The crystallization and preliminary X-ray diffraction analysis of the strawberry PR-10 proteins Fra a 1E and Fra a 3 in the presence of the natural flavonoid (+)-catechin are presented.
Keywords: PR-10 proteins, Fra a 1E, Fra a 3, catechin
Abstract
The strawberry Fra a proteins belong to the pathogenesis-related PR-10 protein family and share a common fold with the Bet v 1 major pollen allergen and the START/PYR/PYL proteins, which are characterized by the presence of a central cavity and are often involved in the binding of a variety of natural compounds. The Fra a proteins play a key role in the control of flavonoid biosynthesis in strawberries and are essential for pigment formation in fruits. In order to understand Fra a protein function, full-length Fra a 1E and Fra a 3 cDNAs were cloned and expressed in Escherichia coli, and the proteins were purified to homogeneity using metal-affinity chromatography. Diffraction-quality crystals of Fra a 1E and of Fra a 3 in the presence of (+)-catechin were obtained by the sitting-drop vapour-diffusion method. X-ray diffraction data from single crystals of Fra a 1E and Fra a 3 were processed to 2.2 and 3.0 Å resolution in space groups P212121 and P2221, with unit-cell parameters a = 70.02, b = 74.42, c = 84.04 Å and a = 137.91, b = 206.61, c = 174.7 Å for Fra a 1E and Fra a 3, respectively.
1. Introduction
Food is the most common origin of allergenic responses and food allergies are the subject of intense research, as they affect up to 6% of young children and 3–4% of adults worldwide (Herman, 2003 ▶; Wang & Sampson, 2011 ▶). Food allergy to vegetables, fruits and berries is often caused by proteins homologous to Bet v 1, the major allergen in birch-tree pollen (Hjernø et al., 2006 ▶; Musidlowska-Persson et al., 2007 ▶). The strawberry Fra a proteins, which are highly expressed during the late steps of fruit development (Muñoz et al., 2010 ▶), show a high degree of sequence similarity to Bet v 1 and have been implicated in allergic reactions to strawberries. Indeed, strawberry varieties showing decreased expression of Fra a proteins were well tolerated by people allergic to normal fruits (Karlsson et al., 2004 ▶; Hjernø et al., 2006 ▶).
Both the Bet v 1 and Fra a proteins belong to the ubiquitous family of plant pathogenesis-related proteins (PR-10), which have been implicated in the response of plants to pathogenic infections and abiotic stress (Marković-Housley et al., 2003 ▶). However, although the allergenic properties of the PR-10 proteins have been widely studied, their physiological function is still poorly understood (Mogensen et al., 2007 ▶). PR-10 proteins share a common fold with the START and PYR/PYL/RCAR proteins which is characterized by the presence of a central hydrophobic cavity (Iyer et al., 2001 ▶; Mogensen et al., 2002 ▶; Radauer et al., 2008 ▶). Proteins with this fold are widespread in eukaryotes and participate in a variety of processes such as nonvesicular lipid transport and steroid-hormone synthesis in mammals (Soccio & Breslow, 2003 ▶) and hormone signalling in plants (Ma et al., 2009 ▶; Melcher et al., 2009 ▶; Miyazono et al., 2009 ▶; Nishimura et al., 2009 ▶; Park et al., 2009 ▶; Santiago et al., 2009 ▶). Elucidation of the structures of these proteins has contributed to gaining an insight into their mechanisms of action at the molecular level, which in all cases involves the binding of specific ligands in their hydrophobic cavity. An NMR structural model of Fra a 1E has recently been described (Seutter von Loetzen et al., 2012 ▶), confirming the presence of a START/Bet v 1 fold and a central cavity. However, the molecular function of Fra a proteins as well as their physiological ligands are still unknown.
