanism is an energy-independent process.71,72
Passive diffusion has the broadest range in substrates out of all uptake mechanisms, and is therefore an attractive mode of uptake for therapeutics. Uptake mediated by passive diffusion is the most difficult to modulate, but it can be done. Uptake of positively charged molecules, such as many inorganic therapeutics, can be driven by the plasma membrane potential of the cell. In mammalian cells, the membrane potential is generated and maintained by a potassium concentration gradient. This potential can be reduced either by using media with a potassium concentration equal to the intracellular potassium concentration, or by adding gramicidin A to the media, a polypeptide that will form transmembrane channels which allow unrestricted potassium traversal, thus destroying the concentration gradient. In contrast, the cell can be hyperpolarized by adding valinomycin to the media, a potassium-specific ionophore that will increase potassium transportation across the membrane.73
Protein-mediated transport, facilitated diffusion and active transport have the capability of being cell-type or tissue-type specific. This allows for the development of targeted drugs, thus attenuating dose-limiting side effects.74 This type of uptake can be repressed by using known inhibitors of specific transport proteins. If uptake of the compound of interest is decreased in the presence of the inhibitor, then the respective transport protein most likely is involved in uptake. Likewise, many efflux transporters have known inhibitors; if uptake of a compound is increased in the presence of an inhibitor, then the compound is likely a substrate for the respective efflux protein.
As with protein-mediated transport, the different types of endocytosis can be mediated by using known inhibitors of the different processes. Ammonium chloride and chloroquine diphosphate are general endocytosis inhibitors.75 Chlorpromazine hydrochloride, monodansylcadaverine (MDC), and phenylarsine oxide are inhibitors of clathrin-mediated endocytosis and macropinocytosis.76 Filipin and nystatin selectively inhibit caveolin-mediated endocytosis via cholesterol sequestration.77 Amiloride, as well as 5-(N,N-dimethyl) amiloride (DMA) and 5-(N-ethyl-N-isopropyl) amiloride (EIPA) inhibit macropinocytosis and phagocytosis by obstructing Na+/H+ exchange.78
Cisplatin is the prototypical medicinal inorganic drug. There have been many studies on the activity, uptake, and subcellular distribution of this molecule and these serve as illustrations of how mechanism may be elucidated. For many years it was widely accepted that cisplatin entered the cell via passive diffusion. In one such study on the mechanism of uptake of cisplatin, Binks and Dobrota measured the amount of uptake of cisplatin into rat small intestines using AAS. They found that uptake of the drug was linear with respect to time and not saturable up to a concentration of 1.0 mM. The authors also found no change in uptake of cisplatin when the experiments were repeated under metabolic inhibition, confirming that the mechanism of uptake is energy-independent and through passive diffusion.79 More recently, however, it was found that several protein-mediated transport pathways can also be responsible for cisplatin uptake. The most important of these pathways are the organic cation transporters and the copper influx transporter CTR1.80 To demonstrate the contribution of CTR1 to cisplatin uptake, Howell and coworkers measures cisplatin uptake in A2780 human ovarian cancer cells using ICP-MS. They increased expression of CTR1 20-fold and found that intracellular platinum levels increased by 55% after 24 hours of platinum incubation.81