Several isoforms of the Fra a protein have been described in strawberry, including Fra a 1E and Fra a 3 (Muñoz et al., 2010 ▶; Musidlowska-Persson et al., 2007 ▶). In addition to their properties as food allergens, it has been shown that the Fra a proteins play an important role in the control of flavonoid biosynthesis and are thus required for the development of colour during fruit ripening (Muñoz et al., 2010 ▶; Hjernø et al., 2006 ▶). Flavonoids are among the most important bioactive secondary metabolites in plants. They are responsible for the colour and flavour of flowers, fruits and other plant organs (Halbwirth et al., 2006 ▶; Fait et al., 2008 ▶), and they also present antioxidative and anticarcinogenic activities in humans when consumed in the diet (Ghasemzadeh & Ghasemzadeh, 2011 ▶). Suppression of the expression of Fra a genes in strawberry fruits leads to decreased expression of the phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS) genes that code for two major enzymes in the flavonoid-biosynthetic pathway and to a decrease in the accumulation of the main flavonoids responsible for the red colour of fruits, as well as other intermediates of the pathway (Muñoz et al., 2010 ▶). The molecular mechanism through which the Fra a proteins control flavonoid biosynthesis is as yet unknown. However, the fact that these proteins are predicted to have cavities for the binding of small ligands suggests that the Fra a proteins might bind metabolic intermediates of the flavonoid pathway. Here, we report the cloning, expression, purification, crystallization and preliminary X-ray analysis of the Fra a 1E and Fra a 3 allergens from strawberry, the latter in the presence of catechin, a natural flavonoid compound. This work could contribute to the structural analysis of these proteins, which would shed light on the molecular function of Fra a proteins and potentially other members of the PR-10 proteins.
2. Materials and methods
2.1. Cloning
The coding regions of Fra a 1E and Fra a 3 (EMBL accession Nos. CAJ85645 and GQ148819) were amplified by PCR using the plasmids pBI-Fraa1ei and pBI-Fraa3i as templates (Muñoz et al., 2010 ▶). Fra a 1E and Fra a 3 open reading frames were PCR-amplified using the primers FwF1 (GGGCCATGGCGGGTGTTTACATTCATGAAAACGAG) and RvF1 (CCCGGATCCTTAGTTGTATTCGCTGGGG), and FwF3 (GGGCCATGGCGGGTGTGTTCACATACGAATCCG) and RvF3 (CCCGGATCCTTAGTTGTATTCCTCAGGATGGG), respectively. The forward and reverse primers contained NcoI and BamHI restriction sites. The amplified sequences were digested with NcoI/BamHI enzymes and were cloned into pETM11 (Dümmler et al., 2005 ▶). The expression constructs, which were named F1-pETM11 and F3-pETM11, included an N-terminal 6×His tag and the TEV cleavage sequence. After TEV cleavage, only three foreign amino acids (Ala-Met-Ala) remained at the N-terminal end of both proteins. DNA sequencing confirmed that the recombinant vectors encoded the expected sequences.
2.2. Protein expression and purification
The F1-pETM11 and F3-pETM11 constructs were introduced into Escherichia coli One Shot BL21(DE3) competent cells (Invitrogen) by the heat-shock method and were grown overnight at 310 K on solid Luria–Bertani (LB) medium supplemented with 50 µg ml−1 kanamycin. Cells were inoculated in 2 l LB medium containing 50 µg ml−1 kanamycin and were grown at 310 K with shaking at 150 rev min−1 until an OD600 of 0.6–0.8 was reached. Protein expression was then induced by the addition of 1 mM IPTG. The cells were incubated overnight at 293 K, harvested by centrifugation at 10 000g for 15 min at 277 K and stored at 193 K before purification.
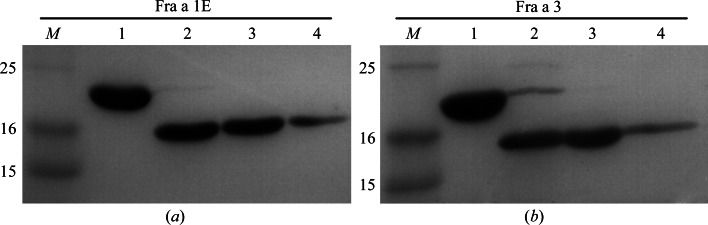
Cell pellets were resuspended in 180 ml lysis buffer (30 mM Tris pH 7.5, 500 mM NaCl, 15 mM imidazole, 1 mM β-mercaptoethanol) containing 20 µg ml−1 DNAse I (Roche) and one EDTA-free protease-cocktail inhibitor tablet (Roche) and lysed with a microfluidizer (Microfluidics). The lysate was centrifuged at 35 000g and 277 K for 45 min. The clear supernatant was incubated for 2 h in a 25 ml nickel–nitrilotriacetic acid (Ni–NTA) agarose column (Qiagen) equilibrated with lysis buffer. Unbound proteins were removed by washing with five column volumes of buffer A1 (30 mM Tris pH 7.5, 300 mM NaCl, 15 mM imidazole, 1 mM β-mercaptoethanol) and buffer W (30 mM Tris pH 7.5, 300 mM NaCl, 30 mM imidazole, 1 mM β-mercaptoethanol). The bound proteins were finally eluted with buffer B (30 mM Tris pH 7.5, 300 mM NaCl, 250 mM imidazole, 1 mM β-mercaptoethanol). The 6×His tag of the purified proteins was removed by digestion with TEV protease. During digestion, samples were extensively dialyzed against buffer A2 (30 mM Tris–HCl pH 7.5, 300 mM NaCl, 1 mM β-mercaptoethanol). The dialyzed samples were kept at 277 K until TEV cleavage was complete (typically overnight). The samples were incubated with Ni–NTA to remove the undigested proteins, TEV protease and other contaminants. The correct size and purity of the recombinant proteins were verified by SDS–PAGE (Fig. 1 ▶). Purified fractions of Fra a 1E and Fra a 3 were pooled, dialyzed in buffer C (30 mM Tris pH 7.5, 150 mM NaCl, 1 mM β-mercaptoethanol) to remove imidazole, concentrated to 60 mg ml−1 by ultrafiltration with Amicon Ultra-15 3K filter units (Millipore) and flash-frozen without glycerol in liquid nitrogen for storage at 193 K. Protein concentrations were determined by measuring the absorbance at 280 nm under denaturing conditions using a UV–Vis biophotometer (Eppendorf). The predicted molecular weights and extinction coefficients based on the amino-acid sequence were 17.8 kDa and 14 900 M −1 cm−1, respectively, for Fra a 1E and 17.5 kDa and 11 920 M −1 cm−1, respectively, for Fra a 3.
Figure 1.
12% SDS–PAGE analysis of purified Fra a 1E (a) and Fra a 3 (b). Lane M, molecular-mass marker (labelled in kDa); lane 1, purified Fra a 1E and Fra a 3; lane 2, the same samples after TEV cleavage; lanes 3 and 4, the same samples as in lane 2 after reverse purification with Ni–NTA. As can be observed, the samples used for crystallization (lanes 3 and 4) were highly pure and had a molecular weight close to the expected value (18 kDa).
2.3. Protein characterization: size-exclusion chromatography–multiple-angle laser-light scattering (SEC–MALLS)
Size-exclusion chromatography (SEC) combined with multi-angle laser-light scattering (MALLS) and refractometry (RI) is a powerful method for measuring the absolute molecular mass of macromolecules and macromolecular complexes (Wyatt, 1998 ▶; Gerard et al., 2007 ▶). Determination of the molecular-mass variation across the chromatographic peak also provides an estimate of the dispersity of the compound. SEC was performed on an S200 Superdex column (GE Healthcare) equilibrated with buffer consisting of 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM β-mercaptoethanol. Proteins were injected at a concentration of 3.6 mg ml−1 (200 µM). All separations were performed at 293 K with a flow rate of 0.5 ml min−1. Online MALLS detection was performed with a DAWN EOS detector (Wyatt Technology Corp., Santa Barbara, California, USA) using a laser emitting at 690 nm and by refractive-index measurements using an RI2000 detector (Schambeck SFD). Weight-averaged molar masses were calculated using the ASTRA software (Wyatt Technology Corp.) as described previously (Wyatt, 1998 ▶).