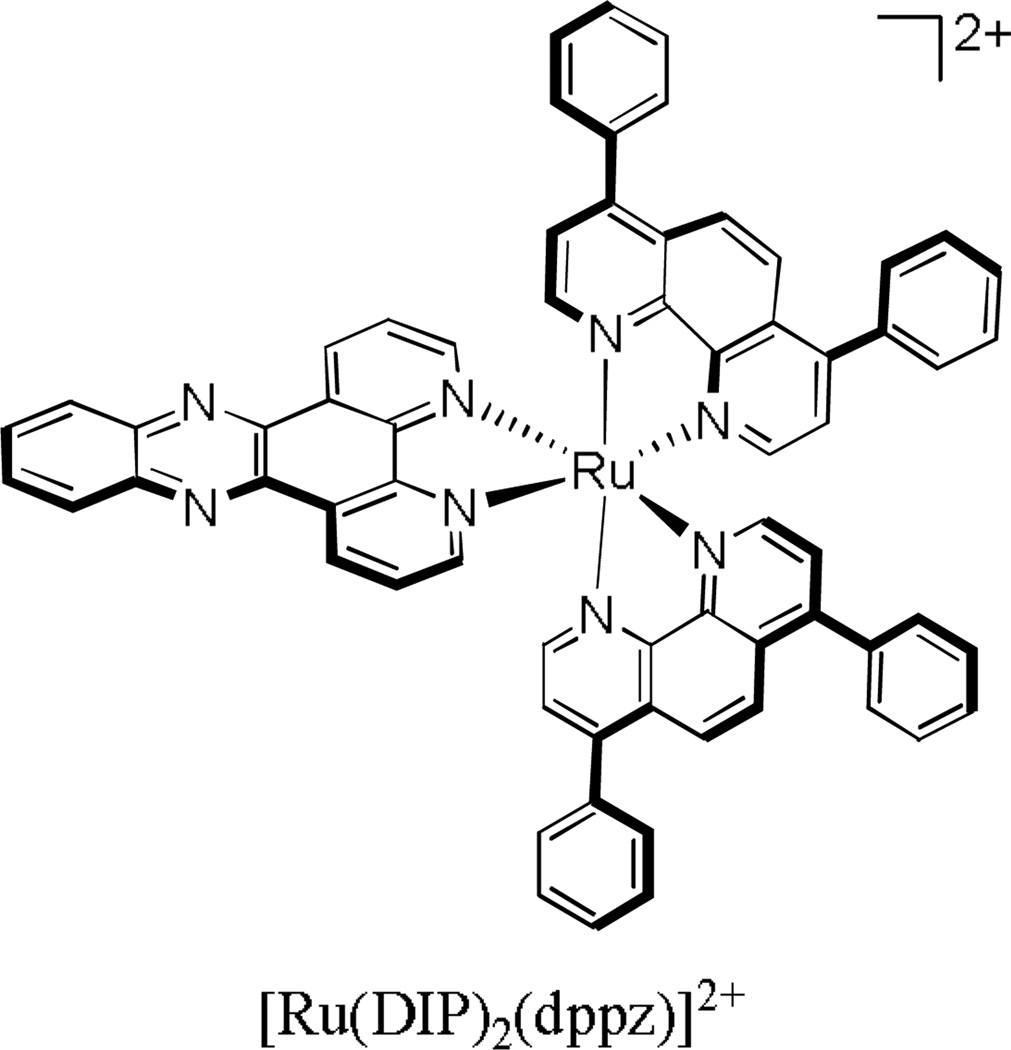
Many ruthenium complexes are easy to monitor in uptake studies owing to their strong luminescence. For example, we have used flow cytometry to study the uptake of the luminescent lipophilic ruthenium complex [Ru(DIP)2(dppz)]2+ into HeLa cells (Figure 14). It was found that under metabolic inhibition, uptake of the ruthenium complex remained unchanged. Additionally, increasing the incubation temperature from 4° C to 37° C had no effect on uptake. These experiments showed that the mechanism of uptake was energy-independent. To rule out facilitated diffusion by organic cation transporters (OCTs), uptake of the ruthenium complex was also studied in the absence and presence of a variety of OCT inhibitors (tetra-n-alkylammonium salts, procainamide, and cimetidine), and was found to be unaffected by the presence of these compounds. Lastly, we examined the effect of changes in the membrane potential on uptake of the ruthenium compound. When incubated with a 170 mM potassium buffer (which reduces the membrane potential to zero), uptake decreased substantially. Furthermore, in the presence of valinomycin (which will increase the membrane potential), uptake of the compound increased substantially. Taken together, it was concluded that uptake of [Ru(DIP)2(dppz)]2+ was solely via passive diffusion.82 It is interesting to consider this complex being taken up at all efficiently. The complex has a diameter of approximately 21 Å, not unlike many small proteins, yet it is efficiently taken up through the cell membrane to the cytoplasm and to some extent to the nucleus of the cell.
Figure 14.
[Ru(DIP)2(dppz)]2+, a luminescent polypyridyl ruthenium complex that enters HeLa cells via passive diffusion.82
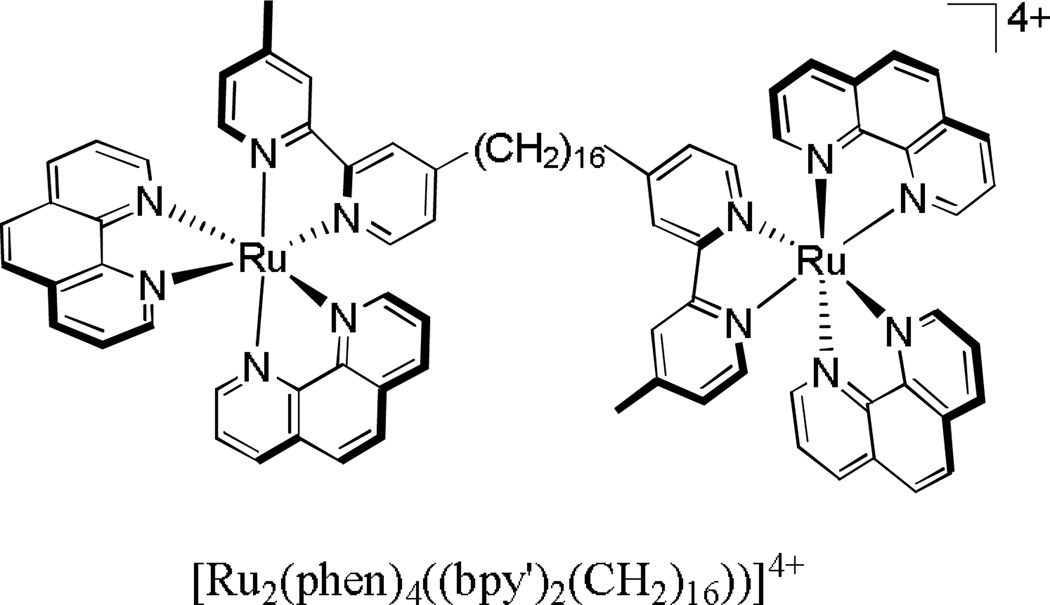
In one last study by Pisani and coworkers, the mechanism of cellular uptake of a dinuclear polypyridylruthenium(II) complex (Figure 15) into L1210 murine leukemia cells was determined through flow cytometry experiments. In this study, the authors found that varying the temperature of incubation from 4° C to 20° C to 37° C had an effect on uptake, with the 4° C sample having the least amount of intracellular ruthenium. Furthermore, when the cells were incubated with an increased amount of glucose, uptake of the ruthenium complex was significantly enhanced.
Figure 15.
[Ru2(phen)4((bpy')2(CH2)16))]4+, a compound shown to enter L1210 murine leukemia cells via an energy-dependent mechanism as well as passive diffusion.83
Conversely, under metabolic inhibition, uptake marginally decreased. In order to rule out endocytosis, the authors measured ruthenium uptake in the presence of a variety of different endocytosis inhibitors (chloroquine diphosphate, filipin, dimethyl amiloride, ammonium chloride, and chlorpromazine hydrochloride), and found that uptake was either unchanged or increased in the presence of these inhibitors. Furthermore, when incubated with a variety of OCT inhibitors (tetra-n-alkylammonium salts or procainamide), uptake remained unchanged. The authors concluded that uptake was in large part due to passive diffusion, with a minor contribution from protein-mediated active transport.83 It is important to consider that complexes so similar in chemical structure need have similar mechanisms of uptake. Experiments need to be determined in each case to establish the uptake mechanism.
Subcellular localization of the Metal Complex
Once a therapeutic has entered the cell, there are a plethora of different organelles in which it can localize. Moreover, activities and targets depend on where within the cell the complex becomes localized. This localization can also determine unwanted toxicities associated with a given complex.