2.4. Crystallization
Initial crystallization conditions for Fra a 1E and Fra a 3–catechin were identified at the High Throughput Crystallization Laboratory of the EMBL Grenoble Outstation (https://htxlab.embl.fr; Dimasi et al., 2007 ▶).
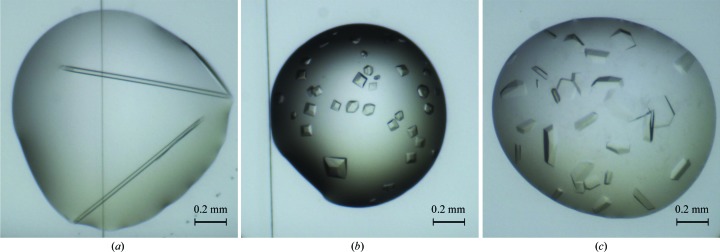
Crystallization experiments were carried out at 293 K in a 96-well plate using the sitting-drop vapour-diffusion method and the screens Crystal Screen, Crystal Screen 2, Crystal Screen Lite, PEG/Ion, MembFac, Natrix, Quick Screen, Grid Screen Ammonium Sulfate, Grid Screen Sodium Malonate, Grid Screen PEG 6000, Grid Screen PEG/LiCl, Grid Screen MPD, Screen MME and The Classics II Suite. Droplets of 200 nl volume (with a 1:1 protein:precipitant ratio) were set up using a 16-channel Cartesian PixSys robot (Cartesian Technologies) and equilibrated against 80 µl reservoir solution (Dimasi et al., 2007 ▶). For these experiments, Fra a 1E and Fra a 3 proteins were diluted in buffer C (30 mM Tris pH 7.5, 150 mM NaCl, 1 mM β-mercaptoethanol) and tested at 50, 26 and 15 mg ml−1. Both proteins were assayed in the presence and the absence of the natural flavonoid compound (+)-catechin. Interestingly, Fra a 1E produced crystals only in the absence of (+)-catechin, even when the chemical was added to the sample in the form of a powder (see below), while Fra a 3 produced crystals exclusively in its presence. In both cases, crystals of Fra a 1E and Fra a 3–catechin appeared within 48 h after setting up the crystallization experiments (Fig. 2 ▶) at a concentration of 26 mg ml−1. The Fra a 1E protein produced crystals in two different crystal forms. Rod-shaped Fra a 1E crystals (Fig. 2 ▶ a) were obtained using 0.2 M ammonium sulfate, 0.1 M sodium cacodylate pH 6.5, 15% PEG 8000; Fra a 1E crystals in the shape of hexagonal prisms (Fig. 3 ▶ b) were obtained using 0.2 M ammonium sulfate, 0.1 M HEPES pH 7.5, 25% PEG 3350 as precipitant. The Fra a 3 protein produced flat hexagon-like crystals (Fig. 2 ▶ c) that were obtained by adding 5 mM (+)-catechin diluted in buffer C supplemented with 10% DMSO to 26 mg ml−1 Fra a 3 solution in 2.9 M sodium malonate pH 7.0. Further optimization of this condition was required to obtain diffraction-quality crystals. The final Fra a 3–catechin crystallization protocol was as follows. Purified Fra a 3 protein was diluted to a concentration of 26 mg ml−1 as described above. To compensate for the limited solubility of (+)-catechin in aqueous solution, solid (+)-catechin was added in excess to the protein solution as a powder. The sample was incubated at 277 K overnight in an overhead shaker. After centrifugation at 14 000g the supernatant was used to set up sitting-drop crystallization experiments at room temperature by mixing 1 µl protein solution and 1 µl precipitant solution (2.25 M sodium malonate pH 7.0) and equilibrating against 0.5 ml precipitant solution. Crystals reached their final size within 48 h. To determine whether Fra a 1E could produce crystals under these conditions, the same protocol was assayed. Again, crystals of Fra a 1E were not obtained in the presence of (+)-catechin.
Figure 2.
Fra a 1E and Fra a 3–catechin crystals. Two different crystallization conditions were identified for Fra a 1E; crystals were obtained in the shapes of rods (a) and trigonal prisms (b). Crystals of Fra a 3 with the shape of hexagonal prisms were only obtained in the presence of catechin (c).
2.5. Data collection
For X-ray data collection, Fra a 1E crystals were mounted on CryoLoops (Hampton Research), soaked in cryoprotectant solution (15% glycerol) and flash-cooled directly in a nitrogen stream at 100 K. The crystallization condition for Fra a 3–catechin crystals was directly compatible with cryofreezing; therefore, no addition of cryoprotecting agents was required and the crystals were directly flash-cooled in the liquid-nitrogen stream prior to data collection.
Diffraction experiments were performed using the Fra a 1E and Fra a 3–catechin crystals on the ID14-4 and ID14-1 beamlines of the European Synchrotron Radiation Facility (ESRF) equipped with ADSC Q315r CCD and ADSC Q210 CCD detectors, respectively (McCarthy et al., 2009 ▶). XDS was used for data reduction and integration (Kabsch, 2010 ▶). After conversion to CCP4 format using COMBAT (Winn et al., 2011 ▶), the data were scaled using SCALA (Evans, 2006 ▶). Matthews coefficient and solvent-content estimations were performed using the PHENIX software (Winn et al., 2011 ▶).
3. Results and discussion
Cloning, expression, purification and identification of crystallization conditions for Fra a 1E and Fra a 3 have successfully been undertaken. Fra a 1E and Fra a 3 open reading frames were cloned into pETM11 expression vectors. The resultant plasmids encoded Fra a 1E and Fra a 3 proteins fused to an N-terminal His6-TEV tag that was removed by digestion with TEV protease during purification. The recombinant proteins showed very high expression levels in E. coli BL21(DE3) cells. Fra a 1E and Fra a 3 were purified to homogeneity by two-step Ni–NTA metal-affinity chromatography, as described in §2. SDS–PAGE analysis of the purified samples (Fig. 1 ▶) indicated migration consistent with the expected molecular weights and a high degree of purity. The typical yield for both proteins was 75–150 mg per litre of culture. SEC–MALLS experiments (Fig. 3 ▶) indicated estimated molecular weights of 18.0 and 17.5 kDa for Fra a 1E and Fra a 3, respectively, which are in good agreement with their expected molecular masses (17.8 and 17.5 kDa, respectively), indicating that both proteins are monomeric in solution.
Figure 3.
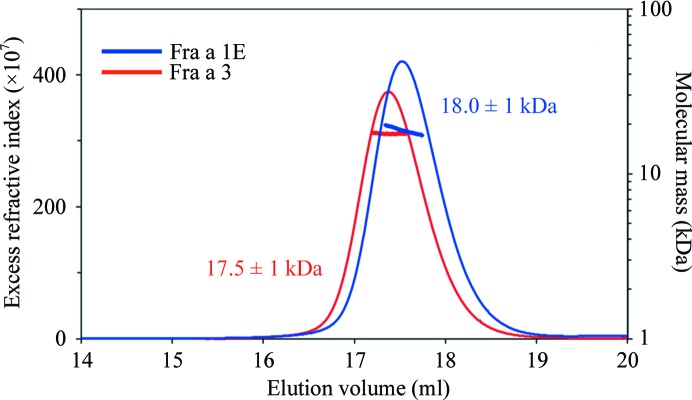
SEC–MALLS analysis of purified Fra a 1E (blue) and Fra a 3 (red). The experiments were performed at protein concentrations of 200 µM, as described in §2. Both the SEC elution profiles (monitored by the excess refractive index, which is proportional to the protein concentration) and the molecular size calculated by MALLS (blue and red crosses shown on the peaks for each species) indicate that the Fra a 1E and Fra a 3 proteins are monomeric in solution (the expected masses of the monomeric forms are 17.8 and 17.5 kDa, respectively).