Methods to monitor Localization
The most commonly exploited technique for the subcellular mapping of inorganic therapeutics is fluorescence microscopy. However, this technique can only be used if the complex of interest is inherently luminescent. Cells can be treated with the luminescent drug of interest concomitantly with any of various fluorescent organelle-specific probes. The extent of overlap between the drug and the probe will provide information on the localization of the drug. For example, Matson and coworkers synthesized a variety of dppz complexes of Ru derivatized with alkyl ether chains of various lengths and used confocal laser scanning microscopy to study their subcellular localization in CHO-K1 cells.84 Using various RNA and membrane-specific dyes, they found, perhaps not surprisingly, that the least lipophilic compound localized in the nucleus, while the most lipophilic localized in membranes.
While fluorescence microscopy has the capability of providing a qualitative assessment of the localization of therapeutics in living cells, due to differences in the quantum yields among different luminescent compounds, this technique cannot be utilized to quantify the amount of drug localized inside the cell. Furthermore, many metal-based luminescent compounds have been shown to exhibit different quantum yields depending on the environment surrounding the compound,85 which further complicates the quantitation of the localization of luminescent metal complexes using this technique.
In a study by Groessl and co-workers, ICP-MS was used to track the uptake and subcellular localization of cisplatin as well as two ruthenium-based chemotherapeutics currently in clinical trials, NAMI-A and KP1019. Reduced mitochondrial accumulation of cisplatin was observed in cisplatin resistant cells, while the ruthenium-based drugs were found to have different localization patterns than cisplatin, which did not change from one cell type to the other. 86
While AAS and ICP-MS can provide quantitative information on the subcellular localization of inorganic complexes, the process of sample preparation for these methods involves the destruction of the cells, and therefore no structural information is obtained. One method for the visualization of the subcellular localization of metal-based therapeutics while obtaining the structural integrity of the cell is electron microscopy. Electron microscopy has the advantage of providing spatial resolution that is almost three orders of magnitude better than conventional light microscopy, allowing for the resolution of structural details in the nanometer range.87 Furthermore, the electron dense property of metal ions can be detected inside cells by electron microscopy due to mass contrast. In transmission electron microscopy (TEM), cells are fixed, dehydrated with organic solvent, embedded in resin, and then thinly sliced (50 to 400 nm thick). These thin samples are then exposed to an electron beam, which will be either scattered by regions of high electron density (metal ions), or transmitted through low electron density regions of the sample to a detector, which then constructs a “contrast image” of the sample, where areas of high electron density have higher contrast. In a study by van Rijt and coworkers, the distribution of a osmium(II) arene complex in ovarian cancer cells was determined by TEM. It was observed that upon treatment of A2780 cells with 5 µM osmium complex, increased contrast was observed in the mitochondria, nucleolus, and nuclear membrane. The morphological changes associated with apoptosis were able to be observed at the same time, illustrating the utility of electron microscopy. 88
Electron microscopy can in certain cases be combined with elemental mapping to obtain a technique called energy-filtered transmission electron microscopy (EFTEM). In this technique, as the electrons from the electron beam hit the sample, some will undergo an inelastic collision, losing an amount of energy that is equivalent to the core atomic level of the element that it just collided with. In this way, not only will an image will be created with the resolution of electron microscopy, but also element distribution maps can be obtained of the sample.89
The last technique to consider here is nano-scale secondary ion mass spectrometry (NanoSIMS). In this technique, a high-energy primary ion beam bombards the surface of a sample, “sputtering” secondary ions which are then detected and analyzed by a mass spectrometer.90 In this way, NanoSIMS can provide spatial resolution up to 50 nm, as well as elemental and isotopic information of the sample. This technique is in its infancy and as such has not been widely utilized in the subcellular mapping of metal based therapeutics. However, in one study by the Berners-Price lab, a complementary EFTEM and NanoSIMS study was performed on an antitumor gold(I) phosphine complex.91 In this study, human breast cancer cells (MDA) were treated with the gold complex and analyzed by both EFTEM and NanoSIMS for subcellular gold localization. Using EFTEM, the localization of gold could be observed, as well as the morphological changes accompanying gold treatment. NanoSIMS allowed for the mapping of 12C14N− (to show cell morphology), 31P− (to show the location of nucleic acids), 197Au− (to show localization of the therapeutic), and 34S− (to show the localization of thiols). The gold signal clearly co-localized with the sulfur signal, thus supporting the idea that the mechanism of action of Au(I) compounds involves the inhibition of thiol-containing protein families.
Peptide Conjugation
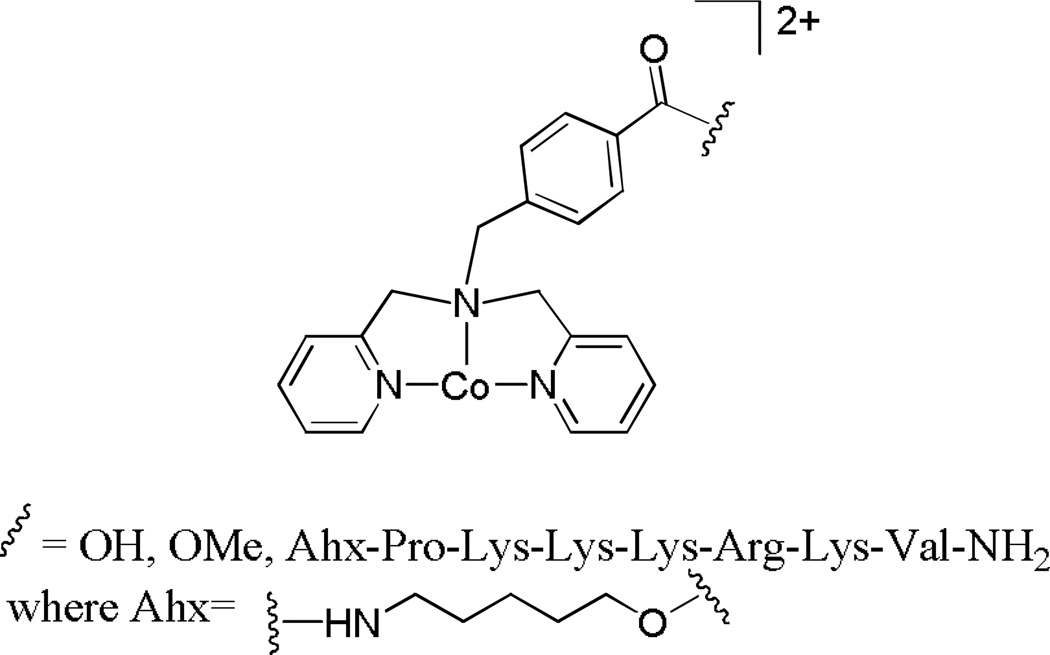
DNA is the cellular target of many inorganic chemotherapeutic agents. As such, it is important for these complexes to localize mainly in the nucleus of cells. One strategy to alter the subcellular localization of a compound is through peptide conjugation. Nuclear localization sequences (NLS) are small peptides which, when appended to a protein, will in essence allow the protein to be imported into the nucleus by nuclear transport.92 As an example, Kirin and coworkers utilized the NLS Pro-Lys-Lys-Lys-Arg-Lys-Val to enhance the uptake and nuclear localization of the cobalt complex shown in Figure 16 into HT-29 cells. AAS experiments revealed that not only does the conjugate have enhanced uptake compared to the unconjugated cobalt complex, but also the intracellular cobalt concentration of the conjugate is higher than the extracellular cobalt concentration. Furthermore, the fraction of intracellular cobalt localized in the nucleus increased for the conjugates compared to the unconjugated cobalt complex.93
Figure 16.