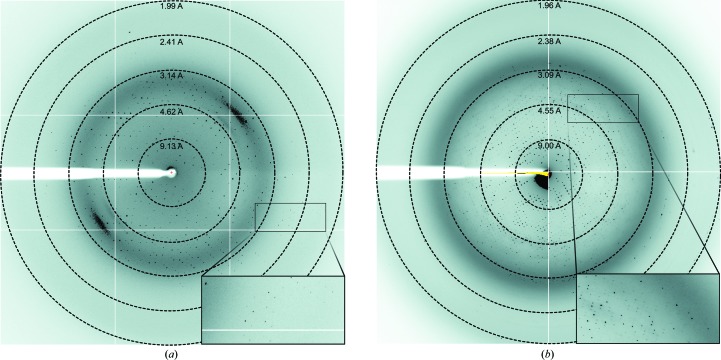
Diffraction-quality crystals of Fra a 1E in the shape of rods (Fig. 2 ▶ a) were obtained by the sitting-drop vapour-diffusion technique. The crystals diffracted reproducibly to 2.2 Å resolution (Fig. 4 ▶ a). Hexagonal prism-like Fra a 1E crystals (Fig. 2 ▶ b) diffracted to 6 Å resolution. However, efforts to obtain better diffracting crystals for this second condition were not successful. Fra a 3 crystals were obtained after optimization by the sitting-drop method and only in the presence of catechin, a natural flavonoid compound (Fig. 2 ▶ c). The crystals diffracted to 3.0 Å resolution (Fig. 4 ▶ b). A complete data set was collected for both the rod-shaped crystals of Fra a 1E and the Fra a 3–catechin crystals with good completeness and crystallographic statistics (see Table 1 ▶). The Fra a 1E and Fra a 3–catechin crystals belonged to space groups P212121 and C2221, with unit-cell parameters a = 70.02, b = 74.42, c = 84.04 Å and a = 137.91, b = 206.61, c = 174.7 Å, respectively. After data processing with XDS (Kabsch, 2010 ▶) and scaling with SCALA (Evans, 2006 ▶), the resulting data sets extended to resolutions of 2.2 and 3.0 Å, with R p.i.m. values of 0.034 and 0.030 and completenesses of 99.9 and 99.5% for Fra a 1E and Fra a 3–catechin, respectively. Data-collection statistics are summarized in Table 1 ▶.
Figure 4.
Diffraction patterns of Fra a 1E and Fra a 3–catechin crystals. The concentric circles show the resolution limits. The Fra a 1E (a) and Fra a 3–catechin (b) crystals diffracted to resolutions of 2.2 nd 3.0 Å, respectively. The high-resolution spots are highlighted in boxes.
Table 1. Crystallographic data-collection statistics for Fra a 1E and Fra a 3–catechin.
Values in parentheses are for the highest resolution shell.
| Fra a 1E | Fra a 3–catechin | |
|---|---|---|
| X-ray source | ID14-4 | ID14-1 |
| Space group | P212121 | C2221 |
| Unit-cell parameters (Å) | a = 70.02, b = 74.42, c = 84.04 | a = 137.91, b = 206.61, c = 174.70 |
| Resolution | 36.03–2.20 (2.32–2.20) | 30–3.00 (3.16–3.00) |
| No. of observations (overall/unique) | 165127/24107 | 371478/49904 |
| Average multiplicity | 7.2 (7.3) | 7.4 (7.5) |
| R p.i.m. † | 0.034 (0.183) | 0.03 (0.23) |
| Completeness (%) | 99.9 (100) | 99.5 (100) |
| 〈I/σ(I)〉 | 14.0 (4.2) | 21.7 (3.6) |
R
p.i.m. = 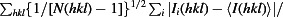
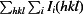 , where Ii(hkl) is the intensity of observation i of reflection hkl and N is the redundancy of reflection hkl (Einspahr & Weiss, 2012 ▶).