The peptide conjugate studied by Kirin and coworkers. Upon conjugation of the cobalt complex to the nuclear localization peptide, uptake of the complex into the nucleus of HT-29 cells increased significantly.93
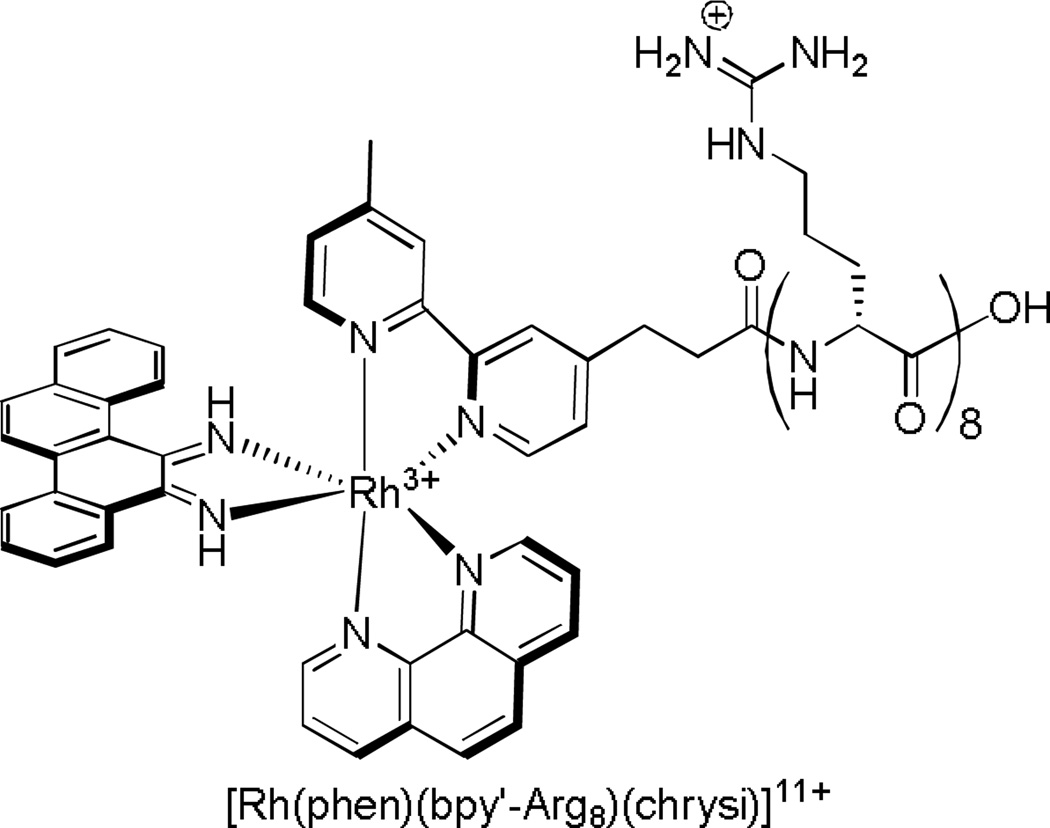
However, in the design of peptide-therapeutic conjugates, care must be taken to ensure that the conjugate itself still has the same biological target as the unconjugated molecule. For example, in an attempt to increase and accelerate cellular uptake of our rhodium metalloinsertors, the [Rh(phen)(bpy’-Arg8)(chrysi)]11+ complex shown in Figure 17 was synthesized. Uptake studies conducted on the fluorescein-appended analogue confirmed fast (within 60 min) nuclear uptake into HeLa cells. However, DNA binding studies revealed that, with the octaarginine appendage, the nonspecific binding affinity of both complexes for mismatched and matched DNA increased, due to the substantial added positive charge of the peptide.94 In fact, studies of cellular proliferation with the metalloinsertor-peptide conjugate confirmed that the complexes no longer showed the preferential inhibition of MMR-deficient cells, as expected if specific binding only to mismatched DNA was lost. One route to restore specificity would be to include a self-cleavable linker so that the conjugate would be removed once inside the nucleus. The work thus illustrates the utility but also subtle new issues that arise with peptide conjugates for therapeutic applications.
Figure 17.
The rhodium metalloinsertor-peptide conjugate synthesized in order to accelerate uptake. While uptake is accelerated upon conjugation of the metalloinsertor to the octaarginine peptide, the presence of the octaarginine increases the nonspecific binding affinity of the complex for matched and mismatched DNA.
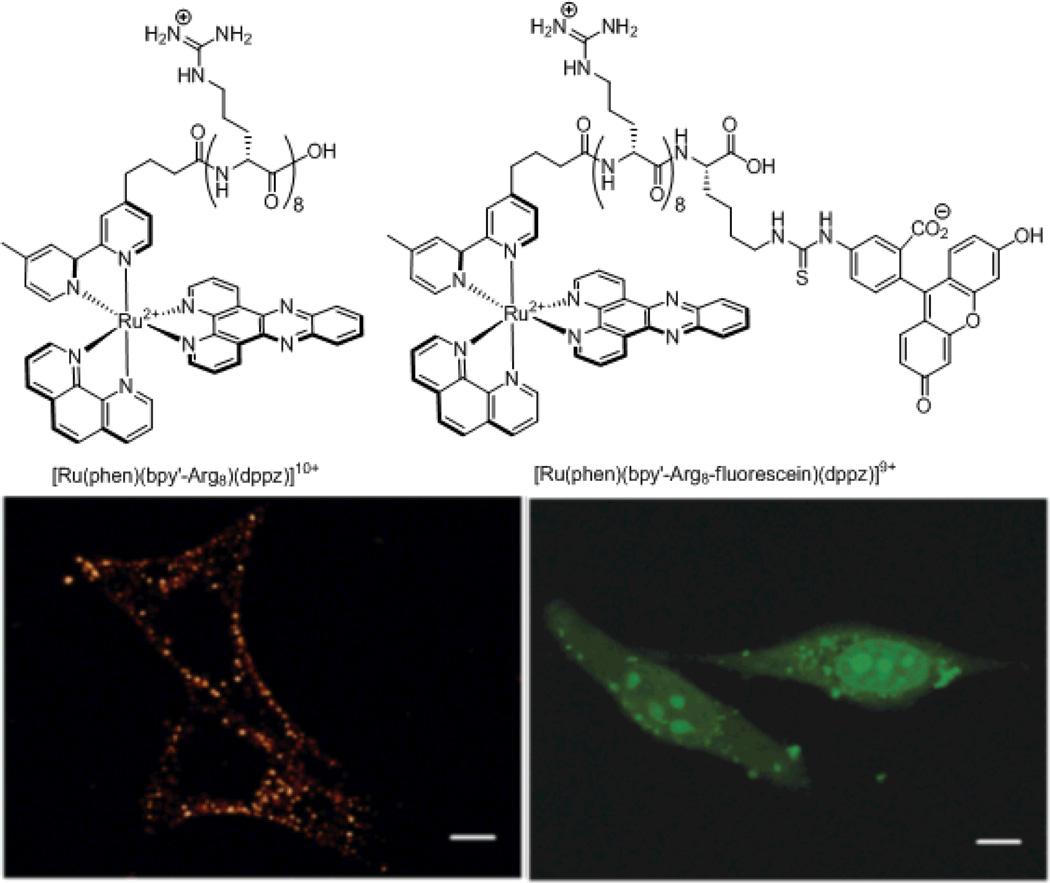
Many laboratories append fluorescent tags onto molecules of interest to follow their subcellular localization. The process of appending a fluorescent tag to the molecule of interest can, however, also alter the subcellular localization.95 As an illustration, we examined the localization properties of two Ru(dppz)-peptide conjugates, one with and one without a fluorescent tag (Figure 18). The conjugate with only the octaarginine peptide was localized throughout the cytoplasm in punctate distributions and was completely absent from the nucleus. On the other hand, the conjugate with both octaarginine and fluorescein exhibited nuclear staining when incubated under the same conditions as the previous conjugate. This study clearly shows the consequences of appending fluorescent tags to non-fluorescent molecules in order to study their subcellular localizations.
Figure 18.
Chemical structures (top) and confocal microscopy images (bottom) of the two metal-peptide fluorophore conjugates examined in our laboratory. The octaarginine conjugate (left) displays only cytosolic localization, while the octaarginine fluorescein conjugate (right) exhibit nuclear localization.
Combination of Techniques
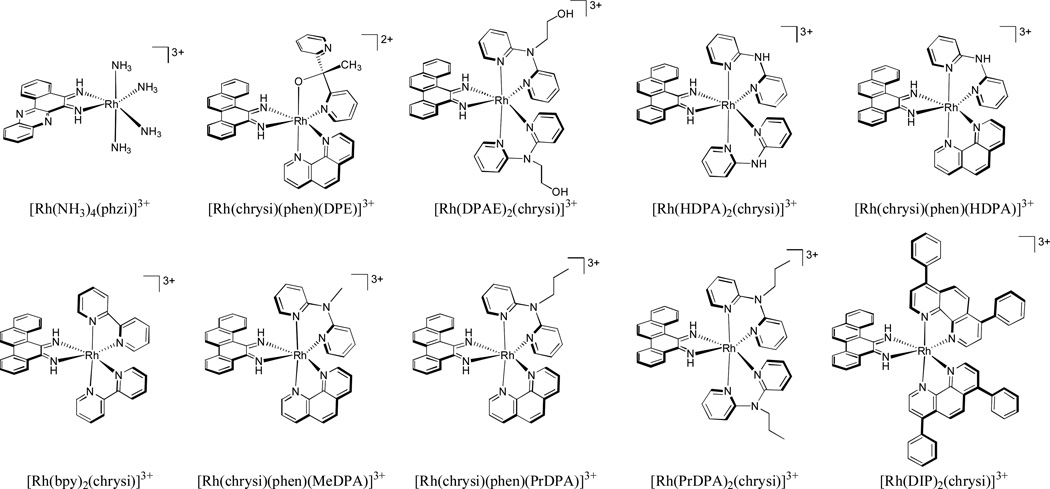
In order to gain a complete understanding of the mechanism of action of a given complex, the techniques discussed here must be combined. For example, in a recent study in our laboratory, the activity, uptake, and subcellular localization of ten different rhodium metalloinsertors were examined (Figure 19). While the binding affinities of all ten compounds were found to be within an order of magnitude of each other, the abilities of the compounds to selectively target MMR-deficient cells over MMR-proficient cells varied dramatically. The more lipophilic compounds showed the least selectivity for the MMR-deficient cells. ICP-MS was then used to determine the intracellular rhodium concentrations of all ten compounds over a 24 hour time course in human colorectal HCT116 cells. The different compounds had drastically different patterns of rhodium uptake over time, reflecting different mechanisms of uptake. However, the amount of intracellular rhodium did not correlate at all with their biological activities. ICP-MS was then combined with organelle fractionation techniques in order to provide us with quantitative information about the subcellular localization of the rhodium metalloinsertors. After treating HCT116 cells with the various compounds, the nuclei and mitochondria were isolated and tested for rhodium content using ICP-MS. It was found that while all compounds tested were localized in the nucleus at concentrations sufficient for DNA mismatch binding, those with higher mitochondrial rhodium accumulation showed lower specificity for MMR-deficient cells over MMR-proficient cells. Binding to DNA in the mitochondria was deleterious to the unique biological activity. Thus this study established clearly that it is mismatches in genomic DNA that are the ultimate target of rhodium metalloinsertors and that are responsible for their unique biological activity.96
Figure 19.