, where Ii(hkl) is the intensity of observation i of reflection hkl and N is the redundancy of reflection hkl (Einspahr & Weiss, 2012 ▶).
The diffraction data were analysed using phenix.xtriage (Adams et al., 2010 ▶). Matthews coefficients of 3.04 and 2.028 Å3 Da−1 were calculated for Fra a 1E, corresponding to two and three molecules in the asymmetric unit, with estimated solvent contents of 59.6 and 39.4%, respectively. For Fra a 3, the calculation suggests the presence of 14 molecules per asymmetric unit, which corresponds to a solvent content of 50.5% and a Matthews coefficient of 2.484 Å3 Da−1. However, assuming solvent contents between 27 and 78% for protein crystals, a number of solutions ranging from seven to 20 molecules per asymmetric unit would be also possible (solvent contents of 75.3–29.3% and Matthews coefficients in the range 5.769–1.739 Å3 Da−1).
In conclusion, the results presented here provide methods for the large-scale production and purification of the Fra a proteins, which seem to be monomeric in solution. Conditions for producing crystals for X-ray diffraction experiments have been established for both proteins. The crystallographic data sets obtained so far are currently being analyzed in order to obtain atomic structures of Fra a 1E and Fra a 3–catechin. Determination of the Fra a 1E and Fra a 3–catechin structures would be an important step towards understanding their mechanisms of action, not only in the control of secondary metabolism in plants but also the origin of their immunogenic properties.
Acknowledgments
We would like to thank the European Synchrotron Radiation Facility and the Grenoble Outstation of the European Molecular Biology Laboratory for access to the Macromolecular Crystallography beamlines. This work was partially supported by the P-CUBE project of the European Commission (FP7/2007-2013; grant No. 227764) and the MICINN (grant BIO2010-15630; Spain). ACS received a FPI fellowship from the Spanish Government.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Dimasi, N., Flot, D., Dupeux, F. & Márquez, J. A. (2007). Acta Cryst. F63, 204–208. [DOI] [PMC free article] [PubMed]
- Dümmler, A., Lawrence, A. M. & de Marco, A. (2005). Microb. Cell Fact. 4, 34. [DOI] [PMC free article] [PubMed]
- Einspahr, H. M. & Weiss, M. (2012). International Tables for Crystallography, Vol. F, edited by E. Arnold, D. M. Himmel & M. G. Rossmann, pp. 64–74. New York: Wiley.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fait, A., Hanhineva, K., Beleggia, R., Dai, N., Rogachev, I., Nikiforova, V. J., Fernie, A. R. & Aharoni, A. (2008). Plant Physiol. 148,730–750. [DOI] [PMC free article] [PubMed]
- Gerard, F. C., Ribeiro Ede, A. Jr, Albertini, A. A., Gutsche, I., Zaccai, G., Ruigrok, R. W. & Jamin, M. (2007). Biochemistry, 46, 10328–10338. [DOI] [PubMed]
- Ghasemzadeh, A. & Ghasemzadeh, N. (2011). J. Med. Plants Res. 5, 6697–6703.