Chemical structures of ten rhodium metalloinsertors studied in our laboratory. While all compounds were localized in the nucleus of HCT116 cells at sufficient concentrations for mismatch binding, only the compounds with low mitochondrial rhodium accumulation exhibited preferential targeting of MMR-deficient cells over MMR-proficient cells.96
Conclusion
The development of new cell-selective therapeutic agents is imperative, and metal complexes offer a wealth of possibilities for new design. Structural characterization of the interaction between a given complex and its target is critical, providing insight as to what changes can be made to increase affinity and specificity for the target. Likewise, structure-function relationships can provide critical information. Also important is developing an understanding of the relationship between uptake and activity. The knowledge of where within the cell the complex is being shuttled is a powerful tool, and a possible driver in the design of new therapeutics with improved effectiveness. In general, metal complexes offer the tools, flexibility in ligand substitution and varied techniques to monitor their path and biological fate within the cell.
Acknowledgements
We are grateful to the NIH (GM33309) for their support of this research.
Biographies

Alexis C. Komor
Alexis C. Komor received her B.S. in chemistry with highest honors at the University of California, Berkeley in 2008. While at Berkeley, she worked on the design of first-row transition metal catalysts for dioxygen activation and group transfer in the laboratory of Christopher J. Chang. She is currently an NSF predoctoral fellow in Dr. Barton’s laboratory studying the design and synthesis of rhodium metalloinsertors with improved biological activity.

Dr Jacqueline K. Barton is the Arthur and Marian Hanisch Memorial Professor of Chemistry and Chair of the Division of Chemistry and Chemical Engineering at the California Institute of Technology. Barton was awarded the A.B. at Barnard College and a PhD in inorganic chemistry at Columbia University. After a postdoctoral fellowship at Bell Laboratories and Yale University, she became an assistant professor at Hunter College, City University of New York. In 1983, she returned to Columbia University, becoming a professor in 1986. In the fall of 1989, she joined the faculty at Caltech. Professor Barton has pioneered the application of transition metal complexes to probe recognition and reactions of double helical DNA. Barton has also carried out seminal studies to elucidate electron transfer chemistry mediated by the DNA helix. Barton has received numerous awards, including a MacArthur Foundation Fellowship and election to the National Academy of Sciences and Institute of Medicine. Last year she was awarded the National Medal of Science
Notes and references
- 1.Zhang CX, Lippard SJ. Curr. Opin. Chem. Biol. 2003;7:481. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 2.Boulikas T, Vougiouka M. Oncol. Rep. 2004;11:559. [PubMed] [Google Scholar]
- 3.Alessio E, Mestroni G, Bergamo A, Sava G. Curr. Top. Med. Chem. 2004;4:1525. doi: 10.2174/1568026043387421. [DOI] [PubMed] [Google Scholar]
- 4.Hartinger GC, Zorbas-Seifried S, Jakupec MA, Kynast B, Zorbas H, Keppler BK. J. Inorg. Biochem. 2006;100:891. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Gianferrara T, Bratsos I, Alessio E. Dalton Trans. 2009:7588. doi: 10.1039/b905798f. [DOI] [PubMed] [Google Scholar]
- 6.Reslova S. Chem. Biol. Interact. 1971;4:66. doi: 10.1016/0009-2797(71)90034-2. [DOI] [PubMed] [Google Scholar]
- 7.Harder HC, Rosenberg B. Int. J. Cancer. 1970;6:207. doi: 10.1002/ijc.2910060207. [DOI] [PubMed] [Google Scholar]
- 8.Stone PJ, Kelman AD, Sinex FM. J. Mol. Biol. 1976;104:793. doi: 10.1016/0022-2836(76)90182-0. [DOI] [PubMed] [Google Scholar]
- 9.Ushy HM, Tullius TD, Lippard SJ. Biochemistry. 1982;21:3744. doi: 10.1021/bi00516a012. [DOI] [PubMed] [Google Scholar]
- 10.Pinto AL, Lippard SJ. Biochim. Biophys. Acta. 1985;780:167. doi: 10.1016/0304-419x(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 11.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Lippard SJ. Nature Rev. Drug Discovery. 2005;4:307. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 13.Bond PJ, Langridge R, Jennette KW, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 1972;75:4825. doi: 10.1073/pnas.72.12.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang AHJ, Nathans J, van der Marel G, van Boom JH, Rich A. Nature. 1978;276:471. doi: 10.1038/276471a0. [DOI] [PubMed] [Google Scholar]
- 15.Barton JK, Danishefsky AT, Goldberg JM. J. Am. Chem. Soc. 1984;106:2172. [Google Scholar]
- 16.Kielkopf CL, Erkkila KE, Hudson RP, Barton JK, Rees DC. Nat. Struct. Biol. 2000;7:117. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 17.Odom DT, Parker CS, Barton JK. Biochemistry. 1999;38:5155. doi: 10.1021/bi9827969. [DOI] [PubMed] [Google Scholar]
- 18.Song H, Kaiser JT, Barton JK. Nat. Chem. 2012;4:615. doi: 10.1038/nchem.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupureur CM, Barton JK. J. Am. Chem. Soc. 1994;116:10286. [Google Scholar]
- 20.Dupureur CM, Barton JK. Inorg. Chem. 1997;36:33. [Google Scholar]
- 21.Holmlin RE, Stemp EDA, Barton JK. Inorg. Chem. 1998;37:29. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]
- 22.Niyazi H, Hall JP, O’Sullivan K, Winter G, Sorensen T, Kelly JM, Cardin CJ. Nat. Chem. 2012;4:621. doi: 10.1038/nchem.1397. [DOI] [PubMed] [Google Scholar]
- 23.Hall JP, O’Sullivan K, Naseer A, Smith JA, Kelly JM, Cardin CJ. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17610. doi: 10.1073/pnas.1108685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson BA, Barton JK. J. Am. Chem. Soc. 1997;119:12986. [Google Scholar]
- 25.Jackson BA, Alekseyev VY, Barton JK. Biochemistry. 1999;38:4655. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 26.Jackson BA, Barton JK. Biochemistry. 2000;39:6176. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 27.Pierre VC, Kaiser JT, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2007;104:429. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barton JK. Science. 1986;233:727. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 29.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. Biochemistry. 2009;48:4247. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong E, Giandomenico CM. Chem. Rev. 1999;99:2451. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 31.Guo Z, Sadler PJ. Angew. Chem. Int. Ed. 1999;38:1512. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1512::AID-ANIE1512>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 32.Gill MR, Thomas JA. Chem. Soc. Rev. 2012;41:3179. doi: 10.1039/c2cs15299a. [DOI] [PubMed] [Google Scholar]
- 33.Dwyer FP, Gyarfas EC, Rogers WP, Koch JH. Nature. 1952;170:190. doi: 10.1038/170190a0. [DOI] [PubMed] [Google Scholar]
- 34.Novakova O, Kasparkova J, Vrana O, van Vliet PM, Reedijk J, Brabec V. Biochemistry. 1995;34:12369. doi: 10.1021/bi00038a034. [DOI] [PubMed] [Google Scholar]
- 35.Tan C, Lai S, Wu S, Hu S, Zhou L, Chen Y, Wang M, Zhu Y, Lian W, Peng W, Ji L, Xu A. J. Med. Chem. 2010;53:7613. doi: 10.1021/jm1009296. [DOI] [PubMed] [Google Scholar]
- 36.Hackenberg F, Oehninger L, Alborzinia H, Can S, Kitanovic I, Geldmacher Y, Kokoschka M, Wolfl S, Ott I, Sheldrick WS. J. Inorg. Biochem. 2011;105:991. doi: 10.1016/j.jinorgbio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Geldmacher Y, Rubbiani R, Wfelmeier P, Prokop A, Ott I, Sheldrick WS. J. Organomet. Chem. 2011;696:1023. [Google Scholar]
- 38.Bieda R, Ott I, Gust R, Sheldrick WS. Eur. J. Inorg. Chem. 2009:3821. [Google Scholar]
- 39.Bieda R, Ott I, Dobroschke M, Prokop A, Gust R, Sheldrick WS. J. Inorg. Biochem. 2009;103:698. doi: 10.1016/j.jinorgbio.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Scharwitz MA, Ott I, Geldmacher Y, Gust R, Sheldrick WS. J. Organomet. Chem. 2008;693:2299. [Google Scholar]
- 41.Dobroschke M, Geldmacher Y, Ott I, Harlos M, Kater L, Wagner L, Gust R, Sheldrick WS, Prokop A. ChemMedChem. 2009;4:177. doi: 10.1002/cmdc.200800311. [DOI] [PubMed] [Google Scholar]
- 42.Noffke AL, Habtemariam A, Pizarro AM, Sadler PJ. Chem. Commun. 2012;48:5219. doi: 10.1039/c2cc30678f. [DOI] [PubMed] [Google Scholar]
- 43.Yan YK, Melchart M, Habtemariam A, Sadler PJ. Chem. Commun. 2005:4764. doi: 10.1039/b508531b. [DOI] [PubMed] [Google Scholar]
- 44.Morris RE, Aird RE, del S Murdoch P, Chen H, Cummings J, Hughes ND, Parsons S, Parkin A, Boyd G, Jodrell DI, Sadler PJ. J. Med. Chem. 2001;44:3616. doi: 10.1021/jm010051m. [DOI] [PubMed] [Google Scholar]
- 45.Aird RE, Cummings J, Ritchie AA, Muir M, Morris RE, Chen H, Sadler PJ, Jodrell DI. Br. J. Cancer. 2002;86:1652. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Parkinson JA, Morris RE, Sadler PJ. J. Am. Chem. Soc. 2003;125:173. doi: 10.1021/ja027719m. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Bella J, Parkinson JA, Sadler PJ. J. Biol. Inorg. Chem. 2005;10:147. doi: 10.1007/s00775-004-0621-5. [DOI] [PubMed] [Google Scholar]
- 48.Novakova O, Chen H, Vrana O, Rodger A, Sadler PJ, Brabec V. Biochemistry. 2003;42:11544. doi: 10.1021/bi034933u. [DOI] [PubMed] [Google Scholar]
- 49.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15359. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ernst RJ, Song H, Barton JK. J. Am. Chem. Soc. 2009;131:2359. doi: 10.1021/ja8081044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ernst RJ, Komor AC, Barton JK. Biochemistry. 2011;50:10919. doi: 10.1021/bi2015822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carethers JM, Hawn MT, Chauhan DP, Luce MC, Marra G, Koi M, Boland CR. J. Clin. Invest. 1996;98:199. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson JW. Anal. Chem. 1960;32:17. [Google Scholar]
- 54.Robinson JW. Anal. Chem. 1961;33:1067. [Google Scholar]
- 55.Brouwers EEM, Tibben M, Rosing H, Schellens JHM, Beijnen JH. Mass Spec. Rev. 2008;27:67. doi: 10.1002/mas.20159. [DOI] [PubMed] [Google Scholar]