- Halbwirth, H., Puhl, I., Haas, U., Jezik, K., Treutter, D. & Stich, K. (2006). J. Agric. Food Chem. 54, 1479–1485. [DOI] [PubMed]
- Herman, E. M. (2003). J. Exp. Bot. 54, 1317–1319. [DOI] [PubMed]
- Hjernø, K., Alm, R., Canbäck, B., Matthiesen, R., Trajkovski, K., Björk, L., Roepstorff, P. & Emanuelsson, C. (2006). Proteomics, 6, 1574–1587. [DOI] [PubMed]
- Iyer, L. M., Koonin, E. V. & Aravind, L. (2001). Proteins, 43, 134–144. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karlsson, A. L., Alm, R., Ekstrand, B., Fjelkner-Modig, S., Schiött, A., Bengtsson, U., Björk, L., Hjernø, K., Roepstorff, P. & Emanuelsson, C. S. (2004). Allergy, 59, 1277–1284. [DOI] [PubMed]
- Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A. & Grill, E. (2009). Science, 324, 1064–1068. [DOI] [PubMed]
- Marković-Housley, Z., Degano, M., Lamba, D., von Roepenack-Lahaye, E., Clemens, S., Susani, M., Ferreira, F., Scheiner, O. & Breiteneder, H. (2003). J. Mol. Biol. 325, 123–133. [DOI] [PubMed]
- McCarthy, A. A., Brockhauser, S., Nurizzo, D., Theveneau, P., Mairs, T., Spruce, D., Guijarro, M., Lesourd, M., Ravelli, R. B. G. & McSweeney, S. (2009). J. Synchrotron Rad. 16, 803–812. [DOI] [PMC free article] [PubMed]
- Melcher, K. et al. (2009). Nature (London), 462, 602–608.
- Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H.-J., Asano, A., Miyauchi, Y., Takahashi, M., Zhi, Y., Fujita, Y., Yoshida, T., Kodaira, K.-S., Yamaguchi-Shinozaki, K. & Tanokura, M. (2009). Nature (London), 462, 609–614. [DOI] [PubMed]
- Mogensen, J. E., Ferreras, M., Wimmer, R., Petersen, S. V., Enghild, J. J. & Otzen, D. E. (2007). Biochemistry, 46, 3356–3365. [DOI] [PubMed]
- Mogensen, J. E., Wimmer, R., Larsen, J. N., Spangfort, M. D. & Otzen, D. E. (2002). J. Biol. Chem. 277, 23684–23692. [DOI] [PubMed]
- Muñoz, C., Hoffmann, T., Escobar, N. M., Ludemann, F., Botella, M. A., Valpuesta, V. & Schwab, W. (2010). Mol. Plant, 3, 113–124. [DOI] [PubMed]
- Musidlowska-Persson, A., Alm, R. & Emanuelsson, C. (2007). Mol. Immunol. 44, 1245–1252. [DOI] [PubMed]
- Nishimura, N., Hitomi, K., Arvai, A. S., Rambo, R. P., Hitomi, C., Cutler, S. R., Schroeder, J. I. & Getzoff, E. D. (2009). Science, 326, 1373–1379. [DOI] [PMC free article] [PubMed]
- Park, S.-Y. et al. (2009). Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed]
- Radauer, C., Lackner, P. & Breiteneder, H. (2008). BMC Evol. Biol. 8, 286. [DOI] [PMC free article] [PubMed]
- Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S.-Y., Jamin, M., Cutler, S. R., Rodriguez, P. L. & Márquez, J. A. (2009). Nature (London), 462, 665–668. [DOI] [PubMed]
- Seutter von Loetzen, C., Schweimer, K., Schwab, W., Rösch, P. & Hartl-Spiegelhauer, O. (2012). Biosci. Rep. 32, 567–575. [DOI] [PMC free article] [PubMed]
- Soccio, R. E. & Breslow, J. L. (2003). J. Biol. Chem. 278, 22183–22186. [DOI] [PubMed]
- Wang, J. & Sampson, H. A. (2011). J. Clin. Invest. 121, 827–835. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wyatt, P. J. (1998). J. Colloid Interface Sci. 197, 9–20. [DOI] [PubMed]