- 56.Harrison RJE. Aust. J. Chem. 2009;62:90. [Google Scholar]
- 57.Frenandez-Moreira V, Thorp-Greenwood FL, Coogan MP. Chem. Commun. 2010;46:186. doi: 10.1039/b917757d. [DOI] [PubMed] [Google Scholar]
- 58.Gately DP, Howell SB. Br. J. Cancer. 1993;67:1171. doi: 10.1038/bjc.1993.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timmer-Bosscha H, Mulder NH, de Vries EGE. Br. J. Cancer. 1992;66:227. doi: 10.1038/bjc.1992.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews PA, Velury S, Mann SC, Howell SB. Cancer Res. 1988;48:68. [PubMed] [Google Scholar]
- 61.Harlos M, Ott I, Gust R, Alborzinia H, Wolfl S, Kromm A, Sheldrick WS. J. Med. Chem. 2008;51:3924. doi: 10.1021/jm800173s. [DOI] [PubMed] [Google Scholar]
- 62.Bugarcic T, Novakova O, Halamikova A, Zerzankova L, Vrana O, Kasparkova J, Habtemariam A, Parsons S, Sadler PJ, Brabec VJ. J. Med. Chem. 2008;51:5310. doi: 10.1021/jm8003043. [DOI] [PubMed] [Google Scholar]
- 63.Louie M-W, Choi AW-T, Liu H-W, Chan BT-N, Lo KK-W. Organomet. 2012;31:5844. [Google Scholar]
- 64.Puckett CA, Ernst RJ, Barton JK. Dalton Trans. 2010;39:1159. doi: 10.1039/b922209j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aderem A, Underhill DM. Annu. Rev. Immunol. 1999;17:593. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 66.Doherty GJ, McMahon HT. Annu. Rev. Biochem. 2009;78:857. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 67.Conner SD, Schmid SL. Nature. 2003;422:37. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 68.Paulsen IT. Curr. Op. Microbiol. 2003;6:446. doi: 10.1016/j.mib.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Schinkel AH, Jonker JW. Adv. Drug Deliver. Rev. 2003;55:3. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 70.Neyfakh AA. Mol.Microbiol. 2002;44:1123. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 71.Barban S, Schulze H. J. Biol. Chem. 1961;236:1887. [PubMed] [Google Scholar]
- 72.Slater C. Methods Enzymol. 1967;10:48. [Google Scholar]
- 73.Shapiro HM. Methods. 2000;21:271. doi: 10.1006/meth.2000.1007. [DOI] [PubMed] [Google Scholar]
- 74.Mizuno N, Sugiyama Y. Drug Metabol. Pharmacokin. 2002;17:93. doi: 10.2133/dmpk.17.93. [DOI] [PubMed] [Google Scholar]
- 75.Steinman RM, Mellman IS, Mullman WA, Cohn ZA. J. Cell Biol. 1983;96:1. doi: 10.1083/jcb.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cooper A, Shaul Y. J. Biol. Chem. 2006;281:16563. doi: 10.1074/jbc.M601418200. [DOI] [PubMed] [Google Scholar]
- 77.Smart J, Anderson RG. Methods Enzymol. 2002;353:131. doi: 10.1016/s0076-6879(02)53043-3. [DOI] [PubMed] [Google Scholar]
- 78.West MA, Bretscher MS, Watts C. J. Cell. Biol. 1989;109:2731. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Binks SP, Dobrota M. Biochem. Pharmacol. 1990;40:1329. doi: 10.1016/0006-2952(90)90400-f. [DOI] [PubMed] [Google Scholar]
- 80.Arnesano F, Natile G. Coord. Chem. Rev. 2009;253:2070. [Google Scholar]
- 81.Holzer AK, Samimi G, Katano K, Naerdemann W, Lin X, Safaei R, Howell SB. Mol. Pharmacol. 2004;66:817. doi: 10.1124/mol.104.001198. [DOI] [PubMed] [Google Scholar]
- 82.Puckett CA, Barton JK. Biochemistry. 2008;47:11711. doi: 10.1021/bi800856t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pisani MJ, Fromm PD, Mulyana Y, Clarke RJ, Korner H, Heimann K, Collins JG, Keene FR. ChemMedChem. 2011;6:848. doi: 10.1002/cmdc.201100053. [DOI] [PubMed] [Google Scholar]
- 84.Matson M, Svensson FR, Norden B, Lincoln P. J. Phys. Chem. B. 2011;115:1706. doi: 10.1021/jp109530f. [DOI] [PubMed] [Google Scholar]
- 85.Friedman AE, Chambron J-C, Sauvage J-P, Turro NJ, Barton JK. J. Am. Chem. Soc. 1990;112:4960. [Google Scholar]
- 86.Groessl M, Zava O, Dyson PJ. Metallomics. 2011;3:591. doi: 10.1039/c0mt00101e. [DOI] [PubMed] [Google Scholar]
- 87.Hoenger A, McIntosh JR. Curr. Op. Cell Biol. 2009;21:89. doi: 10.1016/j.ceb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Rijt SH, Mukherjee A, Pizarro AM, Sadler PJ. J. Med. Chem. 2010;53:840. doi: 10.1021/jm901556u. [DOI] [PubMed] [Google Scholar]
- 89.Thomas PJ, Midgley PA. Top. Catal. 2002;21:109. [Google Scholar]
- 90.Werner HW. Surf. Interface Anal. 2003;35:859. [Google Scholar]
- 91.Wedlock LE, Kilburn MR, Cliff JB, Filgueira L, Saunders M, Berners-Price SJ. Metallomics. 2011;3:917. doi: 10.1039/c1mt00053e. [DOI] [PubMed] [Google Scholar]
- 92.Gorlich D, Mattaj IW. Science. 1996;271:1513. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 93.Kirin SI, Ott I, Gust R, Mier W, Weyhermuller T, Metzler-Nolte N. Angew. Chem. Int. Ed. 2008;47:955. doi: 10.1002/anie.200703994. [DOI] [PubMed] [Google Scholar]
- 94.Brunner J, Barton JK. Biochem. 2006;45:12295. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 95.Puckett CA, Barton JK. J. Am. Chem. Soc. 2009;131:8738. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Komor AC, Schneider CJ, Weidmann AG, Barton JK. J. Am. Chem. Soc. 2012;134:19223. doi: 10.1021/ja3090687. [DOI] [PMC free article] [PubMed] [Google Scholar]